Optimized Extrapolation Methods Enhance Prediction of Elsholtzia densa Distribution on the Tibetan Plateau
Abstract
1. Introduction
2. Materials and Methods
2.1. Sources of Distribution Data
2.2. Environmental Variable Data and Screening
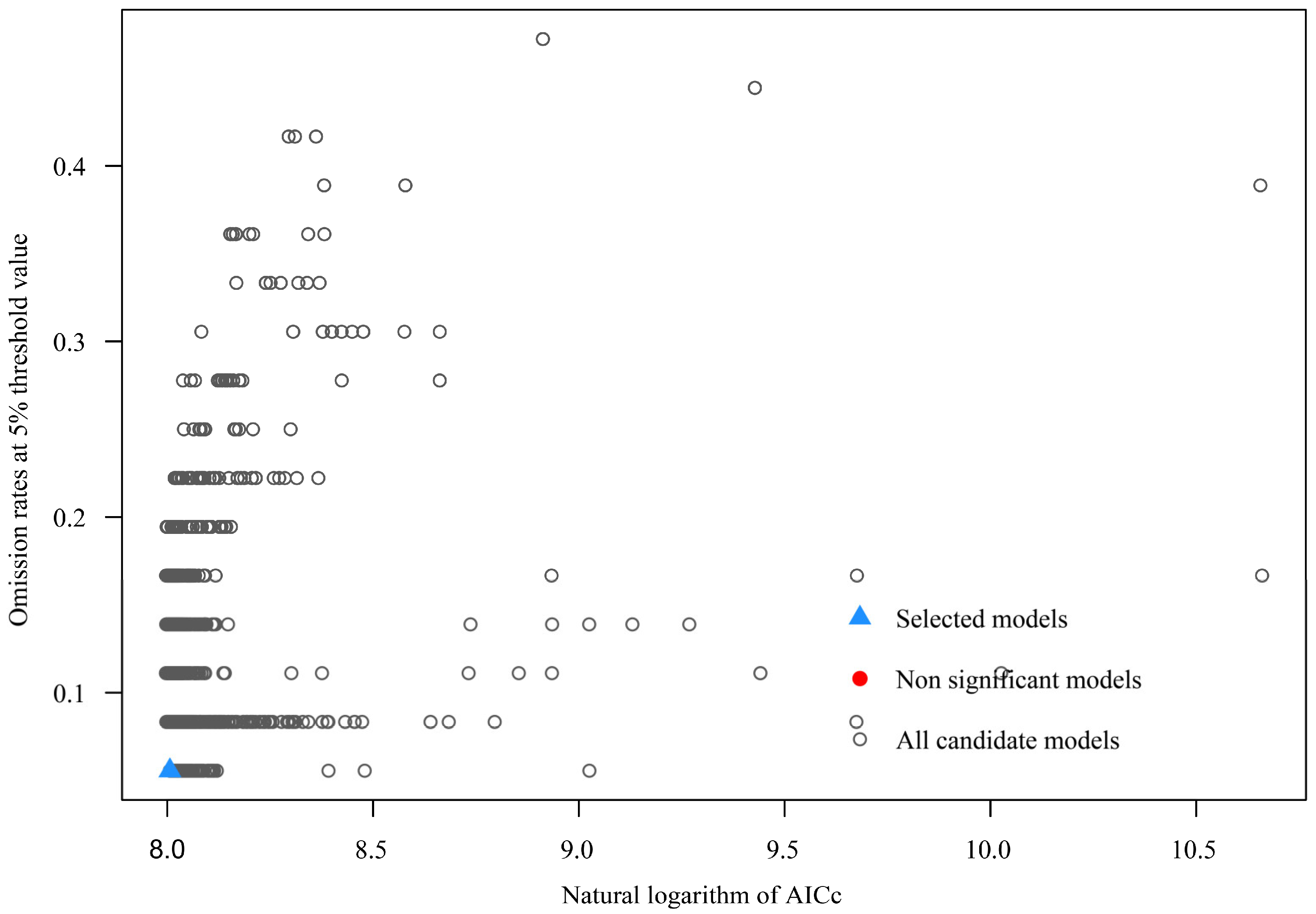
2.3. MaxEnt Parameter Setting and Accuracy Evaluation
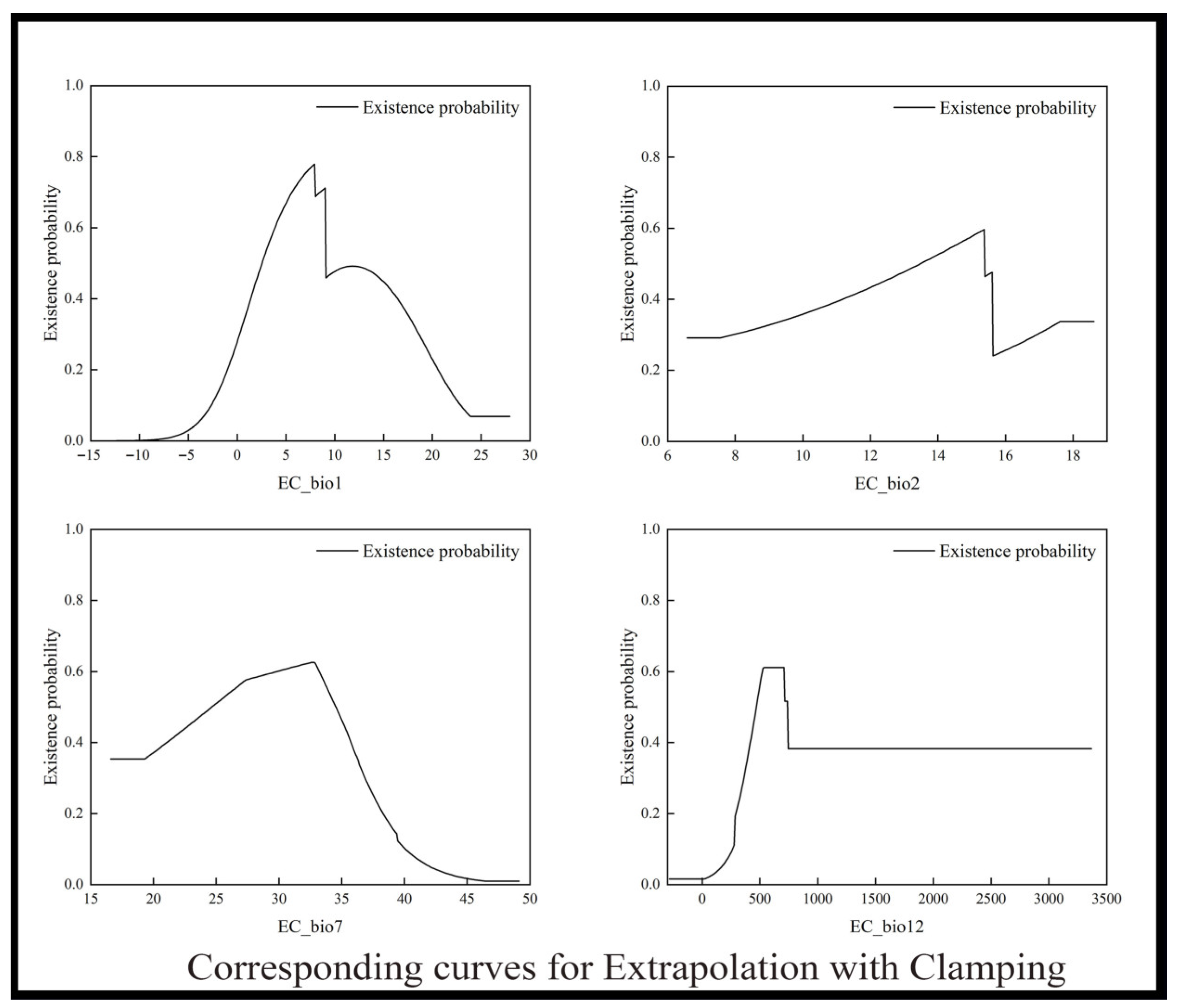
2.4. Model Extrapolation Patterns
2.5. Model Accuracy Assessment and Habitat Suitability Classification
2.6. Multivariate Environmental Similarity Surface and Most Dissimilar Variable
2.7. Integrated Model Uncertainty Analysis
3. Results
3.1. Model Parameter Optimization
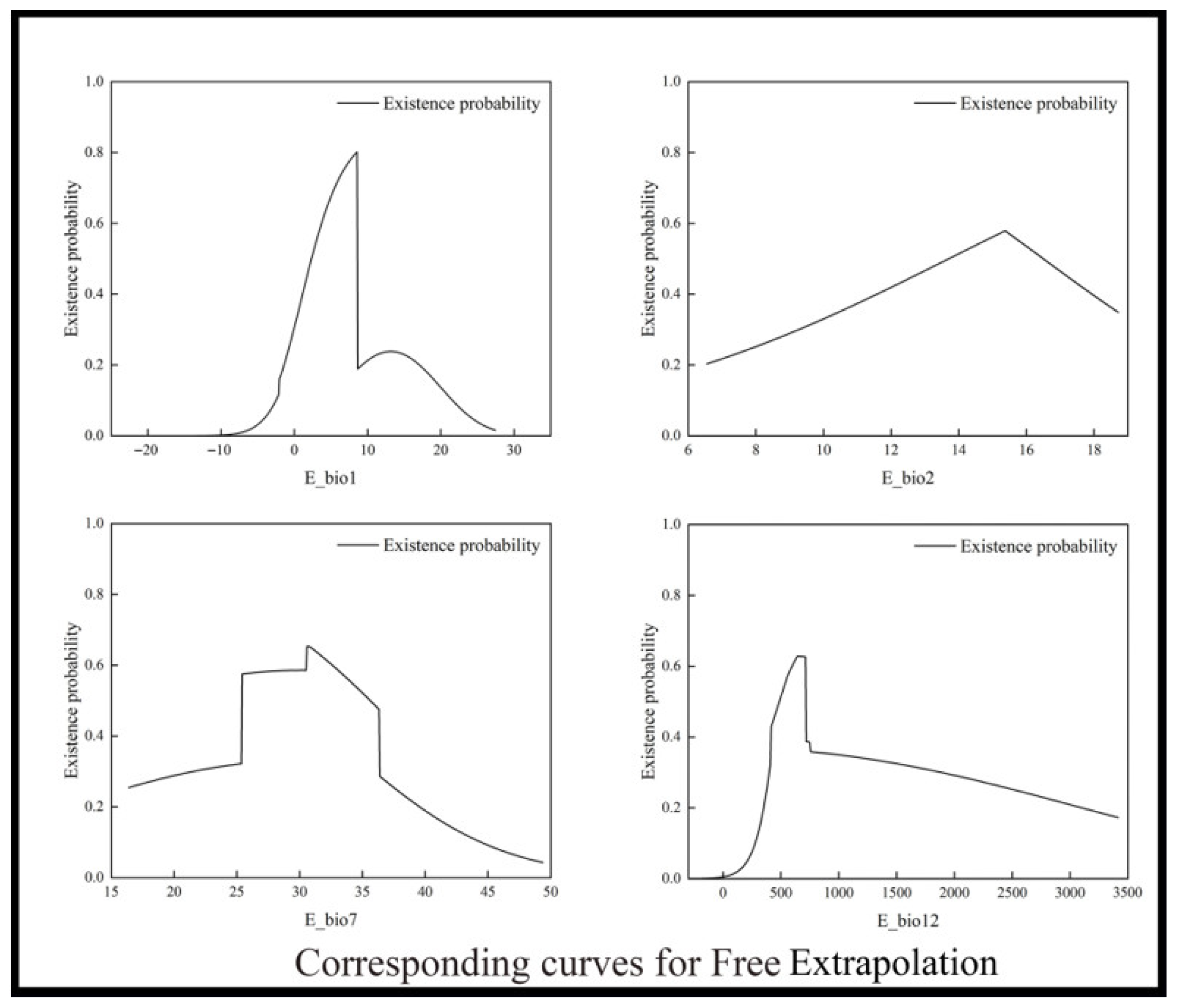
3.2. Dominant Environmental Determinants
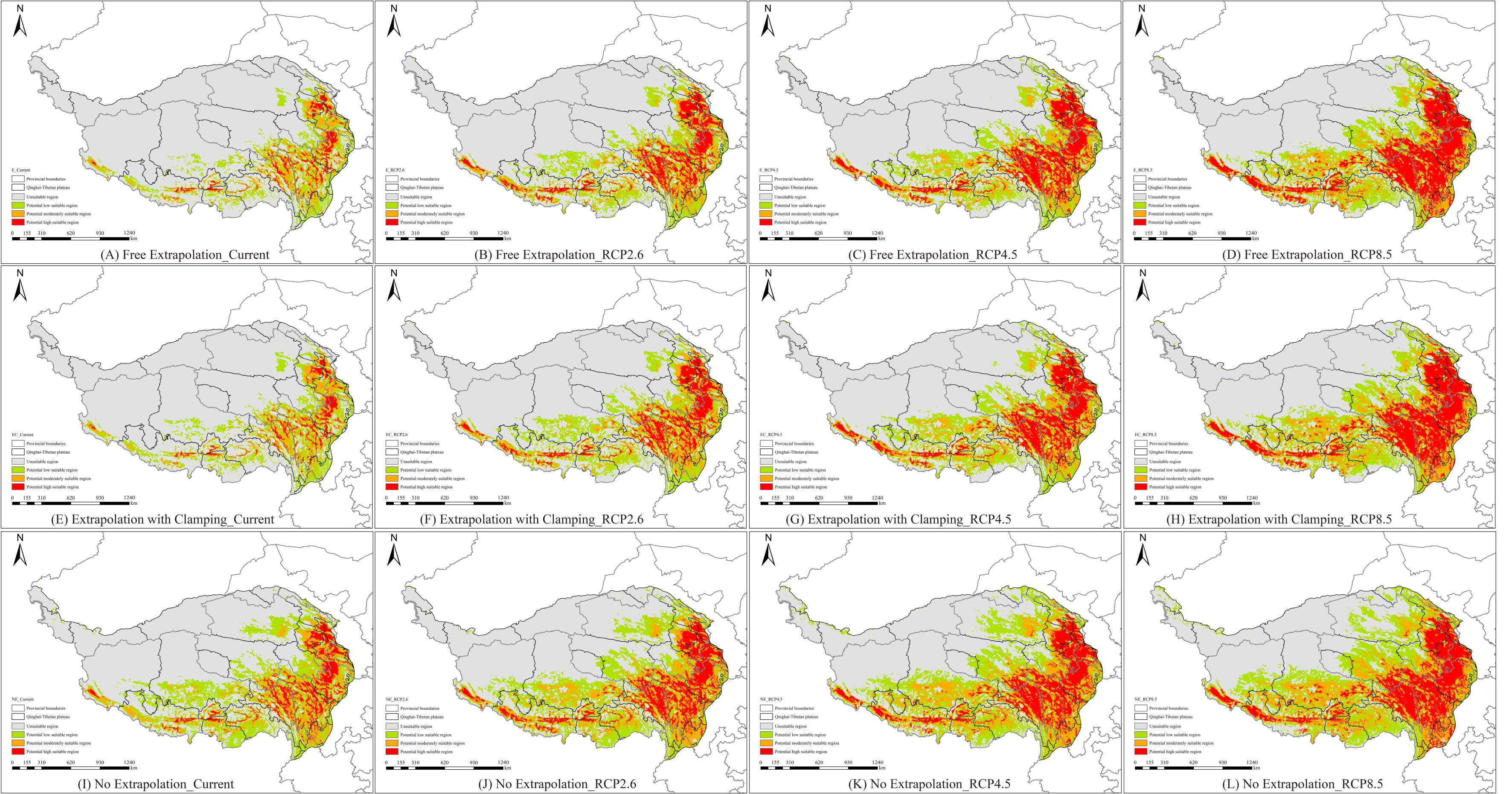
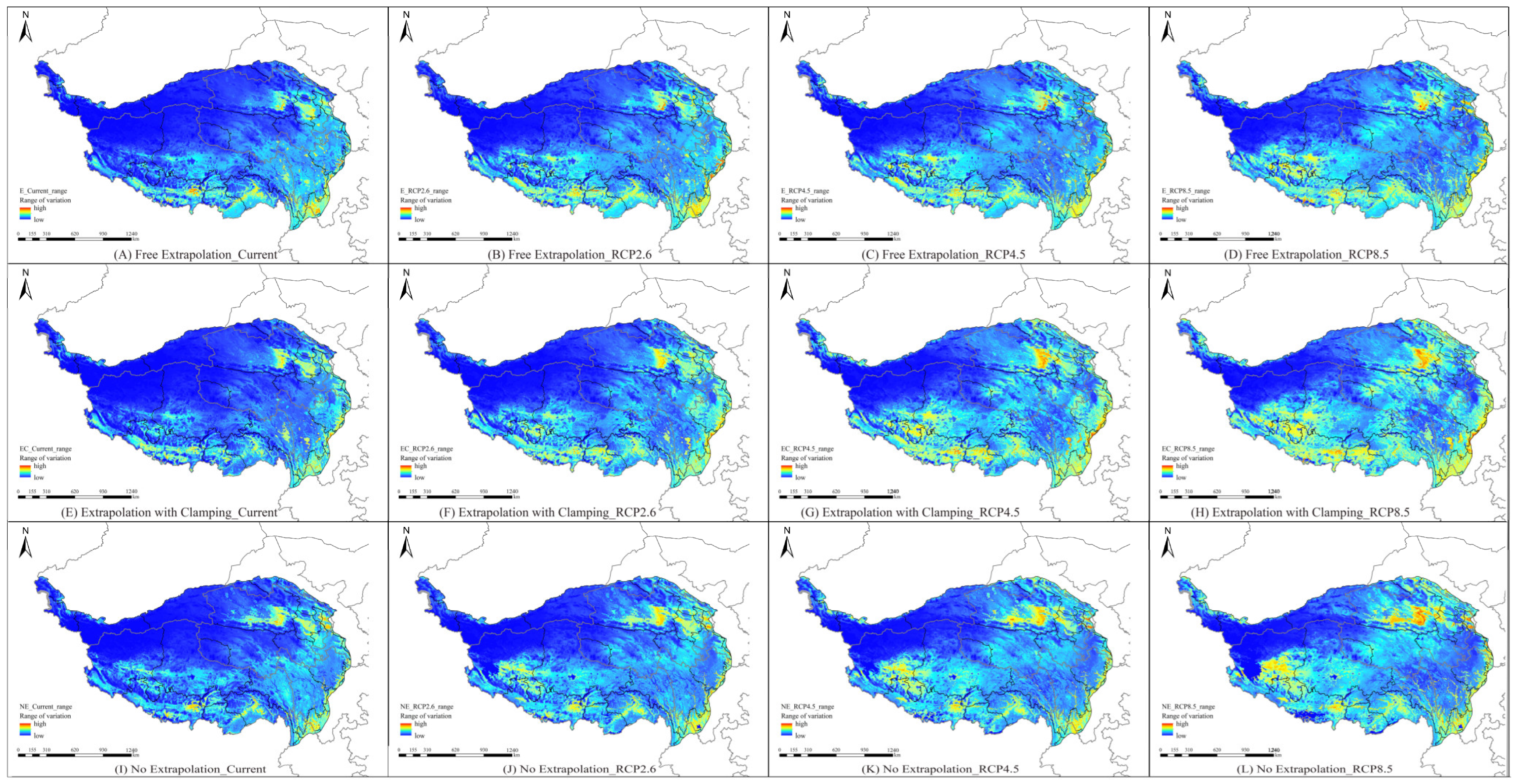
3.3. Comparison of Suitable Areas for Different Periods Under Model Extrapolation
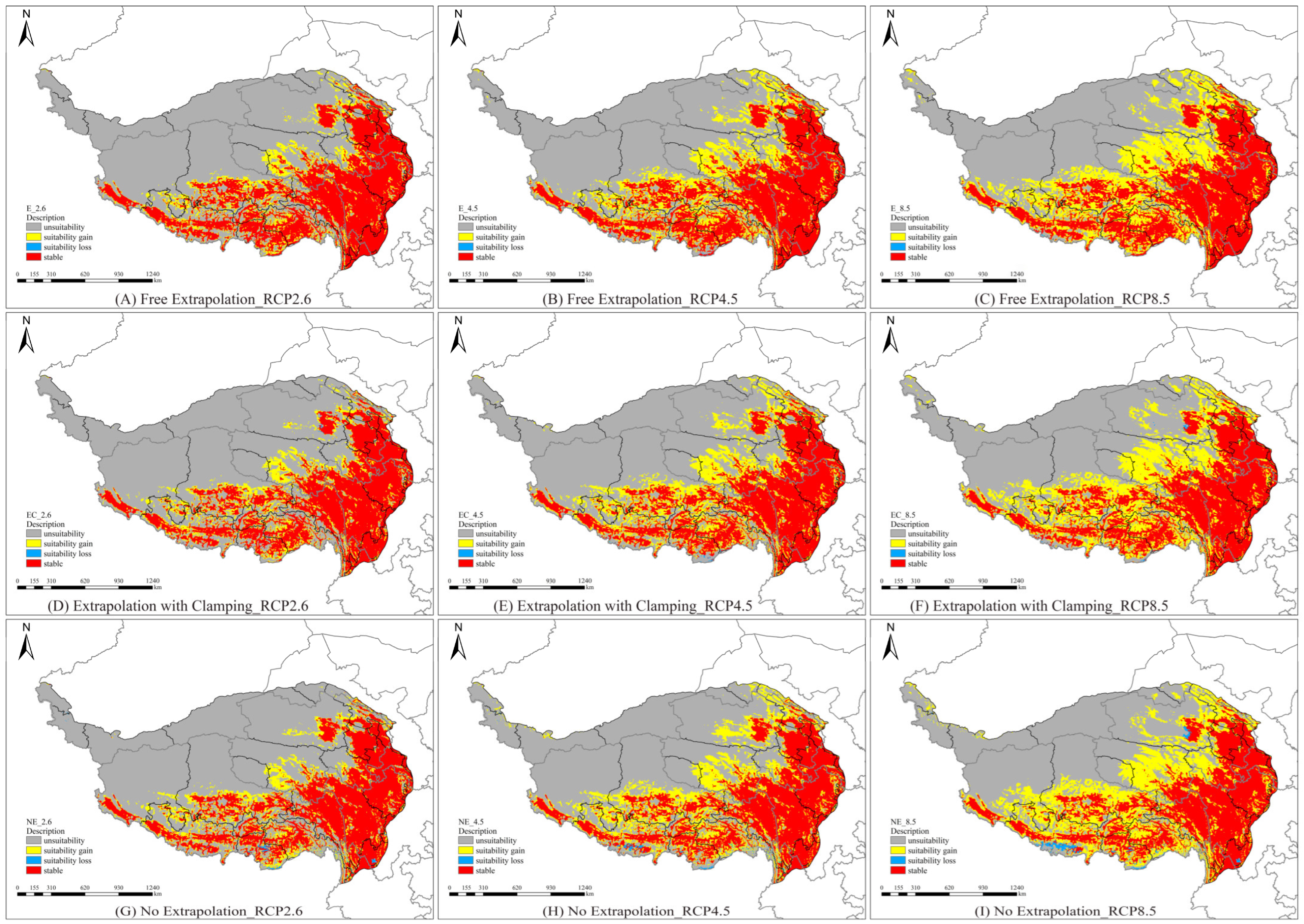
3.4. Spatial Pattern Changes of Potential Habitats Under Future Climate Scenarios
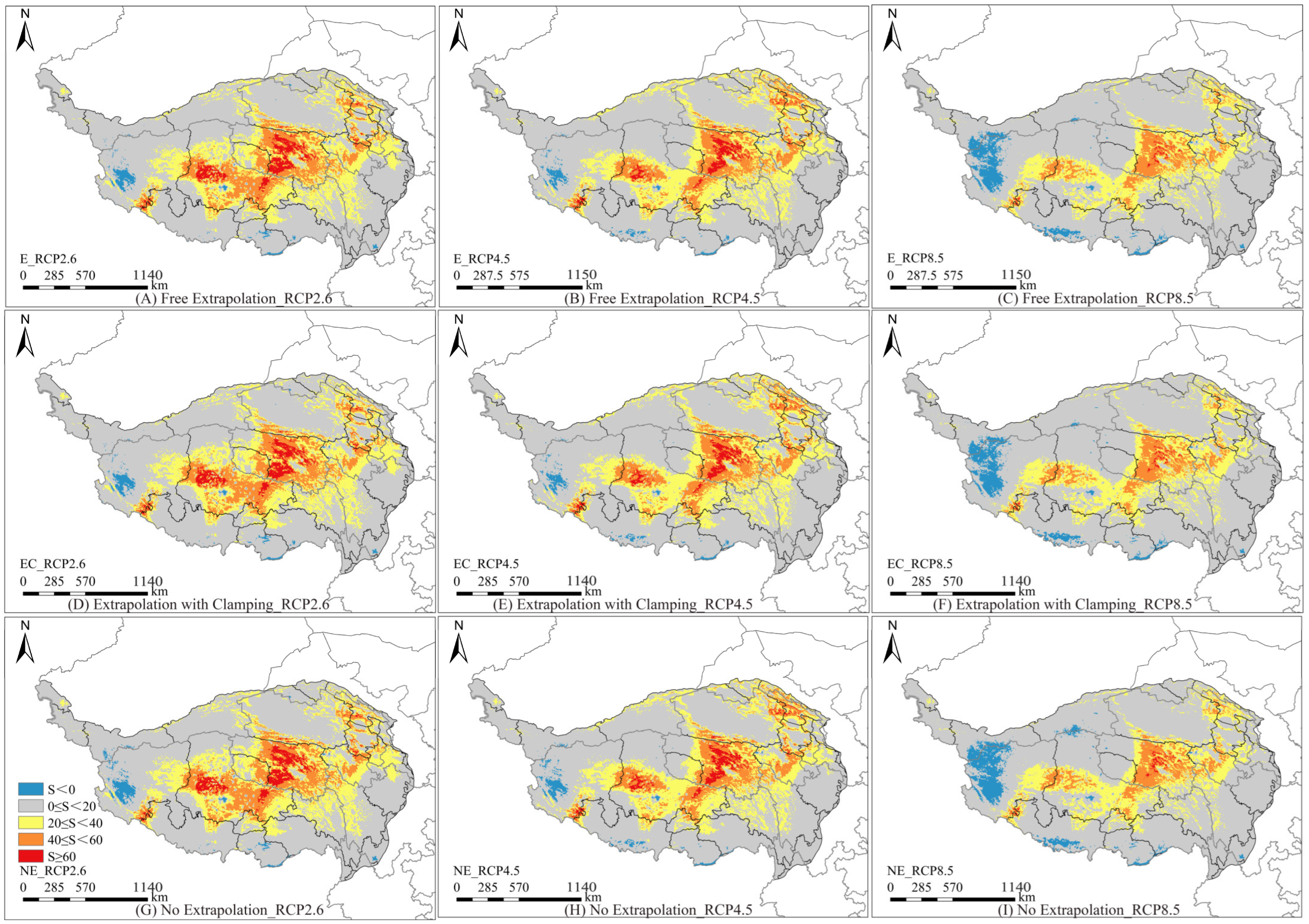
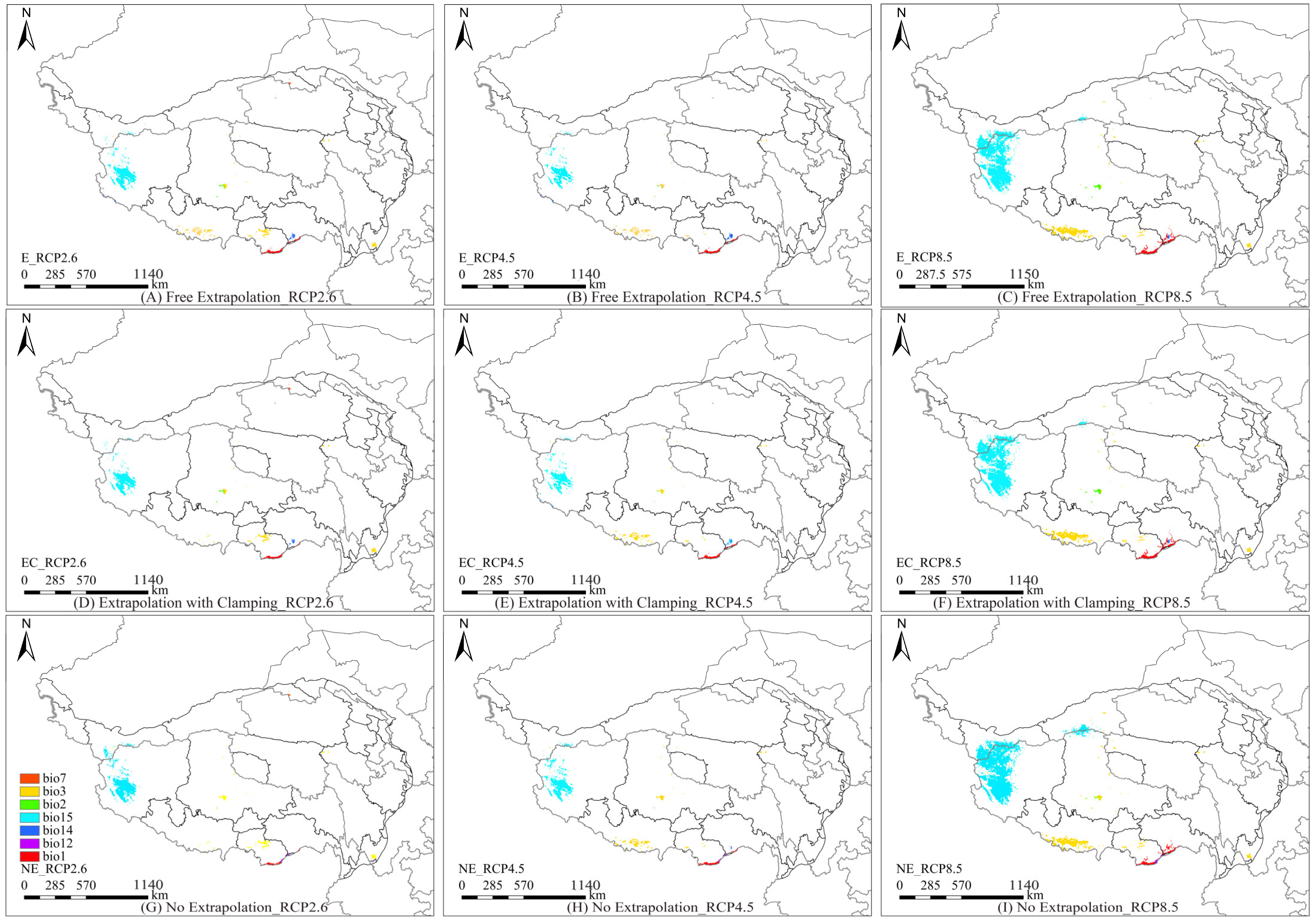
