Tropical Deforestation and Recolonization by Exotic and Native Trees: Spatial Patterns of Tropical Forest Biomass, Functional Groups, and Species Counts and Links to Stand Age, Geoclimate, and Sustainability Goals
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Field Data and Tree Species Characteristics Dataset
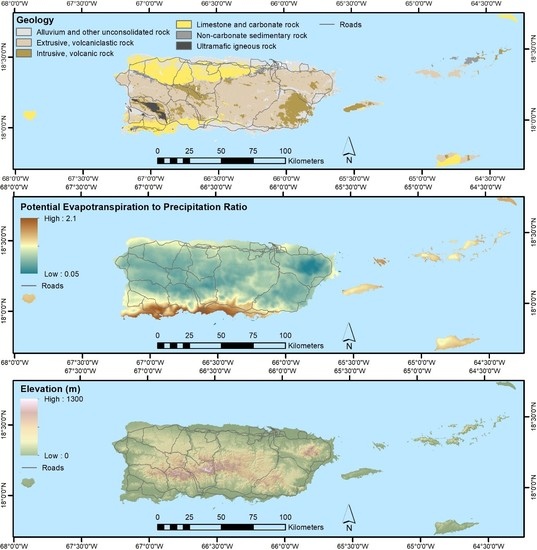
2.3. Mapped Predictor Data
2.3.1. Remotely Sensed Imagery
2.3.2. Other Predictor Data
2.3.3. Background on Modeling Forest Attributes with Cubist and PGNN
2.3.4. Mapping with Forest Inventory Data, Cubist, and PGNN
