Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates
Abstract
1. Introduction
2. Materials and Methods
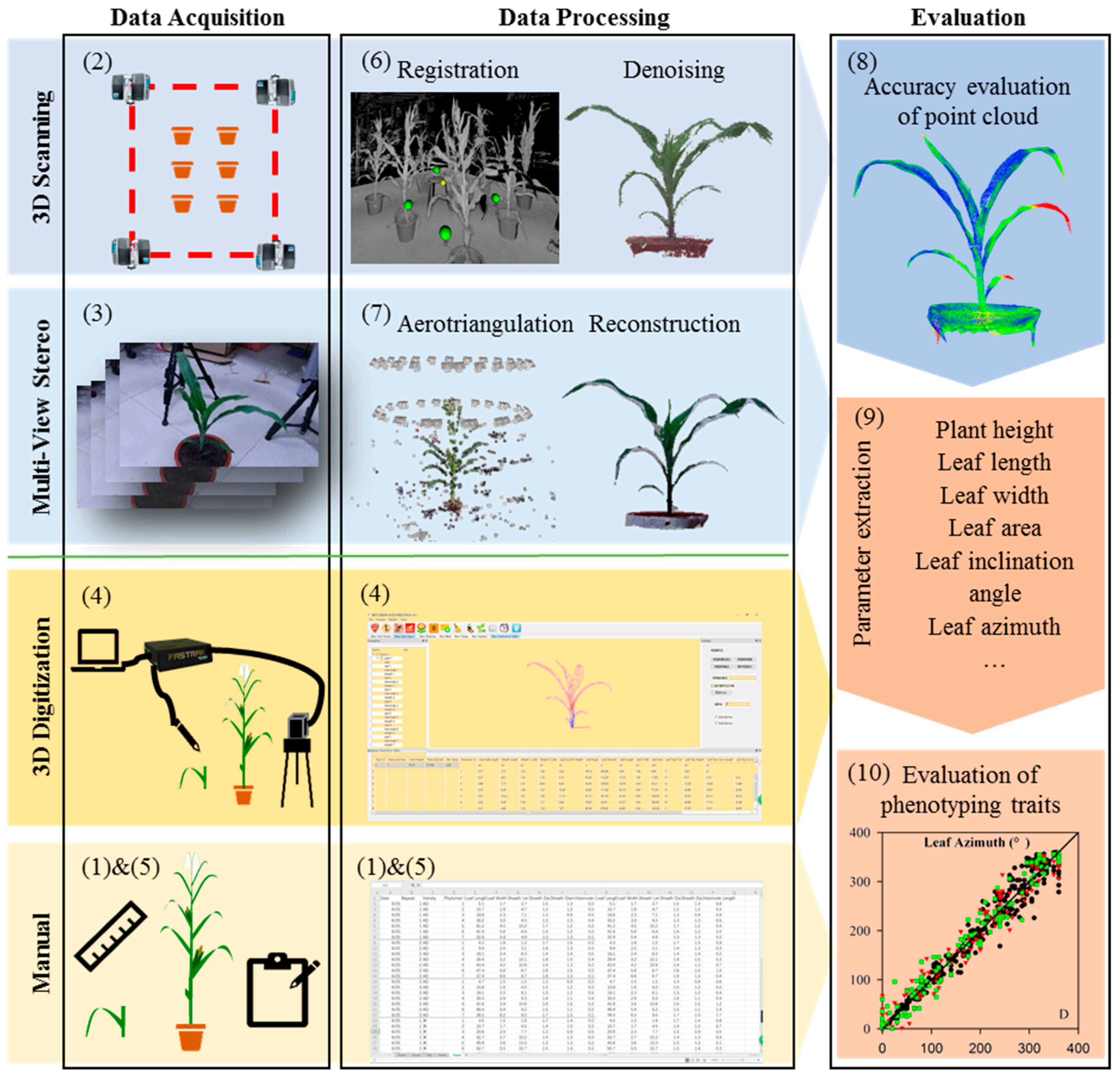
2.1. Overall Process Flow
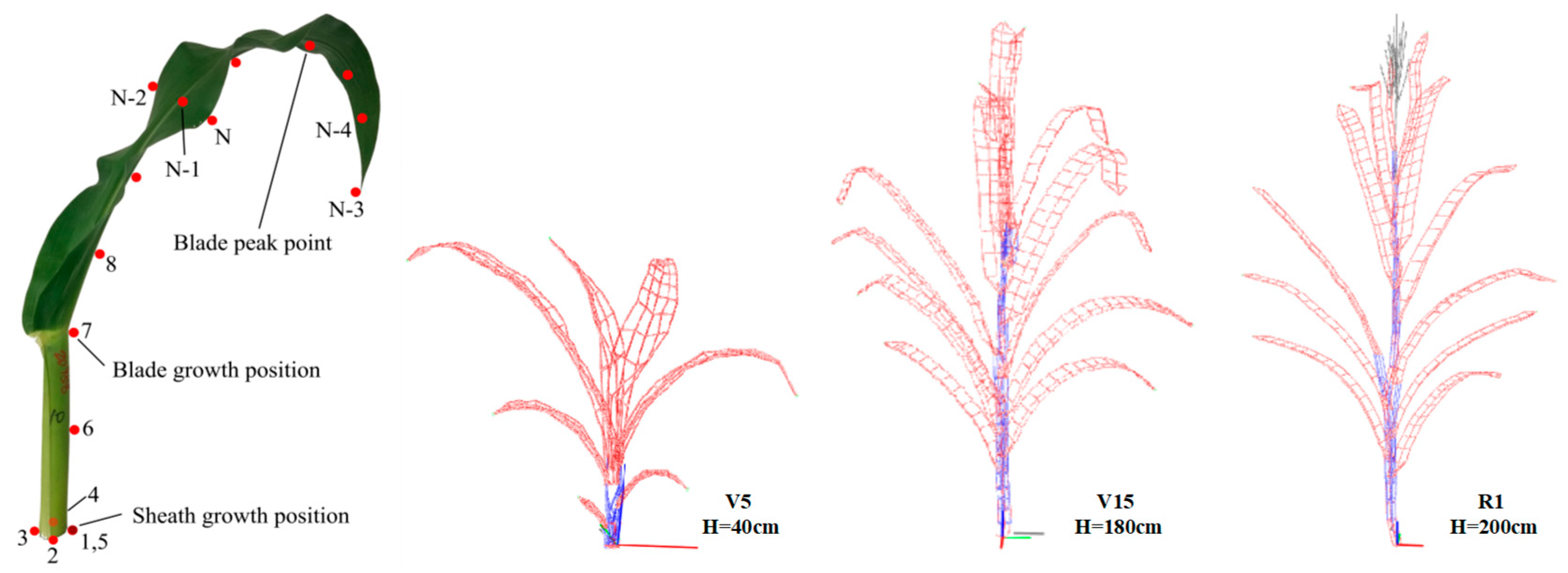
2.2. Plant Material and Sampling
2.3. Data Acquisition and Processing
2.3.1. Three-Dimensional Scanning
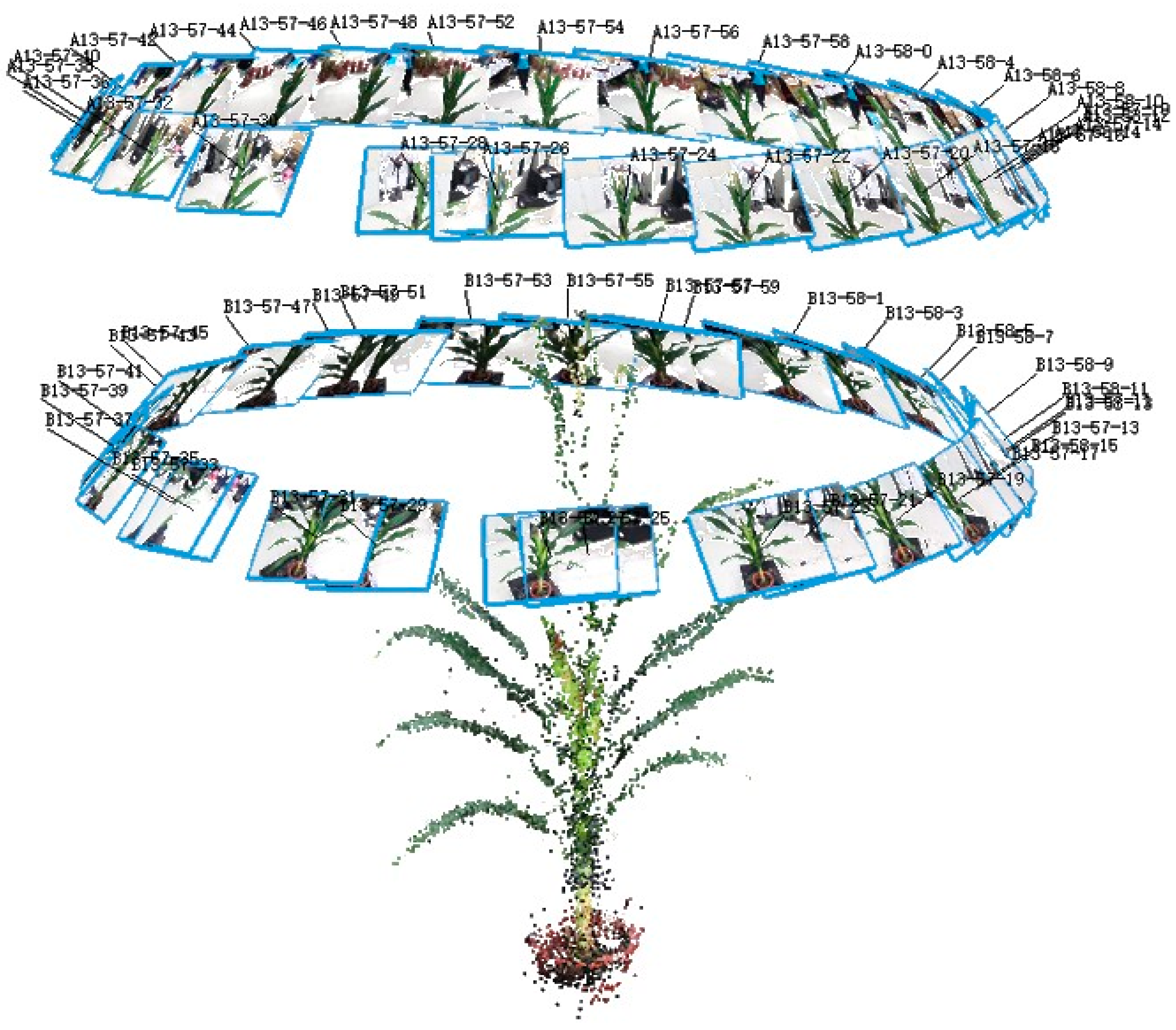
2.3.2. Multi-View Stereo Image Acquisition
2.3.3. Three-Dimensional Digitizing
2.3.4. Manual Measurement
3. Results
3.1. Evaluation of Data Acquisition and Processing Efficiency
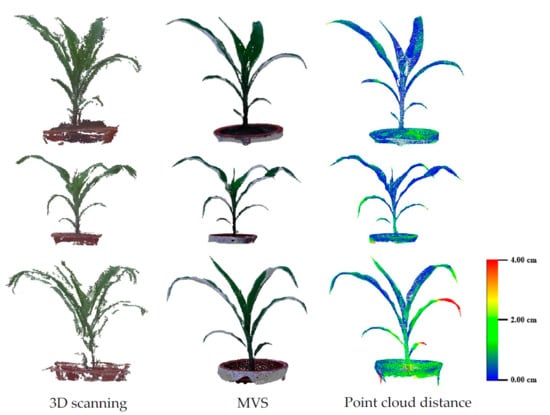
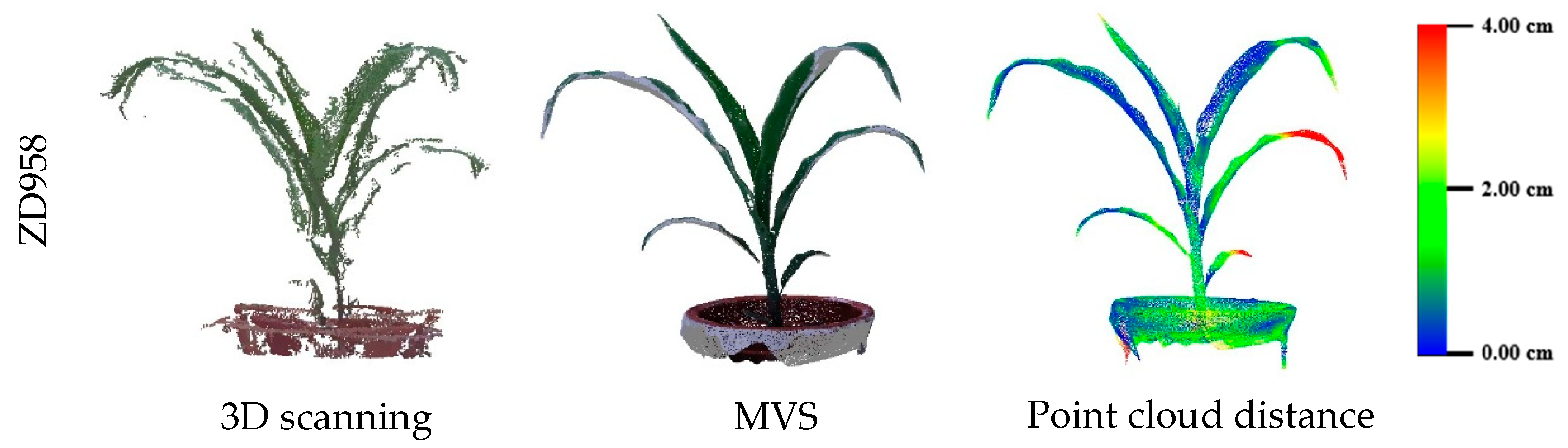
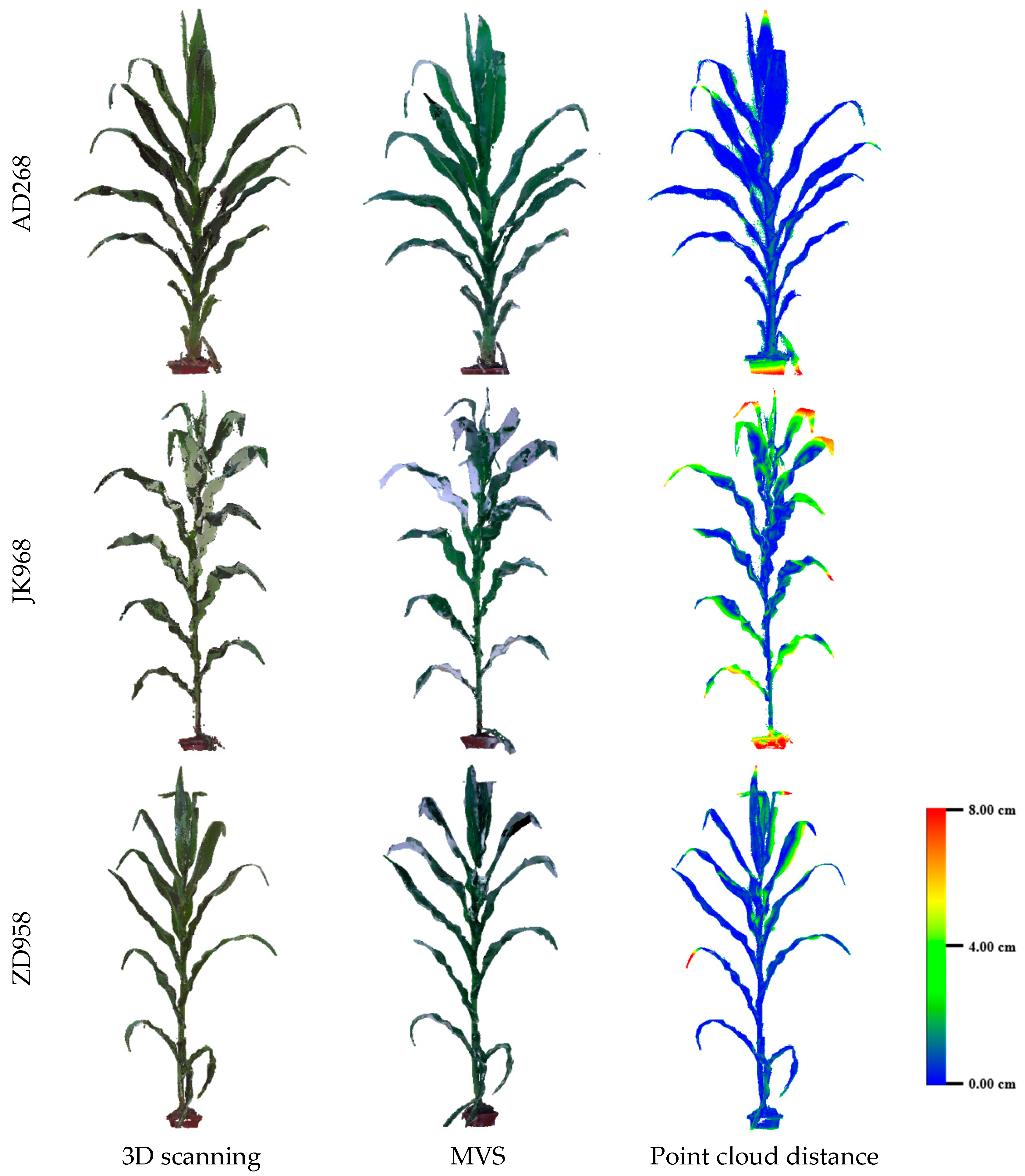
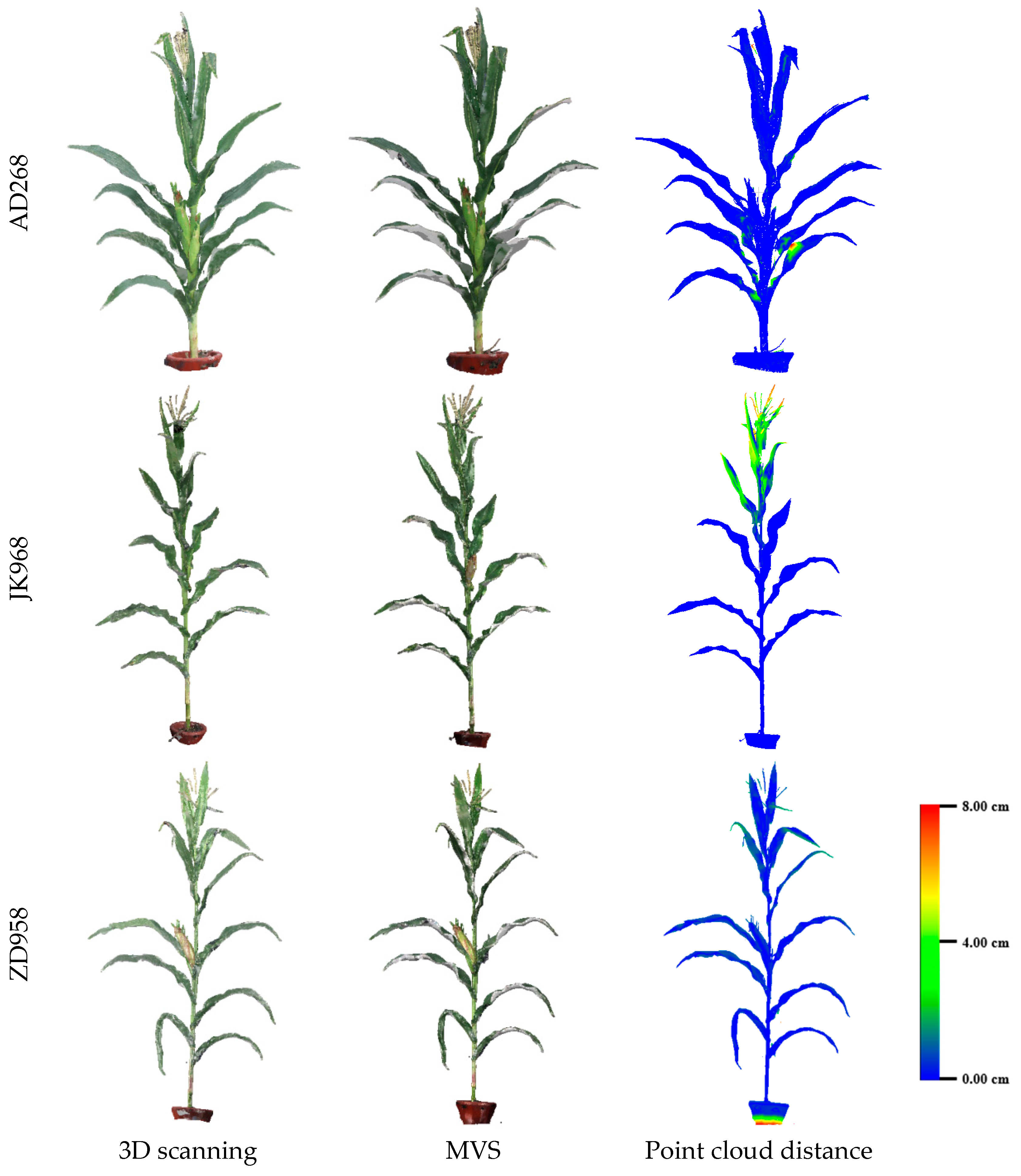
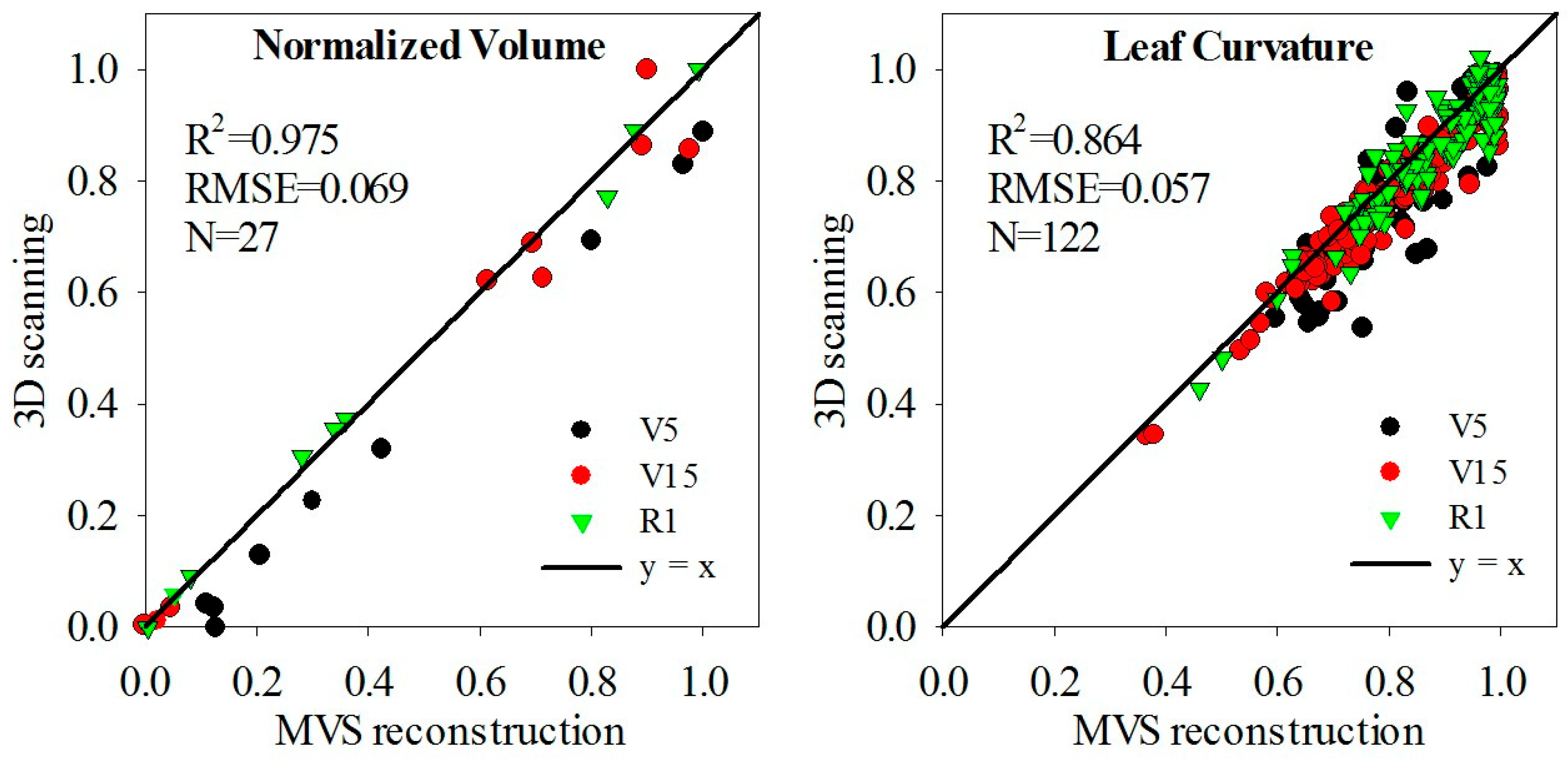
3.2. Evaluation of Three-Dimensional Point Cloud Accuracy of Maize Plants
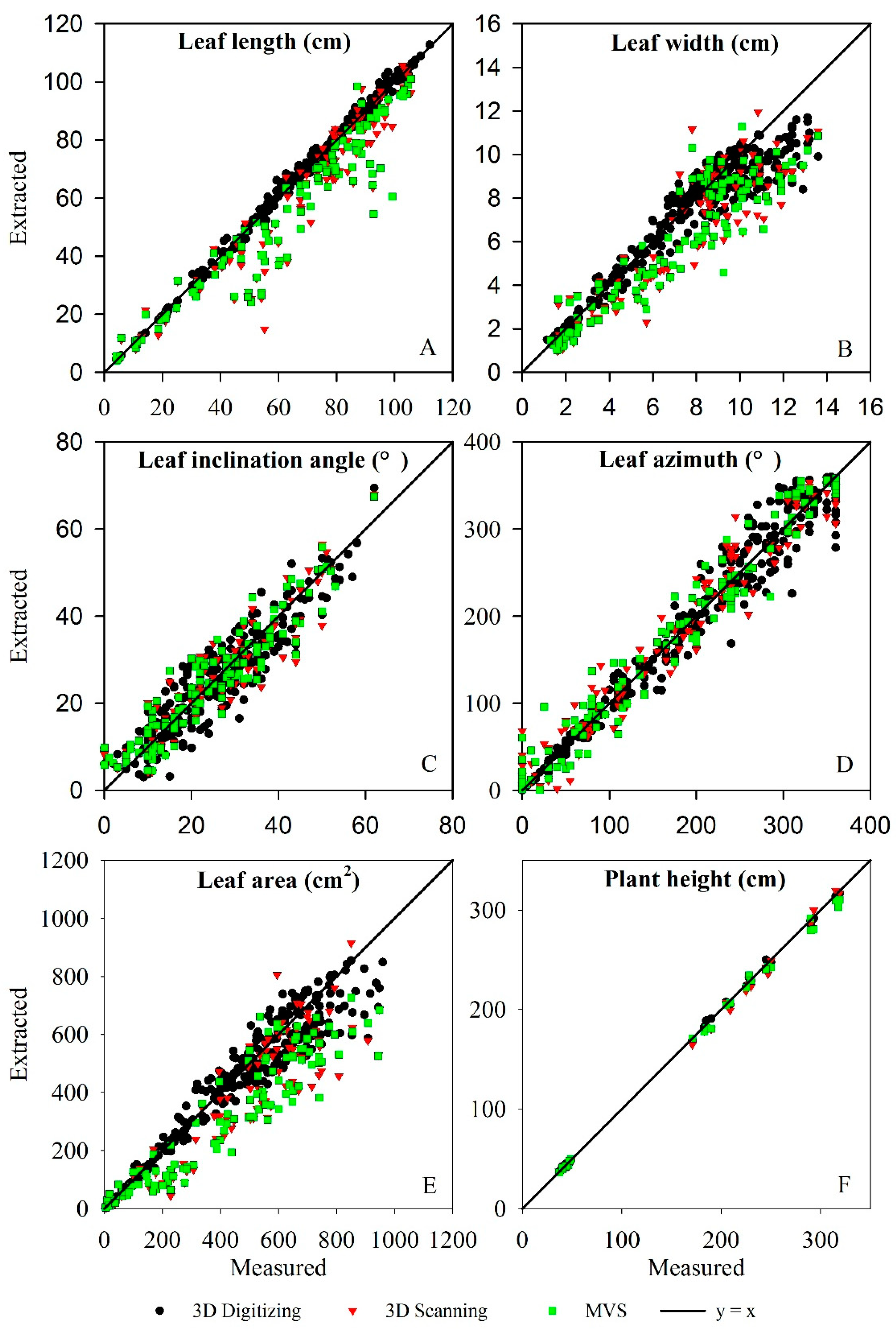
3.3. Evaluation of Three-Dimensional Phenotypic Parameter
4. Discussion
4.1. Evaluation of Efficiency and Accuracy
4.2. Potential Application in Phenotyping Platforms
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating high-throughput phenotyping into genetic gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.; Ghosh, S.; Williams, M.J.; Cuddy, W.S.; Simmonds, J.; Rey, M.D.; Hatta, M.A.M.; Hinchliffe, A.; Steed, A.; Reynolds, D.; et al. Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 2018, 4, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [PubMed]