3.5. Multivariate Environmental Similarity Surface and Most Dissimilar Variable Analysis
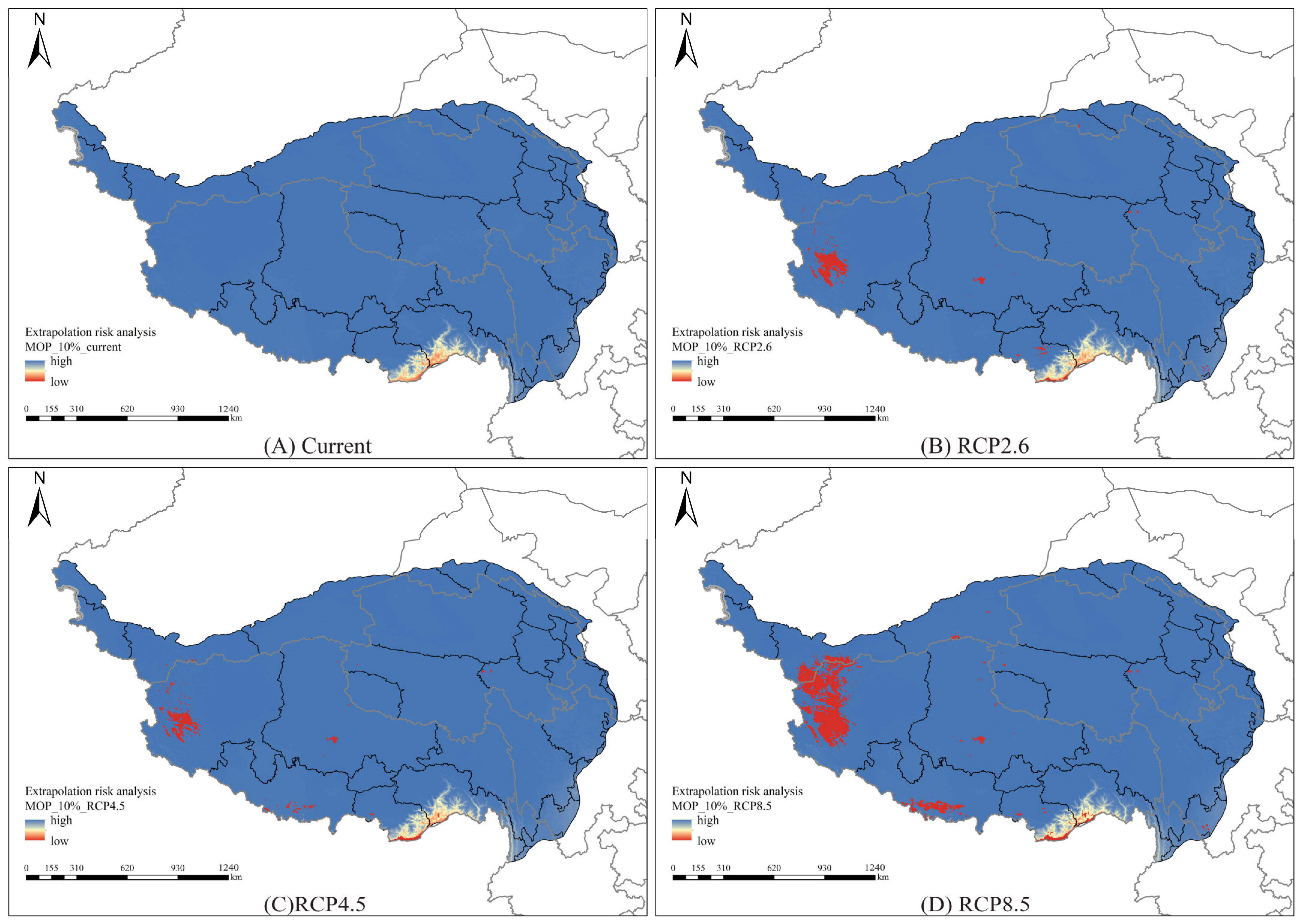
3.6. Model Extrapolation Risk Comparison
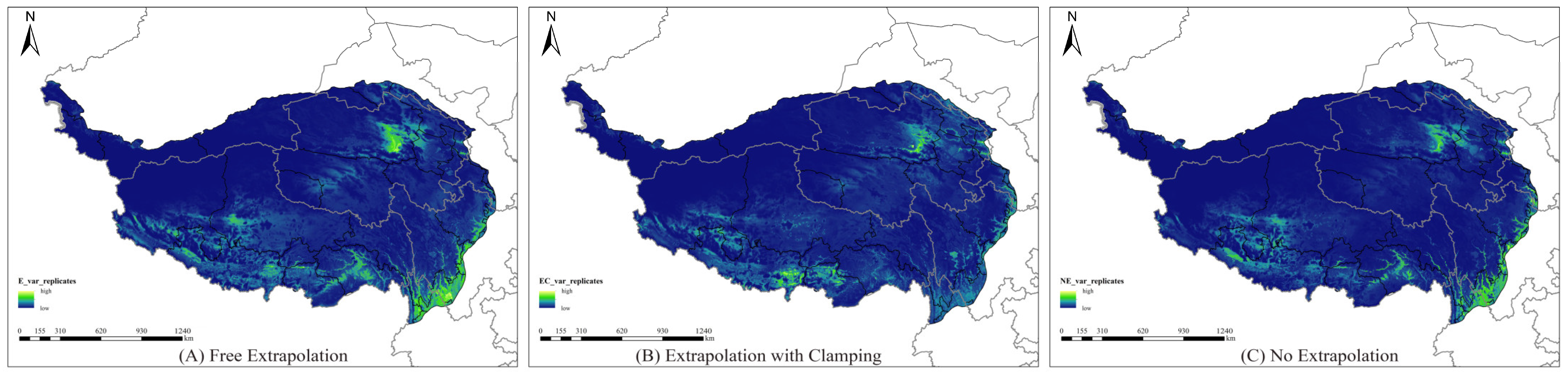
3.7. Range of Differences in Model Replications Under Different Extrapolation Methods
3.8. MaxEnt Model Accuracy Test
4. Discussion
4.1. Species Distribution Modeling and Assessment of Model Effectiveness
4.2. Changes in Potential Suitable Areas over Time
4.3. Model Extrapolation Risks and Uncertainties
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Tian, H.; Wang, Y.; Li, R.; Song, Z.; Zhang, F.; Xu, M.; Li, D. Vulnerability of 208 endemic or endangered species in China to the effects of climate change. Reg. Environ. Change 2013, 4, 843–852. [Google Scholar] [CrossRef]
- IPCC. Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2007. [Google Scholar]
- IPCC. Climate Change 2013: The Physical Science Basis. Contribution of Working Group I to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: New York, NY, USA, 2013. [Google Scholar]