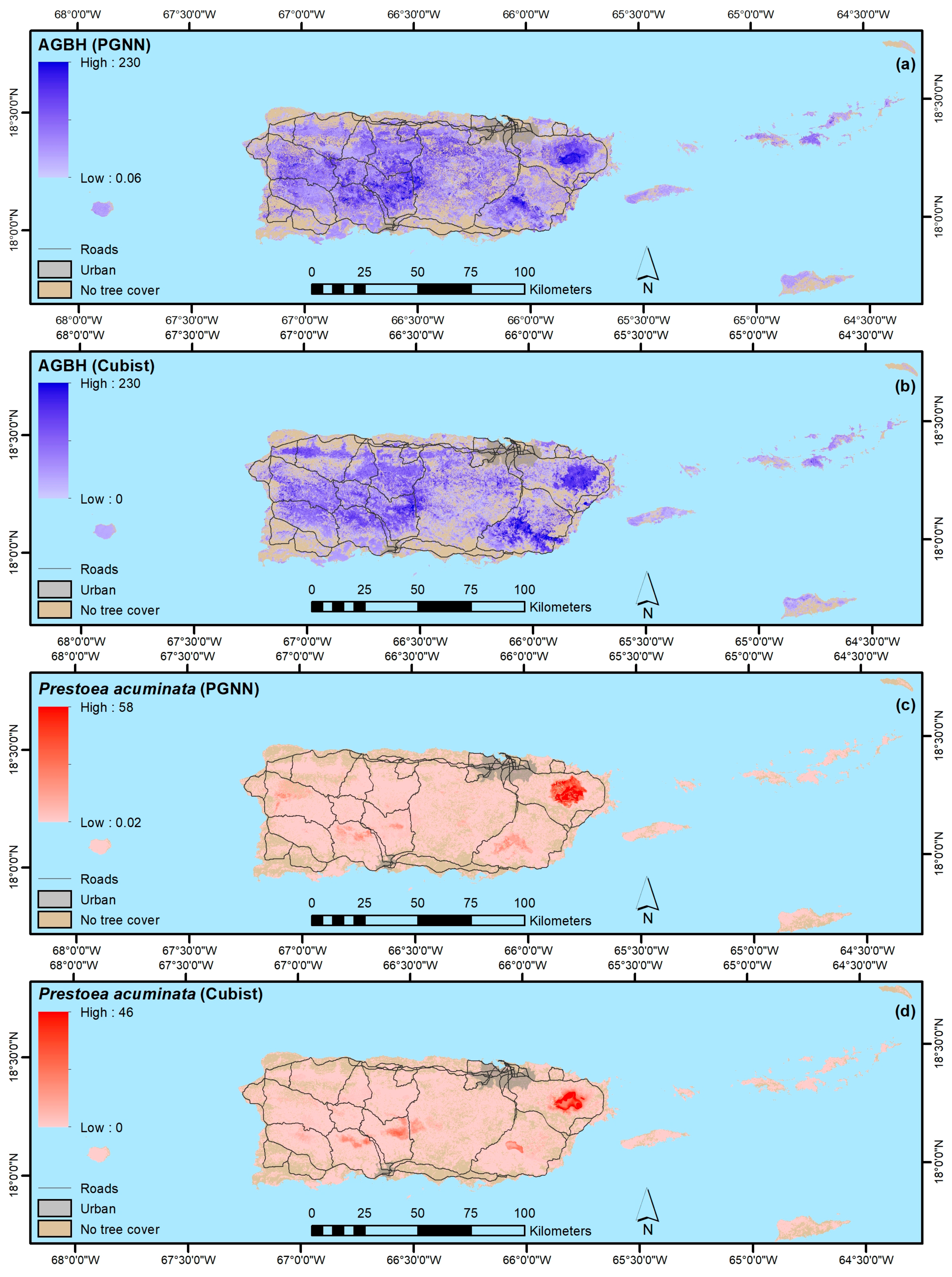
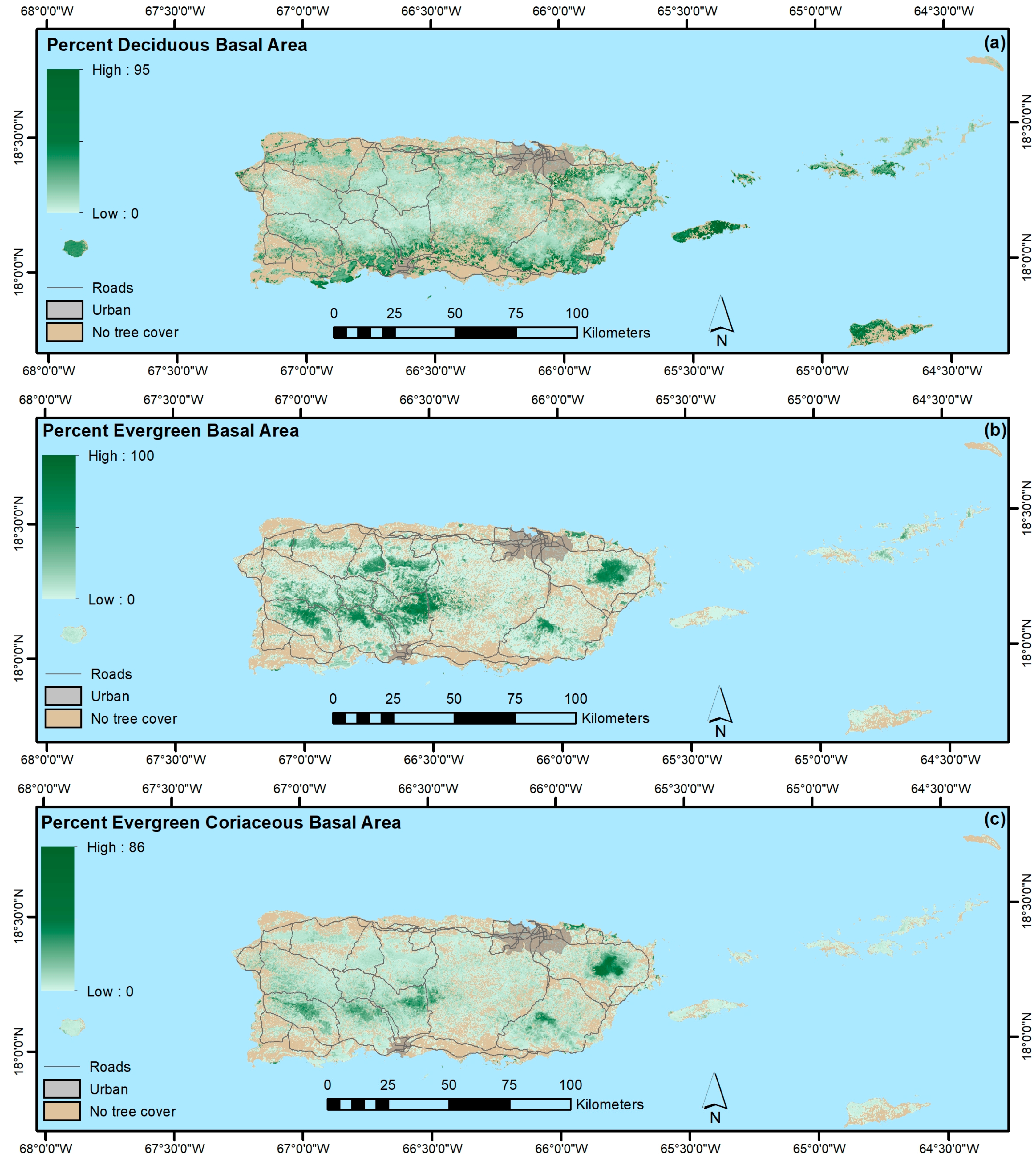
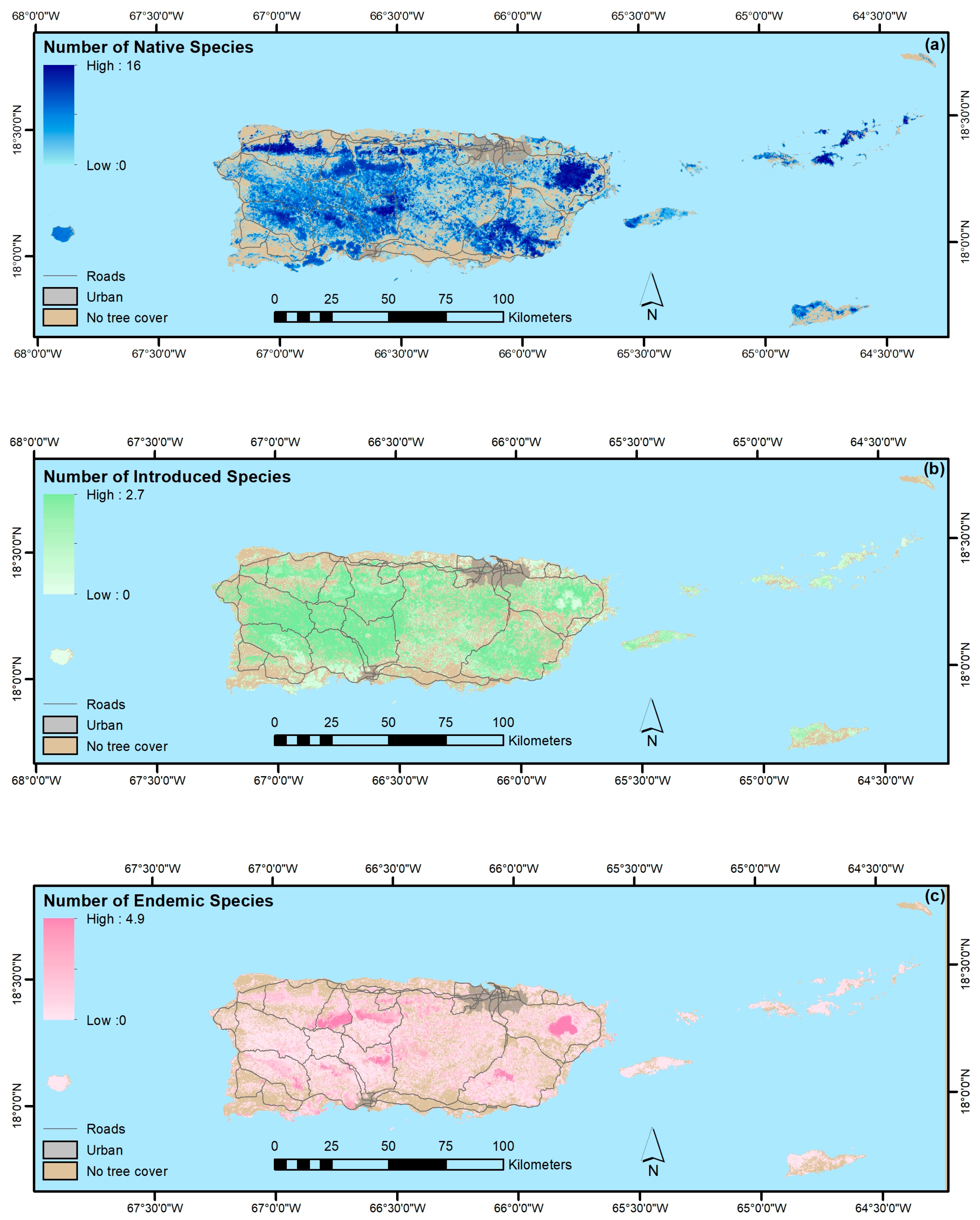
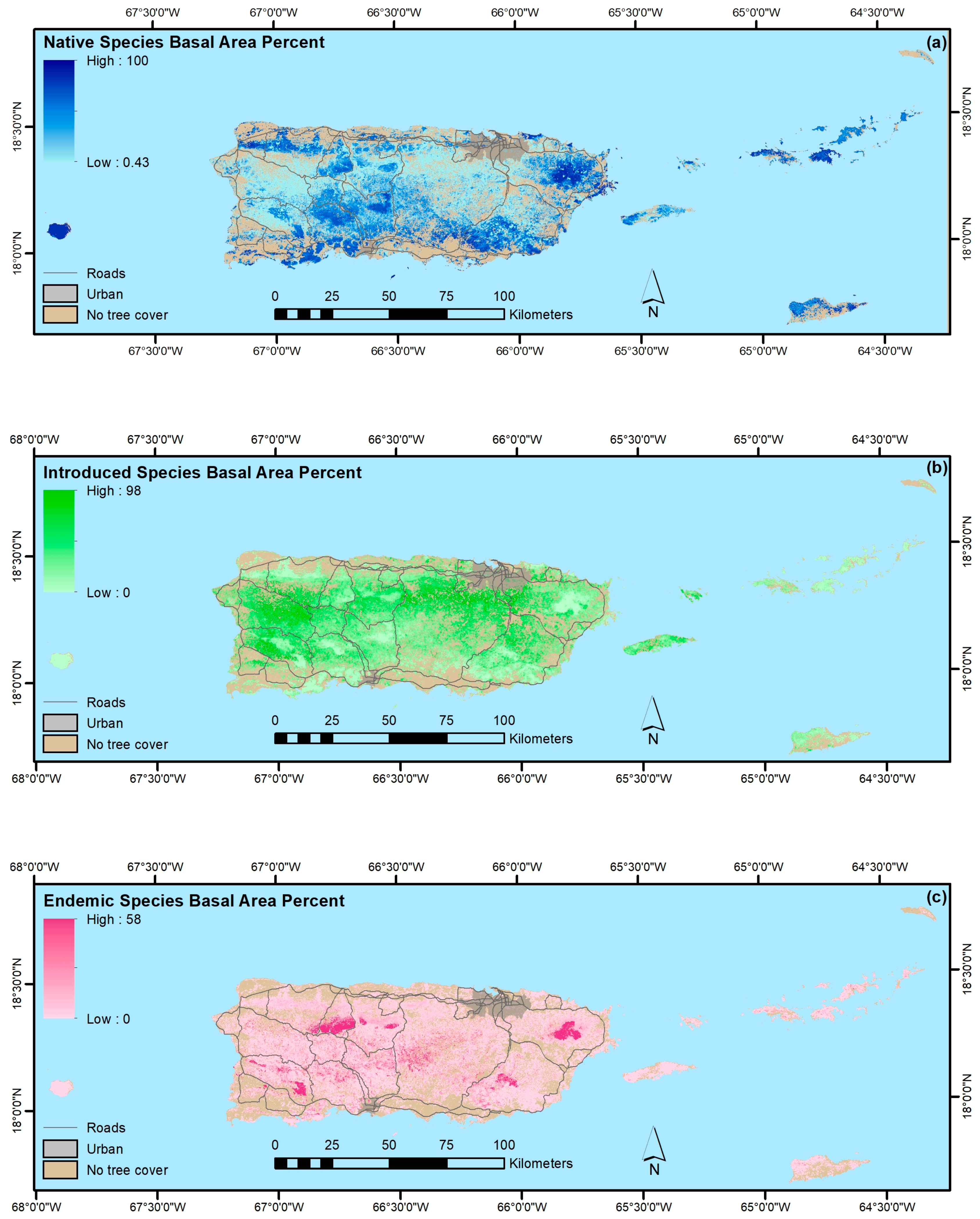
3. Results
4. Discussion
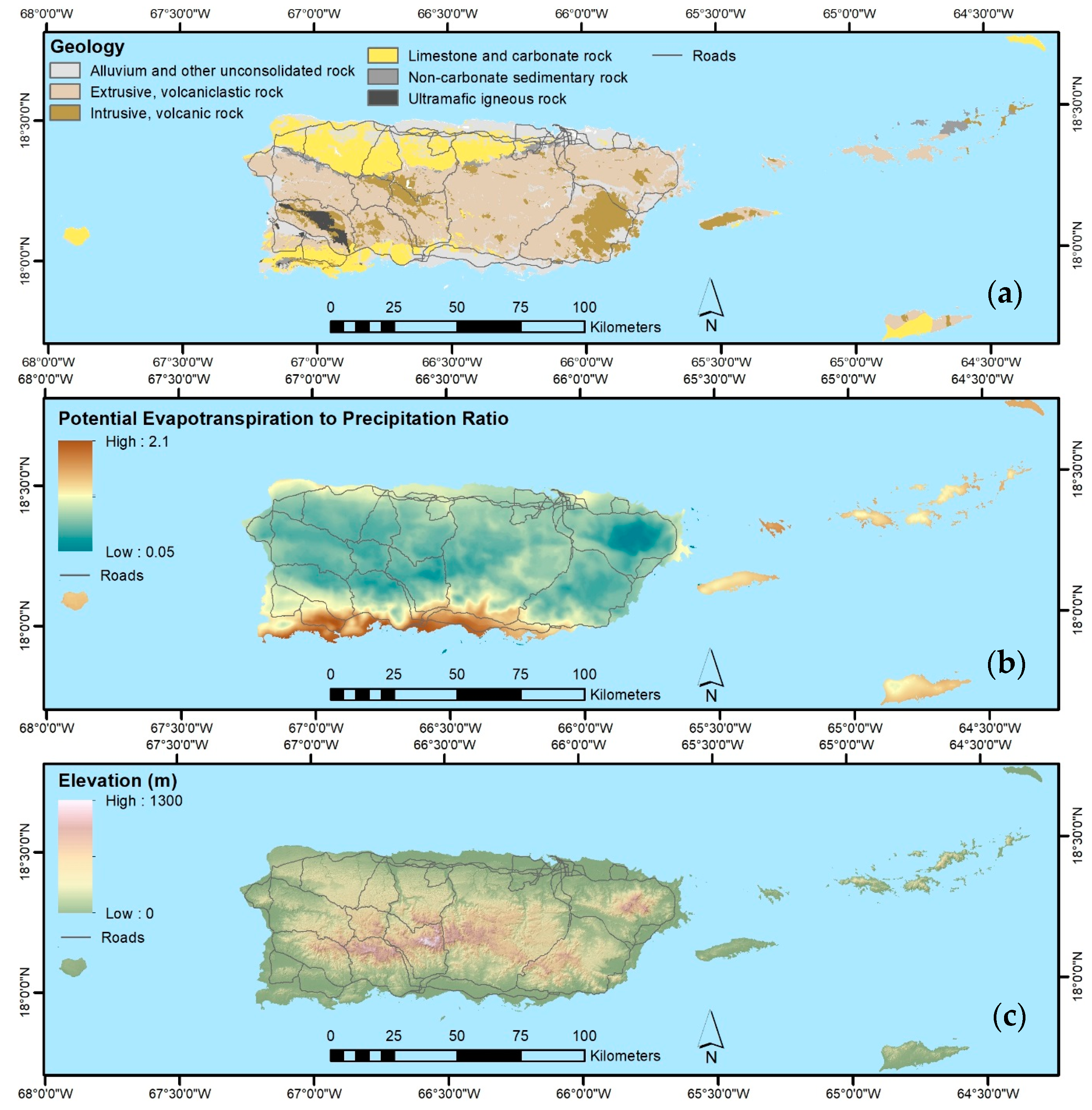
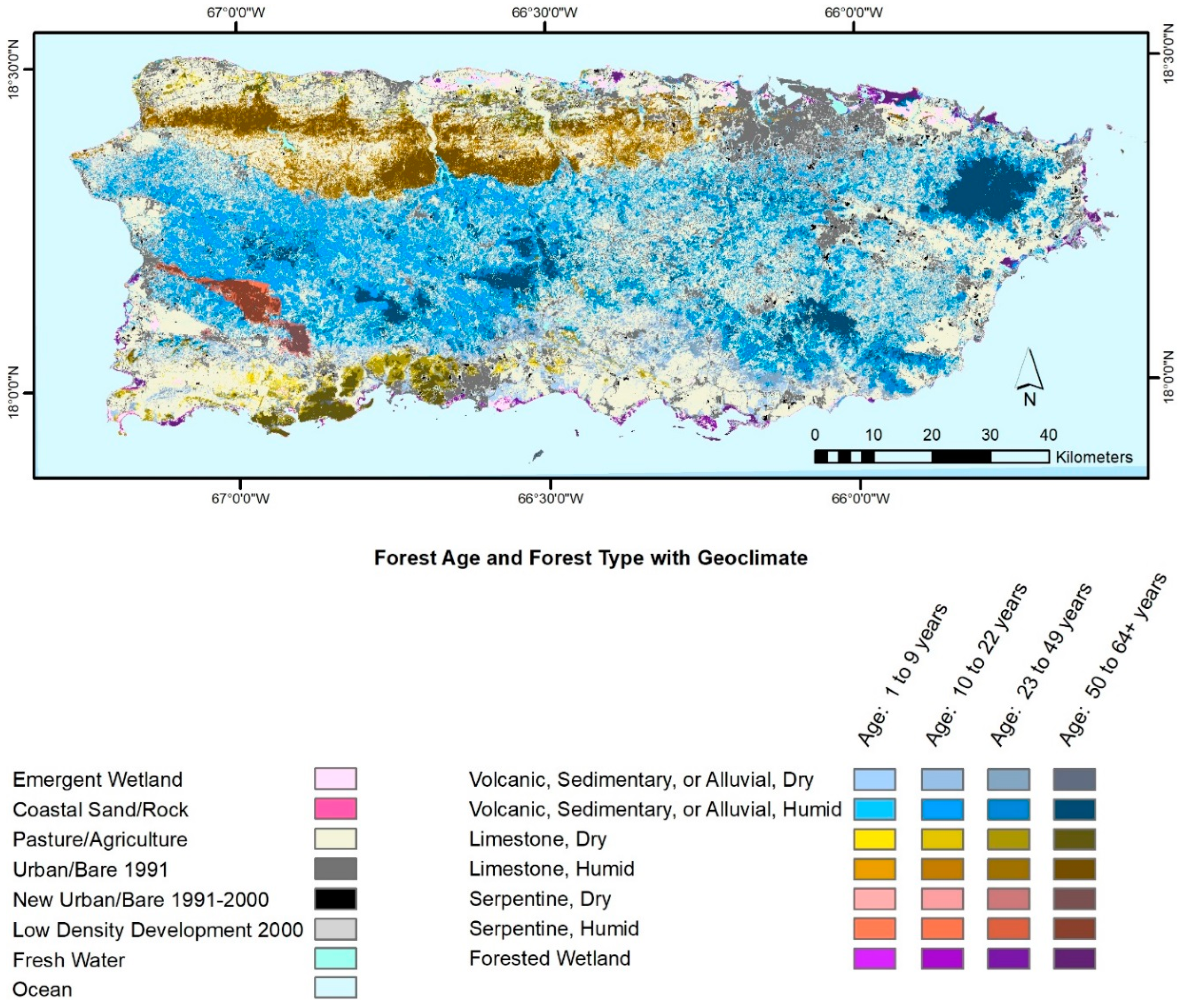
4.1. Forest Spatial Patterns Reflect Stand Age, Geoclimate, and Disturbance History
4.1.1. Biophysical, Socioeconomic, and Landscape Controls on Forest Recovery
4.1.2. Forest Structure and Biomass
4.1.3. Forest Functional Groups
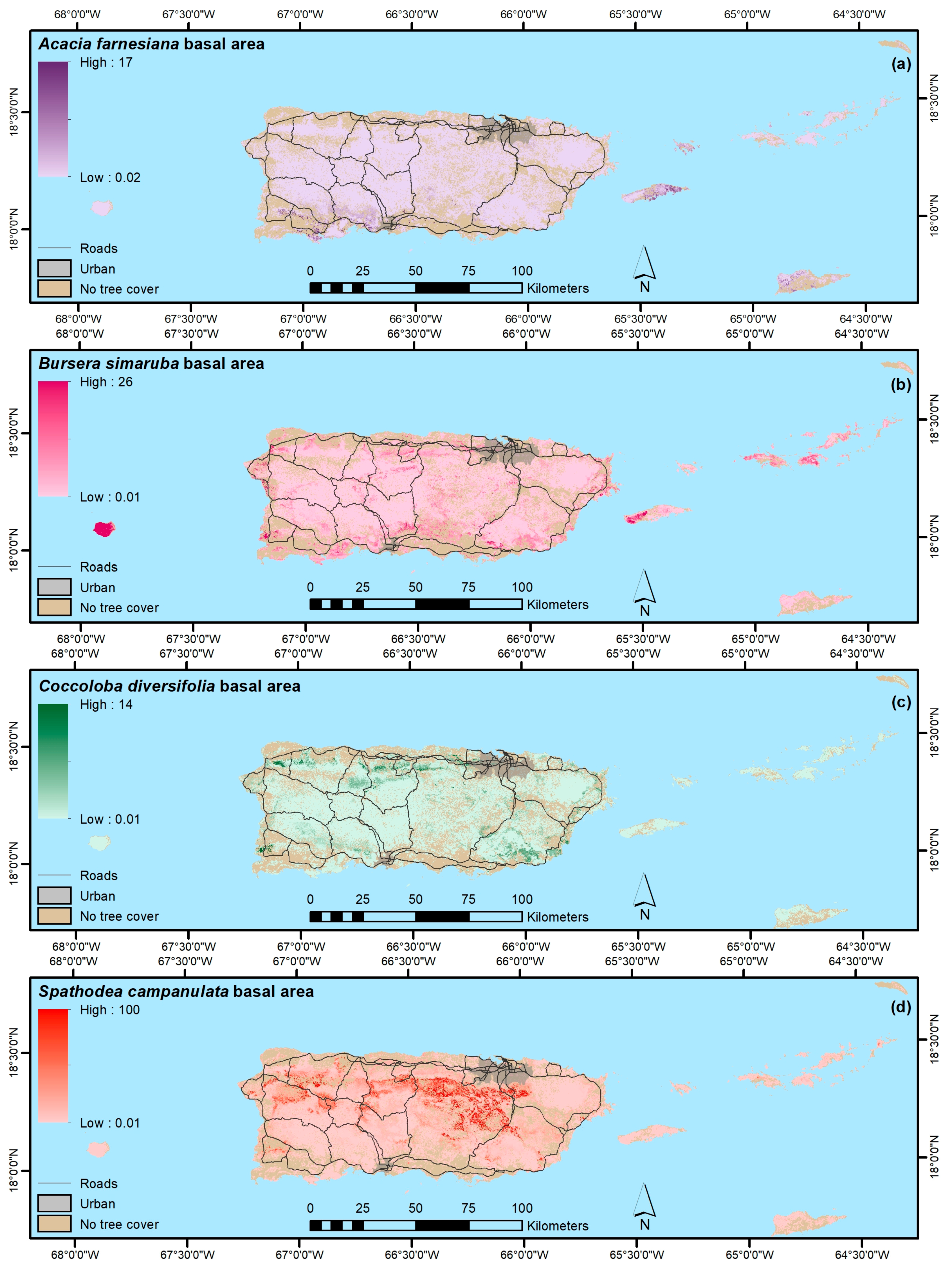
4.1.4. Native, Endemic, Introduced and Individual Species
4.1.5. Implications for Sustainable Development Goal Indicators
4.2. Modeling Forest Characteristics vs. Modeling Individual Tree Species
5. Conclusions
- The legacy of past large-scale clearing and subsequent forest recovery are clear when mapping current forest attributes in this tropical region. Introduced species are widespread across most of Puerto Rico and the Virgin Islands, and they often dominate stands. Moreover, introduced species are most prominent in younger forests. They are less common or rare, however, in the least-arable geoclimatic zones or more remote places.