- Fahlgren, N.; Gehan, M.A.; Baxter, I. Lights, camera, action: High-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 2015, 24, 93–99. [Google Scholar] [CrossRef]
- Andujar, D.; Calle, M.; Fernandez-Quintanilla, C.; Ribeiro, A.; Dorado, J. Three-dimensional modeling of weed plants using low-cost photogrammetry. Sensors 2018, 18, 1077. [Google Scholar] [CrossRef]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant phenomics, from sensors to knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef]
- Yang, G.J.; Liu, J.G.; Zhao, C.J.; Li, Z.H.; Huang, Y.B.; Yu, H.Y.; Xu, B.; Yang, X.D.; Zhu, D.M.; Zhang, X.Y.; et al. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 26. [Google Scholar] [CrossRef]
- Liu, S.Y.; Baret, F.; Abichou, M.; Boudon, F.; Thomas, S.; Zhao, K.G.; Fournier, C.; Andrieu, B.; Irfan, K.; Hemmerle, M.; et al. Estimating wheat green area index from ground-based lidar measurement using a 3d canopy structure model. Agr. For. Meteorol. 2017, 247, 12–20. [Google Scholar] [CrossRef]
- Sun, S.P.; Li, C.Y.; Paterson, A.H.; Jiang, Y.; Xu, R.; Robertson, J.S.; Snider, J.L.; Chee, P.W. In-field high throughput phenotyping and cotton plant growth analysis using lidar. Front. Plant Sci. 2018, 9, 17. [Google Scholar] [CrossRef]
- Jimenez-Berni, J.A.; Deery, D.M.; Rozas-Larraondo, P.; Condon, A.G.; Rebetzke, G.J.; James, R.A.; Bovill, W.D.; Furbank, R.T.; Sirault, X.R.R. High throughput determination of plant height, ground cover, and above-ground biomass in wheat with lidar. Front. Plant Sci. 2018, 9, 18. [Google Scholar] [CrossRef] [PubMed]
- Young, S.N.; Kayacan, E.; Peschel, J.M. Design and field evaluation of a ground robot for high-throughput phenotyping of energy sorghum. Precis. Agric. 2018, 1–26. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2017, 44, 143–153. [Google Scholar] [CrossRef]
- Du, J.J.; Zhang, Y.; Guo, X.Y.; Ma, L.M.; Shao, M.; Pan, X.D.; Zhao, C.J. Micron-scale phenotyping quantification and three-dimensional microstructure reconstruction of vascular bundles within maize stalks based on micro-ct scanning. Funct. Plant Biol. 2017, 44, 10–22. [Google Scholar] [CrossRef]
- Zhang, Y.; Ma, L.; Pan, X.; Wang, J.; Guo, X.; Du, J. Micron-scale phenotyping techniques of maize vascular bundles based on X-ray microcomputed tomography. JoVE 2018, e58501. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.B.; Warburton, M.; Crouch, J. Association mapping for enhancing maize (Zea mays L.) genetic improvement. Crop Sci. 2011, 51, 433–449. [Google Scholar] [CrossRef]