- Bellard, C.; Bertelsmeier, C.; Leadley, P.; Thuiller, W.; Courchamp, F. Impacts of climate change on the future of biodiversity. Ecol. Lett. 2012, 4, 365–377. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhang, Y.; Li, W.-N.; Hu, B.-W.; Zou, J.-B.; Wang, S.-Q.; Niu, J.-F.; Wang, Z.-Z. Predicting the potential distribution of perennial plant Coptis chinensis Franch. in China under multiple climate change scenarios. Forest 2021, 11, 1464. [Google Scholar] [CrossRef]
- Liu, J.; Xin, Z.; Huang, Y.; Yu, J. Climate suitability assessment on the Qinghai-Tibet Plateau. Sci. Total Environ. 2022, 816, 151653. [Google Scholar] [CrossRef]
- Ji, F.; Fan, L.; Kuang, X.; Li, X.; Cao, B.; Cheng, G.; Yao, Y.; Zheng, C. How does soil water content influence permafrost evolution on the Qinghai-Tibet Plateau under climate warming? Environ. Res. Lett. 2022, 6, 064012. [Google Scholar] [CrossRef]
- Araújo, M.B.; Anderson, R.P.; Márcia Barbosa, A.; Beale, C.M.; Dormann, C.F.; Early, R.; Garcia, R.A.; Guisan, A.; Maiorano, L.; Naimi, B.; et al. Standards for distribution models in biodiversity assessments. Sci. Adv. 2019, 1, eaat4858. [Google Scholar] [CrossRef] [PubMed]