- Native species counts are usually larger than counts of introduced species, and their relative basal areas tend to increase with stand age. Endemic species are concentrated in the oldest forests, which are in the least arable and most remote locations, including isolated extreme environments where endemic species often evolve, that were also, probably not coincidentally, protected early and in some places never cleared.
- Our mapping models explained a minority of the variance in forest attributes. Still, our results suggest that PGNN is better than regression tree models for mapping individual species and that, as in others’ work, the most common species are generally not the most accurately mapped ones. Instead, species with ranges limited by climate or other environmental factors are most easily mapped. Further, ordination in particular likely helps map species by virtue of their association with other species, which in effect increases the sample size for model training.
- In contrast, regression tree models appear to be better than ordination-based models for mapping other forest attributes, like forest biomass, functional groups, and species origin, at least when species basal areas are used to fit the ordination-based models.
- Several forest attributes addressed in UN SDGs have spatial patterns that reflect stand age, geoclimate, and aspects of disturbance history other than stand age since abandonment or last disturbance, like past disturbance type or intensity. Forest age, height, biomass, or disturbance-type history can now be mapped more accurately with long time series of globally available imagery [53,124,125,126,127,128]. Our results suggest that such maps, when combined with climate and lithology maps, would be useful for monitoring related to SDGs.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
References
- Achard, F.; Beuchle, R.; Mayaux, P.; Stibig, H.-J.; Bodart, C.; Brink, A.; Carboni, S.; Desclée, B.; Donnay, F.; Eva, H.D.; et al. Determination of tropical deforestation rates and related carbon losses from 1990 to 2010. Glob. Chang. Biol. 2014, 20, 2540–2554. [Google Scholar] [CrossRef] [PubMed]
- Dirzo, R.; Raven, P.H. Global State of Biodiversity and Loss. Annu. Rev. Environ. Resour. 2003, 28, 137–167. [Google Scholar] [CrossRef]
- Bradshaw, C.J.; Sodhi, N.S.; Brook, B.W. Tropical turmoil: A biodiversity tragedy in progress. Front. Ecol. Environ. 2009, 7, 79–87. [Google Scholar] [CrossRef]
- Pimm, S.L.; Raven, P. Extinction by numbers. Nature 2000, 403, 843. [Google Scholar] [CrossRef] [PubMed]
- Berenguer, E.; Ferreira, J.; Gardner, T.A.; Oliveira Cruz Aragão, L.E.; Barbosa De Camargo, P.; Cerri, C.E.; Durigan, M.; Oliveira, R.C.D.; Vieira, I.C.G.; Barlow, J. A large-scale field assessment of carbon stocks in human-modified tropical forests. Glob. Chang. Biol. 2014, 20, 3713–3726. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.A.; Barlow, J.; Sodhi, N.S.; Peres, C.A. A multi-region assessment of tropical forest biodiversity in a human-modified world. Biol. Conserv. 2010, 143, 2293–2300. [Google Scholar] [CrossRef]
- Gardner, T.A.; Barlow, J.; Chazdon, R.; Ewers, R.M.; Harvey, C.A.; Peres, C.A.; Sodhi, N.S. Prospects for tropical forest biodiversity in a human-modified world. Ecol. Lett. 2009, 12, 561–582. [Google Scholar] [CrossRef] [PubMed]
- Gibson, L.; Lee, T.M.; Koh, L.P.; Brook, B.W.; Gardner, T.A.; Barlow, J.; Peres, C.A.; Bradshaw, C.J.A.; Laurance, W.F.; Lovejoy, T.E.; et al. Primary forests are irreplaceable for sustaining tropical biodiversity. Nature 2011, 478, 378. [Google Scholar] [CrossRef] [PubMed]
- Barlow, J.; Gardner, T.A.; Araujo, I.S.; Ávila-Pires, T.C.; Bonaldo, A.B.; Costa, J.E.; Esposito, M.C.; Ferreira, L.V.; Hawes, J.; Hernandez, M.I.M.; et al. Quantifying the biodiversity value of tropical primary, secondary, and plantation forests. Proc. Natl. Acad. Sci. USA 2007, 104, 18555–18560. [Google Scholar] [CrossRef] [PubMed]
- Rulequest. Data Mining with Cubist, Rulequest Research. 2018. Available online: https://www.rulequest.com/cubist-info.html (accessed on 31 October 2018).
- Ohmann, J.L.; Gregory, M.J. Predictive mapping of forest composition and structure with direct gradient analysis and nearest-neighbor imputation in coastal Oregon, U.S.A. Can. J. For. Res. 2002, 32, 725–741. [Google Scholar] [CrossRef]
- Wilson, B.T.; Lister, A.J.; Riemann, R.I. A nearest-neighbor imputation approach to mapping tree species over large areas using forest inventory plots and moderate resolution raster data. For. Ecol. Manag. 2012, 271, 182–198. [Google Scholar] [CrossRef]
- Wilson, B.T.; Woodall, C.W.; Griffith, D.M. Imputing forest carbon stock estimates from inventory plots to a nationally continuous coverage. Carbon Balance Manag. 2013, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, R.H. The Ecology of Serpentine Soils. Ecology 1954, 35, 258–288. [Google Scholar] [CrossRef]
- USDA Forest Service. Forest Service. Forest inventory and analysis national core field guide. In Field Data Collection Procedures for Phase 2 Plots; Version 6.11; U.S. Department of Agriculture Forest Service: Washington, DC, USA, 2014; Volume 1, p. 305. [Google Scholar]
- Acevedo-Rodríguez, P. Flora of St. John, US Virgin Islands; New York Botanical Garden Bronx: New York, NY, USA, 1996. [Google Scholar]
- Little, E.L.; Wadsworth, F.H. Common trees of Puerto Rico and the Virgin Islands, Vol 2; USDA Forest Service, Agriculture Handbook No. 449: Washington, DC, USA, 1974. [Google Scholar]
- Little, E.L.; Woodbury, R.O.; Wadsworth, F.H. Trees of Puerto Rico and the Virgin Islands Vol 1; USDA Forest Service, Agriculture Handbook No. 249: Washington, DC, USA, 1964.
- Liogier, A.H. Descriptive Flora of Puerto Rico and Adjacent Islands. Vol II: Leguminosae to Anacardiaceae; Editorial de la Universidad de Puerto Rico: Río Piedras, Puerto Rico, 1988. [Google Scholar]
- Liogier, A.H. Descriptive Flora of Puerto Rico and Adjacent Islands. Vol. I: Casuarinaceae to Connaraceae; Editorial de la Universidad de Puerto Rico: Río Piedras, Puerto Rico, 1985. [Google Scholar]
- Liogier, A.H. Descriptive Flora of Puerto Rico and Adjacent Islands. Vol. V: Acanthaceae to Compositae; Editorial de la Universidad de Puerto Rico: Río Piedras, Puerto Rico, 1997. [Google Scholar]
- Liogier, A.H. Descriptive Flora of Puerto Rico and Adjacent Islands. Vol. III: Cyrillaceae to Myrtaceae; Editorial de la Universidad de Puerto Rico: Río Piedras, Puerto Rico, 1994. [Google Scholar]
- Liogier, A.H.; Martorell, L.F. Flora of Puerto Rico and Adjacent Islands: A Systematic Synopsis; Editorial de la Universidad de Puerto Rico: Río Piedras, Puerto Rico, 2000. [Google Scholar]
- Gibney, E. A Field Guide to Native Trees & Plants of East End, St. John, US Virgin Islands; Center for the Environment: St. John, U.S. Virgin Islands, USA, 2004; p. 86. [Google Scholar]
- Scurlock, J.P. Native Trees and Shrubs of the Florida Keys: A Field Guide; Laurel Press: Pittsburg, PA, USA, 1987; p. 220. [Google Scholar]
- Weaver, P.L.; Schwagerl, J.J. US Fish and Wildlife Service Refuges and Other Nearby Reserves in Southwestern Puerto Rico; International Institute of Tropical Forestry, USDA Forest Service: Río Piedras, Puerto Rico, 2009.
- Kew. Seed Information Database (SID). Available online: http://data.kew.org/sid/ (accessed on 1 January 2011).