- Zhang, X.H.; Huang, C.L.; Wu, D.; Qiao, F.; Li, W.Q.; Duan, L.F.; Wang, K.; Xiao, Y.J.; Chen, G.X.; Liu, Q.; et al. High-throughput phenotyping and qtl mapping reveals the genetic architecture of maize plant growth. Plant Physiol. 2017, 173, 1554–1564. [Google Scholar] [CrossRef]
- Chen, T.-W.; Cabrera-Bosquet, L.; Alvarez Prado, S.; Perez, R.; Artzet, S.; Pradal, C.; Coupel-Ledru, A.; Fournier, C.; Tardieu, F. Genetic and environmental dissection of biomass accumulation in multi-genotype maize canopies. J. Exp. Bot. 2018, 12. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Fournier, C.; Brichet, N.; Welcker, C.; Suard, B.; Tardieu, F. High-throughput estimation of incident light, light interception and radiation-use efficiency of thousands of plants in a phenotyping platform. New Phytol. 2016, 212, 269–281. [Google Scholar] [CrossRef]
- Junker, A.; Muraya, M.M.; Weigelt-Fischer, K.; Arana-Ceballos, F.; Klukas, C.; Melchinger, A.E.; Meyer, R.C.; Riewe, D.; Altmann, T. Optimizing experimental procedures for quantitative evaluation of crop plant performance in high throughput phenotyping systems. Front. Plant Sci. 2015, 5, 21. [Google Scholar] [CrossRef]