- McCune, J.L.; Baraloto, C. Species distribution models predict rare species occurrences despite significant effects of landscape context. J. Appl. Ecol. 2016, 6, 1871–1879. [Google Scholar] [CrossRef]
- Wang, M.; Peng, H.; Bai, Y.; Bai, Q.; Zhang, Q.; Feng, Y.; Wang, J.; Tang, Y. Impacts of physiological characteristics and human activities on the species distribution models of orchids taking the Hengduan Mountains as a case. Ecol. Evol. 2023, 13, e10566. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Winkler, A.; Viña, A.; Liu, J.; Zhang, Y.; Zhang, X.; Li, X.; Wang, F.; Zhang, J.; Zhao, Z. Uncertainty of future projections of species distributions in mountainous regions. PLoS ONE 2018, 13, e0189496. [Google Scholar] [CrossRef] [PubMed]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudík, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 2011, 1, 43–57. [Google Scholar] [CrossRef]
- Elith, J.; Graham, H.C.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A.; et al. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 2, 129–151. [Google Scholar] [CrossRef]
- Jiang, C.; Zhang, X.; Xie, W.; Wang, R.; Feng, C.; Ma, L.; Li, Q.; Yang, Q.; Wang, H. Predicting the potential distribution of the fall armyworm Spodoptera frugiperda (J.E. Smith) under climate change in China. Glob. Ecol. Conserv. 2022, 33, e01994. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Zhong, Y.; Xue, Z.; Jiang, M.; Liu, B.; Wang, G. The application of species distribution modeling in wetland restoration: A case study in the Songnen Plain, Northeast China. Ecol. Indic. 2021, 121, 107137. [Google Scholar] [CrossRef]
- Cobos, M.E.; Peterson, A.T.; Barve, N.; Osorio-Olvera, L. kuenm: An R package for detailed development of ecological niche models using Maxent. PeerJ 2019, 7, e6281. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zheng, F.; Zhang, W.; Xu, G.; Li, D.; Yang, S.; Jin, G.; Clements, D.R.; Nikkel, E.; Chen, A.; et al. Potential distribution and ecological impacts of Acmella radicans (Jacquin) R.K. Jansen (a new Yunnan invasive species record) in China. BMC Plant Biol. 2024, 24, 494. [Google Scholar] [CrossRef]