- Erickson, H.E.; Helmer, E.H.; Brandeis, T.J.; Lugo, A.E. Controls on fallen leaf chemistry and forest floor element masses in native and novel forests across a tropical island. Ecosphere 2014, 5, art48. [Google Scholar] [CrossRef]
- Gond, V.; Freycon, V.; Molino, J.-F.; Brunaux, O.; Ingrassia, F.; Joubert, P.; Pekel, J.-F.; Prévost, M.-F.; Thierron, V.; Trombe, P.-J.; et al. Broad-scale spatial pattern of forest landscape types in the Guiana Shield. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 357–367. [Google Scholar] [CrossRef]
- Gwenzi, D.; Helmer, E.; Zhu, X.; Lefsky, M.; Marcano-Vega, H. Predictions of Tropical Forest Biomass and Biomass Growth Based on Stand Height or Canopy Area Are Improved by Landsat-Scale Phenology across Puerto Rico and the U.S. Virgin Islands. Remote Sens. 2017, 9, 123. [Google Scholar] [CrossRef]
- Helmer, E.; Goodwin, N.R.; Gond, V.; Souza, C.M., Jr.; Asner, G.P. Characterizing tropical forests with multispectral imagery. Land Resour. 2015, 2, 367–396. [Google Scholar]
- Helmer, E.H.; Ruzycki, T.S.; Benner, J.; Voggesser, S.M.; Scobie, B.P.; Park, C.; Fanning, D.W.; Ramnarine, S. Detailed maps of tropical forest types are within reach: Forest tree communities for Trinidad and Tobago mapped with multiseason Landsat and multiseason fine-resolution imagery. For. Ecol. Manag. 2012, 279, 147–166. [Google Scholar] [CrossRef]
- Helmer, E.H.; Ruefenacht, B. Cloud-Free Satellite Image Mosaics with Regression Trees and Histogram Matching. Photogramm. Eng. Remote Sens. 2005, 71, 1079–1089. [Google Scholar] [CrossRef]
- Helmer, E.H.; Ruefenacht, B. A comparison of radiometric normalization methods when filling cloud gaps in Landsat imagery. Can. J. Remote Sens. 2007, 33, 325–340. [Google Scholar] [CrossRef]
- Helmer, E.H.; Brown, S.; Cohen, W.B. Mapping montane tropical forest successional stage and land use with multi-date Landsat imagery. Int. J. Remote Sens. 2000, 21, 2163–2183. [Google Scholar] [CrossRef]
- Helmer, E.H.; Ruzycki, T.S.; Wunderle, J.M.; Vogesser, S.; Ruefenacht, B.; Kwit, C.; Brandeis, T.J.; Ewert, D.N. Mapping tropical dry forest height, foliage height profiles and disturbance type and age with a time series of cloud-cleared Landsat and ALI image mosaics to characterize avian habitat. Remote Sens. Environ. 2010, 114, 2457–2473. [Google Scholar] [CrossRef]
- Kennaway, T.; Helmer, E.H. The Forest Types and Ages Cleared for Land Development in Puerto Rico. GISci. Remote Sens. 2007, 44, 356–382. [Google Scholar] [CrossRef]
- Kennaway, T.A.; Helmer, E.H.; Lefsky, M.A.; Brandeis, T.J.; Sherrill, K.R. Mapping land cover and estimating forest structure using satellite imagery and coarse resolution lidar in the Virgin Islands. J. Appl. Remote Sens. 2008, 2, 023551. [Google Scholar]
- Daly, C.; Helmer, E.H.; Quiñones, M. Mapping the climate of Puerto Rico, Vieques and Culebra. Int. J. Climatol. 2003, 23, 1359–1381. [Google Scholar] [CrossRef]
- Asner, G.P.; Mascaro, J.; Anderson, C.; Knapp, D.E.; Martin, R.E.; Kennedy-Bowdoin, T.; van Breugel, M.; Davies, S.; Hall, J.S.; Muller-Landau, H.C.; et al. High-fidelity national carbon mapping for resource management and REDD+. Carbon Balance Manag. 2013, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Asner, G.P.; Powell, G.V.N.; Mascaro, J.; Knapp, D.E.; Clark, J.K.; Jacobson, J.; Kennedy-Bowdoin, T.; Balaji, A.; Paez-Acosta, G.; Victoria, E.; et al. High-resolution forest carbon stocks and emissions in the Amazon. Proc. Natl. Acad. Sci. USA 2010, 107, 16738–16742. [Google Scholar] [CrossRef] [PubMed]
- Helmer, E.H. The Landscape Ecology of Tropical Secondary Forest in Montane Costa Rica. Ecosystems 2000, 3, 98–114. [Google Scholar] [CrossRef]
- Helmer, E.H.; Brandeis, T.J.; Lugo, A.E.; Kennaway, T. Factors influencing spatial pattern in tropical forest clearance and stand age: Implications for carbon storage and species diversity. J. Geophys. Res. Biogeosci. 2008, 113. [Google Scholar] [CrossRef]
- Huete, A.; Justice, C.; Van Leeuwen, W. MODIS Vegetation Index (MOD13) Algorithm Theoretical Basis Document; University of Arizona: Tucson, AZ, USA, 1999; p. 213. [Google Scholar]
- Krushensky, R. Generalized geology map of Puerto Rico; US Geological Survey: San Juan, Puerto Rico, 1995.
- Garrison, L.E.; Martin, R.; Berryhill, H.; Buell, M.; Ensminger, H.; Perry, R. Preliminary Tectonic Map of the Eastern Greater Antilles Region; U.S. Department of the Interior—Geological Survey: Washington, DC, USA, 1972.
- Slater, J.A.; Garvey, G.; Johnston, C.; Haase, J.; Heady, B.; Kroenung, G.; Little, J. The SRTM data “finishing” process and products. Photogramm. Eng. Remote Sens. 2006, 72, 237–247. [Google Scholar] [CrossRef]
- Homer, C.; Huang, C.; Yang, L.; Wylie, B.; Coan, M. Development of a 2001 National Land-Cover Database for the United States. Photogramm. Eng. Remote Sens. 2004, 70, 829–840. [Google Scholar] [CrossRef]
- Quinlan, J.R. Improved use of continuous attributes in C4.5. J. Artif. Intell. Res. 1996, 4, 77–90. [Google Scholar] [CrossRef]
- Walton, J.T. Subpixel Urban Land Cover Estimation. Photogramm. Eng. Remote Sens. 2008, 74, 1213–1222. [Google Scholar] [CrossRef]
- Blackard, J.A.; Finco, M.V.; Helmer, E.H.; Holden, G.R.; Hoppus, M.L.; Jacobs, D.M.; Lister, A.J.; Moisen, G.G.; Nelson, M.D.; Riemann, R.; et al. Mapping U.S. forest biomass using nationwide forest inventory data and moderate resolution information. Remote Sens. Environ. 2008, 112, 1658–1677. [Google Scholar] [CrossRef]
- Lefsky, M.A. A global forest canopy height map from the Moderate Resolution Imaging Spectroradiometer and the Geoscience Laser Altimeter System. Geophys. Res. Lett. 2010, 37. [Google Scholar] [CrossRef]
- Li, A.; Huang, C.; Sun, G.; Shi, H.; Toney, C.; Zhu, Z.; Rollins, M.G.; Goward, S.N.; Masek, J.G. Modeling the height of young forests regenerating from recent disturbances in Mississippi using Landsat and ICESat data. Remote Sens. Environ. 2011, 115, 1837–1849. [Google Scholar] [CrossRef]
- Rudiyanto; Budiman, M.; Budi Indra, S.; Chusnul, A.; Saptomo, S.K.; Chadirin, Y. Digital mapping for cost-effective and accurate prediction of the depth and carbon stocks in Indonesian peatlands. Geoderma 2016, 272, 20–31. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Aertsen, W.; Kint, V.; van Orshoven, J.; Özkan, K.; Muys, B. Comparison and ranking of different modelling techniques for prediction of site index in Mediterranean mountain forests. Ecol. Model. 2010, 221, 1119–1130. [Google Scholar] [CrossRef]
- Colditz, R. An Evaluation of Different Training Sample Allocation Schemes for Discrete and Continuous Land Cover Classification Using Decision Tree-Based Algorithms. Remote Sens. 2015, 7, 9655. [Google Scholar] [CrossRef]
- Hudak, A.T.; Crookston, N.L.; Evans, J.S.; Hall, D.E.; Falkowski, M.J. Nearest neighbor imputation of species-level, plot-scale forest structure attributes from LiDAR data. Remote Sens. Environ. 2008, 112, 2232–2245. [Google Scholar] [CrossRef]
- Ter Braak, C.J. The analysis of vegetation-environment relationships by canonical correspondence analysis. Vegetatio 1987, 69, 69–77. [Google Scholar] [CrossRef]
- Ohmann, J.L.; Gregory, M.J.; Roberts, H.M.; Cohen, W.B.; Kennedy, R.E.; Yang, Z. Mapping change of older forest with nearest-neighbor imputation and Landsat time-series. For. Ecol. Manag. 2012, 272, 13–25. [Google Scholar] [CrossRef]
- Pierce, K.B.; Ohmann, J.L.; Wimberly, M.C.; Gregory, M.J.; Fried, J.S. Mapping wildland fuels and forest structure for land management: A comparison of nearest neighbor imputation and other methods. Can. J. For. Res. 2009, 39, 1901–1916. [Google Scholar] [CrossRef]
- Steinberg, D.; Colla, P. CART: Classification and regression trees. Top Ten Algorithms Data Min. 2009, 9, 179. [Google Scholar]
- Ruefenacht, B.; Finco, M.V.; Nelson, M.D.; Czaplewski, R.; Helmer, E.H.; Blackard, J.A.; Holden, G.R.; Lister, A.J.; Salajanu, D.; Weyermann, D.; et al. Conterminuous US and Alaska forest type mapping using forest inventory and analysis data. Photogramm. Eng. Remote Sens. 2008, 74, 1379–1388. [Google Scholar] [CrossRef]
- Dray, S.; Dufour, A.-B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 20. [Google Scholar] [CrossRef]
- Li, M.; Im, J.; Quackenbush, L.J.; Liu, T. Forest Biomass and Carbon Stock Quantification Using Airborne LiDAR Data: A Case Study Over Huntington Wildlife Forest in the Adirondack Park. IEEE J. Sel. Top. Appl. Earth Observ. Remote Sens. 2014, 7, 3143–3156. [Google Scholar] [CrossRef]
- Ohmann, J.L.; Gregory, M.J.; Spies, T.A. Influence of environment, disturbance, and ownership on forest vegetation of coastal Oregon. Ecol. Appl. 2007, 17, 18–33. [Google Scholar] [CrossRef]
- Reese, H.; Nilsson, M.; Pahlén, T.G.; Hagner, O.; Joyce, S.; Tingelöf, U.; Egberth, M.; Olsson, H. Countrywide Estimates of Forest Variables Using Satellite Data and Field Data from the National Forest Inventory. AMBIO 2003, 32, 542–548. [Google Scholar] [CrossRef] [PubMed]
- Sales, M.H.; Souza, C.M.; Kyriakidis, P.C.; Roberts, D.A.; Vidal, E. Improving spatial distribution estimation of forest biomass with geostatistics: A case study for Rondônia, Brazil. Ecol. Model. 2007, 205, 221–230. [Google Scholar] [CrossRef]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A. Novel methods improve prediction of species' distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef]
- Castro-Prieto, J.; Quiñones, M.; Gould, W. Characterization of the Network of Protected Areas in Puerto Rico. Caribb. Nat. 2016, 29, 1–16. [Google Scholar]
- Gould, W.A.; Quinones, M.; Solorzano, M.; Alcobas, W.; Alarcon, C. Protected Natural Areas of Puerto Rico. 1:240,000, Res. Map IITF-RMAP-02; U.S. Department of Agriculture, Forest Service, International Institute of Tropical Forestry: Rio Piedras, Puerto Rico, 2011.