- Brichet, N.; Fournier, C.; Turc, O.; Strauss, O.; Artzet, S.; Pradal, C.; Welcker, C.; Tardieu, F.; Cabrera-Bosquet, L. A robot-assisted imaging pipeline for tracking the growths of maize ear and silks in a high-throughput phenotyping platform. Plant Methods 2017, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, A.; Ward, C.; Talasaz, A.; Ivanov, A.G.; Brophy, M.; Grodzinski, B.; Huner, N.P.A.; Patel, R.V.; Barron, J.L. Machine vision system for 3d plant phenotyping. IEEE/ACM Trans. Comput. Biol. Bioinform. 2017. [Google Scholar] [CrossRef] [PubMed]
- Thapa, S.; Zhu, F.; Walia, H.; Yu, H.; Ge, Y. A novel lidar-based instrument for high-throughput, 3d measurement of morphological traits in maize and sorghum. Sensors 2018, 18, 1187. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Arellano, M.; Griepentrog, H.W.; Reiser, D.; Paraforos, D.S. 3-d imaging systems for agricultural applications-a review. Sensors 2016, 16, 618. [Google Scholar] [CrossRef] [PubMed]
- Chaivivatrakul, S.; Tang, L.; Dailey, M.N.; Nakarmi, A.D. Automatic morphological trait characterization for corn plants via 3d holographic reconstruction. Comput. Electron. Agric. 2014, 109, 109–123. [Google Scholar] [CrossRef]