- Jiao, X.; Long, M.; Li, J.; Yang, Q.; Liu, Z. Reconstructing the Invasive History and Potential Distribution Prediction of Amaranthus palmeri in China. Agronomy 2023, 13, 2498. [Google Scholar] [CrossRef]
- Scherrer, D.; Esperon-Rodriguez, M.; Beaumont, L.J.; Barradas, V.L.; Guisan, A. National assessments of species vulnerability to climate change strongly depend on selected data sources. Divers. Distrib. 2021, 8, 1367–1382. [Google Scholar] [CrossRef]
- Arango Lozano, J.; Patiño Siro, D.; Toro Cardona, F. Reaching new environments through illegal trade: Evidence of a widely traded turtle in Colombia. Aquat. Ecol. 2023, 2, 471–480. [Google Scholar] [CrossRef]
- Yang, J.; Du, J.; Yu, F.; Li, R.; Zhong, J. Two new compounds from the aerial parts of Elsholtzia densa. Phytochem. Lett. 2022, 52, 104–108. [Google Scholar] [CrossRef]
- Editorial Committee of Flora of China, Chinese Academy of Sciences. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1977; p. 332. (In Chinese) [Google Scholar]
- Li, Z.; Ji, Q.; Yang, Y.; Gao, Y.; Xu, M.; Guan, Y. Predicting suitable habitats for Elsholtzia densa on the Qinghai-Xizang (Tibetan) Plateau under climate change using the Biomod2 model. Agric. Res. Arid Areas 2025, 3, 278–286. (In Chinese) [Google Scholar] [CrossRef]
- Zhou, X.; Xiao, Y.; Ma, D.; Xie, Y.; Wang, Y.; Zhang, H.; Wang, Y. The competitive strategies of poisonous weeds Elsholtzia densa Benth. on the Qinghai Tibet Plateau: Allelopathy and improving soil environment. Front. Plant Sci. 2023, 14, 1124139. [Google Scholar] [CrossRef]
- GBIF Occurrence Download. Available online: https://doi.org/10.15468/dl.b8sx2a (accessed on 14 April 2024).
- Warren, D.L.; Matzke, N.J.; Cardillo, M.; Baumgartner, J.B.; Beaumont, L.J.; Turelli, M.; Glor, R.E.; Huron, N.A.; Simões, M.; Iglesias, T.L.; et al. ENMTools 1.0: An R package for comparative ecological biogeography. Ecography 2021, 44, 504–511. [Google Scholar] [CrossRef]
- NGIC. Administrative Boundaries Data at 1:1,000,000 Scale Over the Tibetan Plateau 2017. National Tibetan Plateau/Third Pole Environment Data Center. 2019. Available online: https://cstr.cn/18406.11.Geogra.tpdc.270001 (accessed on 23 June 2024).