- Helmer, E.H. Forest conservation and land development in Puerto Rico. Landsc. Ecol. 2004, 19, 29–40. [Google Scholar] [CrossRef]
- Venter, O.; Magrach, A.; Outram, N.; Klein, C.J.; Possingham, H.P.; Di Marco, M.; Watson, J.E.M. Bias in protected-area location and its effects on long-term aspirations of biodiversity conventions. Conserv. Biol. 2018, 32, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Etter, A.; McAlpine, C.; Pullar, D.; Possingham, H. Modeling the age of tropical moist forest fragments in heavily-cleared lowland landscapes of Colombia. For. Ecol. Manag. 2005, 208, 249–260. [Google Scholar] [CrossRef]
- Newman, M.E.; McLaren, K.P.; Wilson, B.S. Long-term socio-economic and spatial pattern drivers of land cover change in a Caribbean tropical moist forest, the Cockpit Country, Jamaica. Agric. Ecosyst. Environ. 2014, 186, 185–200. [Google Scholar] [CrossRef]
- Nagendra, H.; Southworth, J.; Tucker, C. Accessibility as a determinant of landscape transformation in western Honduras: Linking pattern and process. Landsc. Ecol. 2003, 18, 141–158. [Google Scholar] [CrossRef]
- Aide, T.M.; Zimmerman, J.K.; Pascarella, J.B.; Rivera, L.; Marcano-Vega, H. Forest Regeneration in a Chronosequence of Tropical Abandoned Pastures: Implications for Restoration Ecology. Restor. Ecol. 2000, 8, 328–338. [Google Scholar] [CrossRef]
- Aide, T.M.; Zimmerman, J.K.; Rosario, M.; Marcano, H. Forest recovery in abandoned cattle pastures along an elevational gradient in northeastern Puerto Rico. Biotropica 1996, 28, 537–548. [Google Scholar] [CrossRef]
- Zarin, D.J.; Davidson, E.A.; Brondizio, E.; Vieira, I.C.; Sá, T.; Feldpausch, T.; Schuur, E.A.; Mesquita, R.; Moran, E.; Delamonica, P.; et al. Legacy of fire slows carbon accumulation in Amazonian forest regrowth. Front. Ecol. Environ. 2005, 3, 365–369. [Google Scholar] [CrossRef]
- Pregitzer, K.S.; Euskirchen, E.S. Carbon cycling and storage in world forests: Biome patterns related to forest age. Glob. Chang. Biol. 2004, 10, 2052–2077. [Google Scholar] [CrossRef]
- Luyssaert, S.; Schulze, E.D.; Börner, A.; Knohl, A.; Hessenmöller, D.; Law, B.E.; Ciais, P.; Grace, J. Old-growth forests as global carbon sinks. Nature 2008, 455, 213. [Google Scholar] [CrossRef] [PubMed]
- Stephenson, N.L.; Das, A.J.; Condit, R.; Russo, S.E.; Baker, P.J.; Beckman, N.G.; Coomes, D.A.; Lines, E.R.; Morris, W.K.; Rüger, N.; et al. Rate of tree carbon accumulation increases continuously with tree size. Nature 2014, 507, 90. [Google Scholar] [CrossRef] [PubMed]
- Detto, M.; Muller-Landau, H.C.; Mascaro, J.; Asner, G.P. Hydrological Networks and Associated Topographic Variation as Templates for the Spatial Organization of Tropical Forest Vegetation. PLoS ONE 2013, 8, e76296. [Google Scholar] [CrossRef] [PubMed]
- Finegan, B. Forest succession. Nature 1984, 312, 109. [Google Scholar] [CrossRef]
- Chaer, G.M.; Resende, A.S.; Campello, E.F.C.; de Faria, S.M.; Boddey, R.M. Nitrogen-fixing legume tree species for the reclamation of severely degraded lands in Brazil. Tree Physiol. 2011, 31, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Franco, A.A.; De Faria, S.M. The contribution of N2-fixing tree legumes to land reclamation and sustainability in the tropics. Soil Biol. Biochem. 1997, 29, 897–903. [Google Scholar] [CrossRef]
- Erickson, H.; Davidson, E.A.; Keller, M. Former land-use and tree species affect nitrogen oxide emissions from a tropical dry forest. Oecologia 2002, 130, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Gei, M.; Rozendaal, D.M.A.; Poorter, L.; Bongers, F.; Sprent, J.I.; Garner, M.D.; Aide, T.M.; Andrade, J.L.; Balvanera, P.; Becknell, J.M.; et al. Legume abundance along successional and rainfall gradients in Neotropical forests. Nat. Ecol. Evol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Powers, J.S.; Marín-Spiotta, E. Ecosystem Processes and Biogeochemical Cycles in Secondary Tropical Forest Succession. Annu. Rev. Ecol. Evol. Syst. 2017, 48, 497–519. [Google Scholar] [CrossRef]
- Eamus, D. Ecophysiological traits of deciduous and evergreen woody species in the seasonally dry tropics. Trends Ecol. Evol. 1999, 14, 11–16. [Google Scholar] [CrossRef]
- Jin, Y.; Russo, S.E.; Yu, M. Effects of light and topography on regeneration and coexistence of evergreen and deciduous tree species in a Chinese subtropical forest. J. Ecol. 2018, 106, 1634–1645. [Google Scholar] [CrossRef]
- Leigh, E.G., Jr. Structure and climate in tropical rain forest. Annu. Rev. Ecol. Syst. 1975, 6, 67–86. [Google Scholar] [CrossRef]
- Read, J.; Sanson, G.D.; Garine-Wichatitsky, M.; Jaffré, T. Sclerophylly in two contrasting tropical environments: Low nutrients vs. low rainfall. Am. J. Bot. 2006, 93, 1601–1614. [Google Scholar] [CrossRef] [PubMed]
- Abelleira Martínez, O.J. Invasion by native tree species prevents biotic homogenization in novel forests of Puerto Rico. Plant Ecol. 2010, 211, 49–64. [Google Scholar] [CrossRef]
- Brandeis, T.J.; Helmer, E.H.; Marcano-Vega, H.; Lugo, A.E. Climate shapes the novel plant communities that form after deforestation in Puerto Rico and the U.S. Virgin Islands. For. Ecol. Manag. 2009, 258, 1704–1718. [Google Scholar] [CrossRef]
- Chinea, J.D. Tropical forest succession on abandoned farms in the Humacao Municipality of eastern Puerto Rico. For. Ecol. Manag. 2002, 167, 195–207. [Google Scholar] [CrossRef]
- Chinea, J.D.; Helmer, E.H. Diversity and composition of tropical secondary forests recovering from large-scale clearing: Results from the 1990 inventory in Puerto Rico. For. Ecol. Manag. 2003, 180, 227–240. [Google Scholar] [CrossRef]
- Helmer, E.H.; Kennaway, T.A.; Pedreros, D.H.; Clark, M.L.; Marcano-Vega, H.; Tieszen, L.L.; Ruzycki, T.R.; Schill, S.R.; Sean Carrington, C.M. Land Cover and Forest Formation Distributions for St. Kitts, Nevis, St. Eustatius, Grenada and Barbados from Decision Tree Classification of Cloud-Cleared Satellite Imagery. Caribbean J. Sci. 2008, 44, 175–198. [Google Scholar] [CrossRef]
- Rivera, L.W.; Aide, T.M. Forest recovery in the karst region of Puerto Rico. For. Ecol. Manag. 1998, 108, 63–75. [Google Scholar] [CrossRef]
- Wolfe, B.T.; Van Bloem, S.J. Subtropical dry forest regeneration in grass-invaded areas of Puerto Rico: Understanding why Leucaena leucocephala dominates and native species fail. For. Ecol. Manag. 2012, 267, 253–261. [Google Scholar] [CrossRef]
- Larkin, C.C.; Kwit, C.; Wunderle, J.M.; Helmer, E.H.; Stevens, M.H.H.; Roberts, M.T.K.; Ewert, D.N. Disturbance Type and Plant Successional Communities in Bahamian Dry Forests. Biotropica 2012, 44, 10–18. [Google Scholar] [CrossRef]
- Pascarella, J.B.; Aide, T.M.; Serrano, M.I.; Zimmerman, J.K. Land-Use History and Forest Regeneration in the Cayey Mountains, Puerto Rico. Ecosystems 2000, 3, 217–228. [Google Scholar] [CrossRef]