- Vázquez-Arellano, M.; Reiser, D.; Paraforos, D.; Garrido-Izard, M.; Griepentrog, H. Leaf area estimation of reconstructed maize plants using a time-of-flight camera based on different scan directions. Robotics 2018, 7, 63. [Google Scholar] [CrossRef]
- Guan, H.; Liu, M.; Ma, X.; Yu, S. Three-dimensional reconstruction of soybean canopies using multisource imaging for phenotyping analysis. Remote Sens. 2018, 10, 1206. [Google Scholar] [CrossRef]
- Hui, F.; Zhu, J.; Hu, P.; Meng, L.; Zhu, B.; Guo, Y.; Li, B.; Ma, Y. Image-based dynamic quantification and high-accuracy 3d evaluation of canopy structure of plant populations. Ann. Bot. 2018, 121, 1079–1088. [Google Scholar] [CrossRef]
- Burgess, A.J.; Retkute, R.; Pound, M.P.; Mayes, S.; Murchie, E.H. Image-based 3d canopy reconstruction to determine potential productivity in complex multi-species crop systems. Ann. Bot. 2017, 119, 517–532. [Google Scholar] [CrossRef]
- Wen, W.; Guo, X.; Zhao, C.; Xiao, B.; Wang, Y. Research on maize plant type parameter extraction by using three dimensional digitizing data. Sci. Agric. Sin. 2018, 51, 1034–1044. [Google Scholar] [CrossRef]