- Marcano-Vega, H.; Mitchell Aide, T.; Báez, D. Forest regeneration in abandoned coffee plantations and pastures in the Cordillera Central of Puerto Rico. Plant Ecol. 2002, 161, 75–87. [Google Scholar] [CrossRef]
- Parrotta, J.A. The role of plantation forests in rehabilitating degraded tropical ecosystems. Agric. Ecosyst. Environ. 1992, 41, 115–133. [Google Scholar] [CrossRef]
- Parrotta, J.A.; Turnbull, J.W.; Jones, N. Catalyzing native forest regeneration on degraded tropical lands. For. Ecol. Manag. 1997, 99, 1–7. [Google Scholar] [CrossRef]
- Brown, K.A.; Scatena, F.N.; Gurevitch, J. Effects of an invasive tree on community structure and diversity in a tropical forest in Puerto Rico. For. Ecol. Manag. 2006, 226, 145–152. [Google Scholar] [CrossRef]
- Burman, E.; Ackerman, J.D.; Tremblay, R.L. Invasive Syzygium jambos trees in Puerto Rico: No refuge from guava rust. J. Trop. Ecol. 2017, 33, 205–212. [Google Scholar] [CrossRef]
- Francis, J.K.; Parrotta, J.A. Vegetation response to grazing and planting of Leucaena leucocephala in a Urochloa maximum-dominated grassland in Puerto Rico. Caribbean J. Sci. 2006, 42, 67–74. [Google Scholar]
- Parrotta, J.A. Productivity, nutrient cycling, and succession in single- and mixed-species plantations of Casuarina equisetifolia, Eucalyptus robusta, and Leucaena leucocephala in Puerto Rico. For. Ecol. Manag. 1999, 124, 45–77. [Google Scholar] [CrossRef]
- Galanes, I.T.; Thomlinson, J.R. Relationships between spatial configuration of tropical forest patches and woody plant diversity in northeastern Puerto Rico. In Forest Ecology: Recent Advances in Plant Ecology; Van der Valk, A.G., Ed.; Springer: Dordrecht, The Netherlands, 2009; pp. 101–113. [Google Scholar]
- Muñiz-Castro, M.A.; Williams-Linera, G.; Benayas, J.M.R. Distance effect from cloud forest fragments on plant community structure in abandoned pastures in Veracruz, Mexico. J. Trop. Ecol. 2006, 22, 431–440. [Google Scholar] [CrossRef]
- Gould, W.A.; González, G.; Carrero Rivera, G. Structure and composition of vegetation along an elevational gradient in Puerto Rico. J. Veg. Sci. 2006, 17, 653–664. [Google Scholar] [CrossRef]
- Caraballo-Ortiz, M.A.; Trejo-Torres, J.C. Two new endemic tree species from Puerto Rico: Pisonia horneae and Pisonia roqueae (Nyctaginaceae). PhytoKeys 2017, 97–115. [Google Scholar] [CrossRef] [PubMed]
- Cedeño-Maldonado, J.; Breckon, G. Serpentine endemism in the flora of Puerto Rico. Caribb. J. Sci. 1996, 32, 348–356. [Google Scholar]
- Gentry, A.H. Tropical Forest Biodiversity: Distributional Patterns and Their Conservational Significance. Oikos 1992, 63, 19–28. [Google Scholar] [CrossRef]
- Weaver, P.L. Dwarf Forest Recovery after Disturbances in the Luquillo Mountains of Puerto Rico. Caribb. J. Sci. 2008, 44, 150–163. [Google Scholar] [CrossRef]
- UNDESA. SDG indicators metadata repository. Available online: https://unstats.un.org/sdgs/metadata/ (accessed on 16 July 2018).
- Bustamante, M.; Helmer, E.H.; Schill, S.; Belnap, J.; Brugnoli, E.; Compton, J.E.; Coupe, R.H.; Hernández, M.; Isbell, F.; Azcárate, J.P.L.; et al. Chapter 4: Direct and Indirect Drivers of Change in Biodiversity. Americas Regional Assessment; United Nations Environment Programme: Bonn, Germany, 2018. [Google Scholar]
- Steffen, W.; Richardson, K.; Rockström, J.; Cornell, S.E.; Fetzer, I.; Bennett, E.M.; Biggs, R.; Carpenter, S.R.; de Vries, W.; de Wit, C.A.; et al. Planetary boundaries: Guiding human development on a changing planet. Science 2015, 347. [Google Scholar] [CrossRef] [PubMed]
- Nagy, R.C.; Rastetter, E.B.; Neill, C.; Porder, S. Nutrient limitation in tropical secondary forests following different management practices. Ecol. Appl. 2017, 27, 734–755. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.N.; Chazdon, R.L.; Bachelot, B.; Menge, D.N.L. Nitrogen-fixing trees inhibit growth of regenerating Costa Rican rainforests. Proc. Natl. Acad. Sci. USA 2017, 114, 8817–8822. [Google Scholar] [CrossRef] [PubMed]
- Cusack, D.F.; McCleery, T.L. Patterns in understory woody diversity and soil nitrogen across native- and non-native-urban tropical forests. For. Ecol. Manag. 2014, 318, 34–43. [Google Scholar] [CrossRef]
- Masek, J.G.; Wolfe, R.; Hall, F.; Cohen, W.; Kennedy, R.; Powell, S.; Goward, S.; Huang, C.; Healey, S.; Moisen, G. Assessing North American forest disturbance from the Landsat archive. Proceedings of Geoscience and Remote Sensing Symposium, Barcelona, Spain, 23–28 July 2007; pp. 5294–5297. [Google Scholar]
- Gregory, P.A.; Raul, T. Accelerated losses of protected forests from gold mining in the Peruvian Amazon. Environ. Res. Lett. 2016, 12, 094004. [Google Scholar]
- Schroeder, T.A.; Wulder, M.A.; Healey, S.P.; Moisen, G.G. Mapping wildfire and clearcut harvest disturbances in boreal forests with Landsat time series data. Remote Sens. Environ. 2011, 115, 1421–1433. [Google Scholar] [CrossRef]
- Pflugmacher, D.; Cohen, W.B.; Kennedy, R.E.; Yang, Z. Using Landsat-derived disturbance and recovery history and lidar to map forest biomass dynamics. Remote Sens. Environ. 2014, 151, 124–137. [Google Scholar] [CrossRef]
- Helmer, E.H.; Lefsky, M.A.; Roberts, D.A. Biomass accumulation rates of Amazonian secondary forest and biomass of old-growth forests from Landsat time series and the Geoscience Laser Altimeter System. J. Appl. Remote Sens. 2009, 3, 033505. [Google Scholar]
- Kennedy, R.E.; Andréfouët, S.; Cohen, W.B.; Gómez, C.; Griffiths, P.; Hais, M.; Healey, S.P.; Helmer, E.H.; Hostert, P.; Lyons, M.B.; et al. Bringing an ecological view of change to Landsat-based remote sensing. Front. Ecol. Environ. 2014, 12, 339–346. [Google Scholar] [CrossRef]

| Mapped Predictor Data | Source Cell Size (m) | Source |
|---|---|---|
| Landsat MSS—Multispectral bands of cloud-minimized image composites centered on two epochs: the years 1980 and 1985 | 57 | TS 1,2 |
| Landsat TM and ETM+—Multispectral optical bands of cloud-minimized image composites centered on two epochs: the years 1991 and 2000 | 28.5 | TS 1,2 |
| SPOT 3 panchromatic image composite for the year 1995 from normalizing image sections with linear regression including filling cloud and data gaps with the red band from 1991 Landsat | 10 | TS 1,3 |
| Moderate Resolution Imaging Spectroradiometer (MODIS)—Mean + coefficients of Fourier-transformed composites produced from all 16-day Enhanced Vegetation Index (EVI) composites from the year 2001 4 | 250 | [44], TS 1 |
| Precipitation—monthly and annual totals, minimums, maximums; dry and wet season totals; mean + coefficients of Fourier-transformed monthly totals 4 | ~450 | [39], TS 1 |