- Sinoquet, H.; Thanisawanyangkura, S.; Mabrouk, H.; Kasemsap, P. Characterization of the light environment in canopies using 3d digitising and image processing. Ann. Bot. 1998, 82, 203–212. [Google Scholar] [CrossRef]
- Su, W.; Zhu, D.H.; Huang, J.X.; Guo, H. Estimation of the vertical leaf area profile of corn (Zea mays) plants using terrestrial laser scanning (TLS). Comput. Electron. Agric. 2018, 150, 5–13. [Google Scholar] [CrossRef]
- Garrido, M.; Paraforos, D.S.; Reiser, D.; Vazquez Arellano, M.; Griepentrog, H.W.; Valero, C. 3d maize plant reconstruction based on georeferenced overlapping lidar point clouds. Remote Sens. 2015, 7, 17077–17096. [Google Scholar] [CrossRef]
- Abendroth, L.J.; Elmore, R.W.; Matthew, J. Boyer; Marlay, S.K. Corn Growth and Development; PMR 1009; Iowa State University Extension: Ames, IA, USA, 2011. [Google Scholar]
- Lin, Y. Lidar: An important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput. Electron. Agric. 2015, 119, 61–73. [Google Scholar] [CrossRef]
- Wen, W.; Guo, X.; Wang, Y.; Zhao, C.; Liao, W. Constructing a three-dimensional resource database of plants using measured in situ morphological data. Appl. Eng. Agric. 2017, 33, 747–756. [Google Scholar] [CrossRef]
- Wu, C. Visualsfm: A Visual Structure from Motion System. Available online: http://ccwu.me/vsfm/ (accessed on 11 June 2014).