| Temperature—monthly and annual averages, minimums, maximums; dry and wet season averages; mean + coefficients of Fourier-transformed monthly averages 4 | ~450 | [39], TS 1 |
| Distance to nearest secondary, tertiary, and all roads and to the coast | 28.5 | TS 1 |
| Land cover in the year 2000 | 30 | [37,38], TS 1 |
| Forest mask from year 2000 land cover, and forest cover proportion in surrounding 3 × 3, 9 × 9, 17 × 17, and 35 × 35-m windows | [37,38], TS 1 | |
| Geological Substrate | 28.5 | [45,46], TS 1 |
| Soil Orders | 28.5 | NRCS 5 |
| Predictor data zones—discrete variable identifying islands | 28.5 | TS 1 |
| Topography—elevation, aspect, percent slope, moisture index, and topographic shadow for the base scene date of each Landsat composite | ~90 | [47] |
| Lidar-derived topography—elevation and slope position (Puerto Rico, St. John, and St. Thomas; other areas filled in with above topographic data) | 10 | TS 1 |
| Pixel-level percent tree canopy and impervious surface cover (PR only) | 30 | [48] |
| Land cover in 1978 (mainland PR only) | 28.5 | [37] |
| Forest Characteristic 1 | Cubist—Forest Test Data | PGNN 2—Forest Test Data | PGNN 2—Forest and Nonforest Test Data | |||
|---|---|---|---|---|---|---|
| Correl. Coef. | MAE | Correl. Coef. | MAE | Correl. Coef. | MAE | |
| Mean height (m) | 0.65 | 2.5 | 0.37 | 3.6 | 0.51 | 3.9 |
| Basal area (BA) (m2 ha−1) | 0.57 | 7.7 | 0.21 | 10 | 0.35 | 8.7 |
| Evergreen to nearly deciduous species (% BA) | 0.56 | 22 | 0.28 | 30 | 0.46 | 29 |
| Non-nitrogen-fixing (% BA) | 0.53 | 22 | 0.26 | 30 | 0.48 | 30 |
| Aboveground live dry biomass per ha (Mg/ha) | 0.52 | 34 | 0.22 | 42 | 0.35 | 36 |
| Deciduous nitrogen-fixing species | 0.51 | 17 | 0.30 | 19 | 0.29 | 16 |
| Deciduous and facultative species (% BA) | 0.51 | 21 | 0.31 | 24 | 0.36 | 21 |
| Number of native species | 0.51 | 2.9 | 0.10 | 3.9 | 0.34 | 3.5 |
| Number of introduced species | 0.50 | 0.72 | 0.28 | 0.84 | 0.32 | 0.7 |
| Evergreen to near evergreen species (% BA) | 0.50 | 25 | 0.19 | 31 | 0.40 | 28 |
| Evergreen coriaceous species (% BA) | 0.49 | 16 | 0.26 | 19 | 0.31 | 15 |
| Nitrogen-fixing species (% BA) | 0.47 | 20 | 0.30 | 22 | 0.30 | 18 |
| Number of endemic species | 0.47 | 0.3 | 0.23 | 0.36 | 0.25 | 0.3 |
| Coriaceous species (% BA) | 0.45 | 16 | 0.25 | 19 | 0.31 | 15 |
| Introduced species (% BA) | 0.42 | 20 | 0.25 | 22 | 0.27 | 18 |
| Chartaceous species (% BA) | 0.42 | 21 | 0.27 | 22 | 0.29 | 19 |
| Spathodea campanulata (% BA) | 0.38 | 8.6 | 0.26 | 10 | 0.24 | 8.4 |
| Proportion of plot forested | 0.37 | 0.19 | 0.11 | 0.27 | 0.50 | 0.3 |
| Subcoriaceous species (% BA) | 0.34 | 26 | 0.14 | 29 | 0.36 | 27 |
| Native species (% BA) | 0.33 | 25 | 0.14 | 33 | 0.43 | 32 |
| Number of trees per hectare | 0.28 | 1600 | 0.09 | 1900 | 0.29 | 1700 |
| Endemic species basal area (% BA) | 0.24 | 3.1 | 0.06 | 3.3 | 0.07 | 2.4 |
| Species BA (m2ha−1) | Count of Nonzero Observations | Cubist—Forest Test Data | PGNN—Forest Test Data | ||
|---|---|---|---|---|---|
| Correl. Coef. | MAE (m2 ha−1) | Correl. Coef.1 | MAE (m2 ha−1) | ||
| Bursera simaruba | 98 | 0.42 | 1.5 | 0.63 | 1.4 |
| Roystonea borinquena | 21 | −0.01 | 0.2 | 0.60 | 0.21 |
| Coccolobadiversifolia | 26 | 0.03 | 0.13 | 0.59 | 0.16 |
| Guapira fragrans | 52 | 0.35 | 0.67 | 0.59 | 0.57 |
| Acacia farnesiana | 29 | 0.05 | 0.48 | 0.55 | 0.34 |
| Cordia alliodora | 22 | −0.01 | 0.13 | 0.46 | 0.16 |
| Melicoccus bijugatus | 20 | 0.13 | 0.28 | 0.45 | 0.28 |
| Bourreria succulenta | 45 | 0.11 | 0.68 | 0.43 | 0.71 |
| Prestoea acuminata | 25 | 0.34 | 0.56 | 0.40 | 0.53 |
| Cecropia schreberiana | 72 | 0.26 | 0.99 | 0.33 | 1.2 |
| Guarea guidonia | 93 | 0.36 | 1.7 | 0.30 | 2.3 |
| Spathodea campanulata | 90 | 0.29 | 3.9 | 0.28 | 5.4 |
| Inga laurina | 46 | 0.07 | 0.53 | 0.26 | 0.56 |
| Ocotea leucoxylon | 47 | 0.31 | 0.29 | 0.26 | 0.33 |
| Citrus sinensis | 20 | 0.24 | 0.15 | 0.25 | 0.17 |
| Leucaena leucocephala | 92 | 0.12 | 1.4 | 0.21 | 1.4 |
| Tabebuia heterophylla | 74 | 0.07 | 0.97 | 0.21 | 1.1 |
| Mangifera indica | 26 | 0.06 | 1.6 | 0.20 | 1.7 |
| Inga vera | 48 | 0.30 | 0.57 | 0.20 | 0.67 |
| Andira inermis | 98 | 0.04 | 1.3 | 0.18 | 1.3 |
| Citharexylum spinosum | 53 | 0.16 | 0.34 | 0.15 | 0.36 |
| Dendropanax arboreus | 39 | 0.23 | 0.36 | 0.14 | 0.39 |
| Cordia sulcata | 28 | 0.13 | 0.22 | 0.14 | 0.25 |
| Alchornea latifolia | 24 | 0.12 | 0.31 | 0.13 | 0.33 |
| Schefflera morototonii | 31 | 0.17 | 0.3 | 0.13 | 0.35 |
| Ficus citrifolia | 25 | −0.01 | 0.21 | 0.10 | 0.22 |
| Syzygium jambos | 51 | 0.06 | 0.87 | 0.10 | 0.87 |
| Bucida buceras | 44 | 0.03 | 0.7 | 0.10 | 0.74 |
| Thouinia striata | 32 | 0.15 | 0.22 | 0.09 | 0.24 |
| Eugenia biflora | 22 | 0.00 | 0.66 | 0.04 | 0.37 |
| Zanthoxylum martinicense | 45 | 0.05 | 0.4 | 0.00 | 0.49 |
| Cupania americana | 24 | 0.00 | 0.17 | 0.00 | 0.15 |
| Neolaugeria resinosa | 20 | 0.00 | 0.21 | −0.01 | 0.26 |
| Casearia sylvestris | 39 | −0.01 | 0.34 | −0.01 | 0.38 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Helmer, E.H.; Ruzycki, T.S.; Wilson, B.T.; Sherrill, K.R.; Lefsky, M.A.; Marcano-Vega, H.; Brandeis, T.J.; Erickson, H.E.; Ruefenacht, B. Tropical Deforestation and Recolonization by Exotic and Native Trees: Spatial Patterns of Tropical Forest Biomass, Functional Groups, and Species Counts and Links to Stand Age, Geoclimate, and Sustainability Goals. Remote Sens. 2018, 10, 1724. https://doi.org/10.3390/rs10111724
Helmer EH, Ruzycki TS, Wilson BT, Sherrill KR, Lefsky MA, Marcano-Vega H, Brandeis TJ, Erickson HE, Ruefenacht B. Tropical Deforestation and Recolonization by Exotic and Native Trees: Spatial Patterns of Tropical Forest Biomass, Functional Groups, and Species Counts and Links to Stand Age, Geoclimate, and Sustainability Goals. Remote Sensing. 2018; 10(11):1724. https://doi.org/10.3390/rs10111724
Chicago/Turabian StyleHelmer, Eileen H., Thomas S. Ruzycki, Barry T. Wilson, Kirk R. Sherrill, Michael A. Lefsky, Humfredo Marcano-Vega, Thomas J. Brandeis, Heather E. Erickson, and Bonnie Ruefenacht. 2018. "Tropical Deforestation and Recolonization by Exotic and Native Trees: Spatial Patterns of Tropical Forest Biomass, Functional Groups, and Species Counts and Links to Stand Age, Geoclimate, and Sustainability Goals" Remote Sensing 10, no. 11: 1724. https://doi.org/10.3390/rs10111724
APA StyleHelmer, E. H., Ruzycki, T. S., Wilson, B. T., Sherrill, K. R., Lefsky, M. A., Marcano-Vega, H., Brandeis, T. J., Erickson, H. E., & Ruefenacht, B. (2018). Tropical Deforestation and Recolonization by Exotic and Native Trees: Spatial Patterns of Tropical Forest Biomass, Functional Groups, and Species Counts and Links to Stand Age, Geoclimate, and Sustainability Goals. Remote Sensing, 10(11), 1724. https://doi.org/10.3390/rs10111724