- Armoniene, R.; Odilbekov, F.; Vivekanand, V.; Chawade, A. Affordable imaging lab for noninvasive analysis of biomass and early vigour in cereal crops. Biomed. Res. Int. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Li, B.; Li, B.-J.; Guo, X. A leaf modeling and multi-scale remeshing method for visual computation via hierarchical parametric vein and margin representation. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Huang, H.; Wu, S.H.; Cohen-Or, D.; Gong, M.L.; Zhang, H.; Li, G.Q.; Chen, B.Q. L-1-medial skeleton of point cloud. ACM Trans. Graph. 2013, 32, 8. [Google Scholar] [CrossRef]
- Yin, K.X.; Huang, H.; Long, P.X.; Gaissinski, A.; Gong, M.L.; Sharf, A. Full 3d plant reconstruction via intrusive acquisition. Comput. Graph. Forum 2016, 35, 272–284. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, F.; Pang, S.; Zhao, X.; Chen, L.; Liu, J.; Xue, B.; Xu, G.; Li, L.; Jing, H.; et al. Crop 3d: A platform based on lidar for 3d high-throughput crop phenotyping. Sci. Sin. 2016, 46, 1210–1221. [Google Scholar] [CrossRef]
- Xu, Y.B. Envirotyping for deciphering environmental impacts on crop plants. Theor. Appl. Genet. 2016, 129, 653–673. [Google Scholar] [CrossRef] [PubMed]
- Josephs, E.B. Determining the evolutionary forces shaping G × E. New Phytol. 2018, 219, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]

| Hybrids and Growth Stages | AD268 | JK968 | ZD958 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| V5 | V15 | R1 | V5 | V15 | R1 | V5 | V15 | R1 | |
| Averaged total leaf number | 7 | 20 | 22 | 7 | 18 | 20 | 7 | 20 | 22 |
| Averaged plant height (cm) | 40 | 180 | 200 | 43 | 228 | 315 | 45 | 246 | 288 |
| Index | 3D Scanning | MVS | 3D Digitizing | Manual |
|---|---|---|---|---|
| Preparation time | Short | Long | Moderate | Short |
| Preparation difficulty | Easy | Moderate (V5, V15) Hard (R1) | Moderate | Easy |
| Data acquisition device | FARO Focus3D S120 | Canon camera Camera support Wireless shutter | Fastrak, Laptop | Ruler, goniometer |
| Cost of device | $70,000 | $1000 | $15,000 | $10 |
| Raw data format | Point clouds | Image sequences | 3D coordinates of key points | Numbers |
| Data acquisition time | Short (V5), Moderate (V15, R1) | Short | Moderate | Long |
| Data acquisition difficulty | Easy | Easy (V5), moderate (V15), hard (R1) | Moderate | Hard |
| Data processing software | SCENE, Geomagic Studio | PhotoScan Standard Edition | Fastrak | Excel |
| Data processing software costs | SCENE: attached to FARO, Geomagic Studio: $13,000 | $179 | Attached to device | $70 |
| Data processing time | 10 min/plant | 30 min/plant (V5) 40 min/plant (V15, R1) | - | - |
| Data processing difficulty | Moderate | Moderate (V5, V15) Hard (R1) | Easy | Easy |
| Parameters extraction software | CloudCompare, MaizeTypeAna | CloudCompare, MaizeTypeAna | MaizeTypeAna | - |
| Parameters extraction software costs | CloudCompare: open source, MaizeTypeAna: Customized development | CloudCompare: open source, MaizeTypeAna: Customized development | MaizeTypeAna: Customized development | - |
| Parameters extraction time | 10 min/plant (V5) 15 min/plant (V15, R1) | 10 min/plant (V5) 15 min/plant (V15, R1) | 10 s/plant | - |
| Parameters extraction difficulty | Moderate | Moderate | Easy | - |
| Extracted parameter quantity | ≥5 | ≥5 | 17 | 10 |
| Destructiveness | No | No | Yes | Yes |
| Precision | Low (V5), High (V15, R1) | High | Moderate | Moderate |
| Personal error | Low | Low | Moderate | High |
| Expansibility | No | Yes | Yes | No |
| Automation | 80% | 60% | 20% | 0% |
| Labor cost | Low | Moderate | High | High |
| Phenotyping Parameters | 3D Digitizing | 3D Scanning | MVS | ||||||
|---|---|---|---|---|---|---|---|---|---|
| R² | RMSE | N | R² | RMSE | N | R² | RMSE | N | |
| Leaf length | 0.996 | 1.76 cm | 300 | 0.914 | 12.58 cm | 122 | 0.910 | 11.52 cm | 122 |
| Leaf width | 0.902 | 1.06 cm | 300 | 0.843 | 1.71 cm | 122 | 0.852 | 1.77 cm | 122 |
| Leaf angle | 0.852 | 4.89° | 300 | 0.866 | 4.97° | 122 | 0.870 | 4.93° | 122 |
| Leaf azimuth | 0.966 | 20.70° | 300 | 0.948 | 25.37° | 122 | 0.954 | 23.92° | 122 |
| Plant height | 0.999 | 2.41 cm | 27 | 0.998 | 4.99 cm | 27 | 0.998 | 5.69 cm | 27 |
| Leaf area | 0.933 | 69.4 cm2 | 300 | 0.869 | 131.40 cm2 | 122 | 0.887 | 140.88 cm2 | 122 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wen, W.; Wu, S.; Wang, C.; Yu, Z.; Guo, X.; Zhao, C. Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates. Remote Sens. 2019, 11, 63. https://doi.org/10.3390/rs11010063
Wang Y, Wen W, Wu S, Wang C, Yu Z, Guo X, Zhao C. Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates. Remote Sensing. 2019; 11(1):63. https://doi.org/10.3390/rs11010063
Chicago/Turabian StyleWang, Yongjian, Weiliang Wen, Sheng Wu, Chuanyu Wang, Zetao Yu, Xinyu Guo, and Chunjiang Zhao. 2019. "Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates" Remote Sensing 11, no. 1: 63. https://doi.org/10.3390/rs11010063
APA StyleWang, Y., Wen, W., Wu, S., Wang, C., Yu, Z., Guo, X., & Zhao, C. (2019). Maize Plant Phenotyping: Comparing 3D Laser Scanning, Multi-View Stereo Reconstruction, and 3D Digitizing Estimates. Remote Sensing, 11(1), 63. https://doi.org/10.3390/rs11010063