- Jiang, R.; Zou, M.; Qin, Y.; Tan, G.; Huang, S.; Quan, H.; Zhou, J.; Liao, H. Modeling of the Potential Geographical Distribution of Three Fritillaria Species Under Climate Change. Front. Plant Sci. 2022, 12, 749838. [Google Scholar] [CrossRef]
- Zhu, G.; Bu, W.; Gao, Y.; Liu, G. Potential geographic distribution of brown marmorated stink bug invasion (Halyomorpha halys). PLoS ONE 2012, 2, e31246. [Google Scholar] [CrossRef]
- Chen, S.; Bai, X.; Ye, J.; Chen, W.; Xu, G. Prediction of Suitable Habitat of Alien Invasive Plant Ambrosia trifida in Northeast China under Various Climatic Scenarios. Diversity 2024, 16, 322. [Google Scholar] [CrossRef]
- Zhang, B.; Xie, Z.; Ma, Y.; Zhao, Y.; Wei, C.; Fu, Y.; Liu, M.; Xu, F. Analysis on Changes in Suitable Areas for Fagopyrum dibotrys under Climate Change Scenarios in China. Chin. J. Agrometeorol. 2024, 8, 835–848. [Google Scholar] [CrossRef]
- Morales, N.S.; Fernandez, I.C.; Baca-Gonzalez, V. MaxEnt’s parameter configuration and small samples: Are we paying attention to recommendations? A systematic review(Article). PeerJ 2017, 3, e3093. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 1982, 1, 29–36. [Google Scholar] [CrossRef]
- Zhuo, Z.; Xu, D.; Pu, B.; Wang, R.; Ye, M. Predicting distribution of Zanthoxylum bungeanum Maxim. in China. BMC Ecol. 2020, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Motloung, R.F.; Chaisi, M.E.; Sibiya, M.S.; Nyangiwe, N.; Shivambu, T.C. Predicting tick distributions in a changing climate: An ensemble approach for South Africa. Vet. Parasitol. 2025, 338, 110528. [Google Scholar] [CrossRef]
- Elith, J.; Kearney, M.; Phillips, S. The art of modelling range-shifting species. Methods Ecol. Evol. 2010, 4, 330–342. [Google Scholar] [CrossRef]
- Owens, H.L.; Campbell, L.P.; Dornak, L.L.; Saupe, E.E.; Barve, N.; Soberón, J.; Ingenloff, K.; Lira-Noriega, A.; Hensz, C.M.; Myers, C.E.; et al. Constraints on interpretation of ecological niche models by limited environmental ranges on calibration areas. Ecol. Model. 2013, 263, 10–18. [Google Scholar] [CrossRef]
- Quiroga, M.P.; Souto, C.P. Ecological niche modeling, niche overlap, and good old Rabinowitz’s rarities applied to the conservation of gymnosperms in a global biodiversity hotspot. Landsc. Ecol. 2022, 10, 2571–2588. [Google Scholar] [CrossRef]
- He, S.; Gu, C.; Ma, D.; Zhang, H. Regulation of ROS and NO in Chenopodium ambrosioides allelopathic stress-induced death of Vicia faba root border cells. Acta Ecol. Sin. 2022, 2, 818–827. (In Chinese) [Google Scholar] [CrossRef]
- Song, Z.; Li, X.; Li, J.; Ka, Z.; Ma, G. Effects of plateau pika (Ochotona curzoniae) trails on enlargement and connection of degraded patches in alpine meadow. Chin. J. Ecol. 2020, 10, 3276–3284. (In Chinese) [Google Scholar] [CrossRef]
- Shen, Y.; Qiu, P.; Liao, Z.Y.; Wu, X.G.; Sun, X.M.; Zhang, L.; Pan, Z.F.; Pan, K.W. Changes in distribution patterns of weed species richness in agricultural lands on Qinghai-Tibet Plateau under climate change. Chin. J. Appl. Environ. Biol. 2022, 4, 897–908. (In Chinese) [Google Scholar] [CrossRef]
- Wang, C.J.; Wan, J.Z. Assessing the habitat suitability of 10 serious weed species in global croplands. Glob. Ecol. Conserv. 2020, 3, 142–153. [Google Scholar] [CrossRef]
- Root, T.L.; Price, J.T.; Hall, K.R.; Schneider, S.H.; Rosenzweig, C.; Pounds, J.A. Fingerprints of global warming on wild animals and plants. Nature 2003, 421, 57–60. [Google Scholar] [CrossRef] [PubMed]
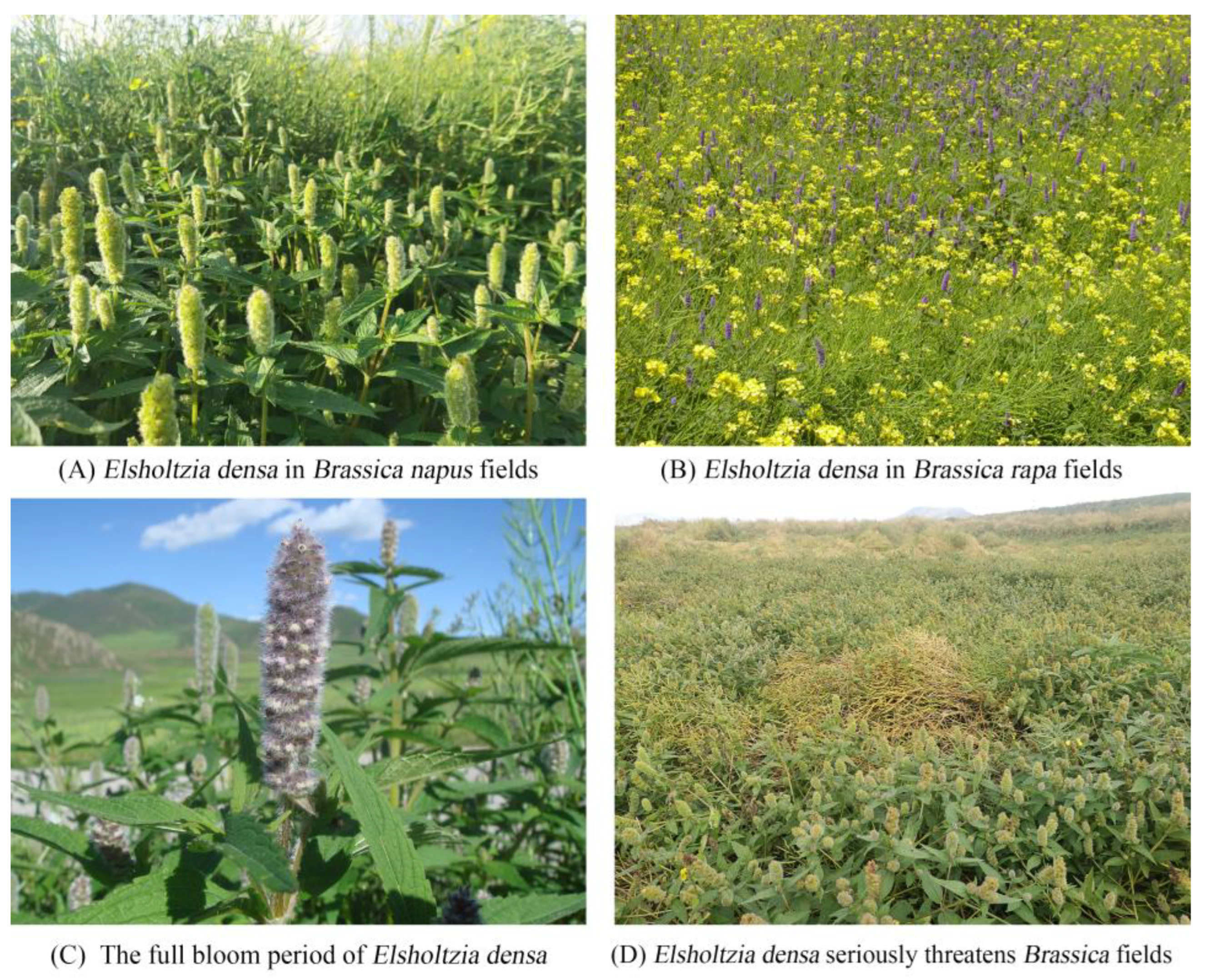
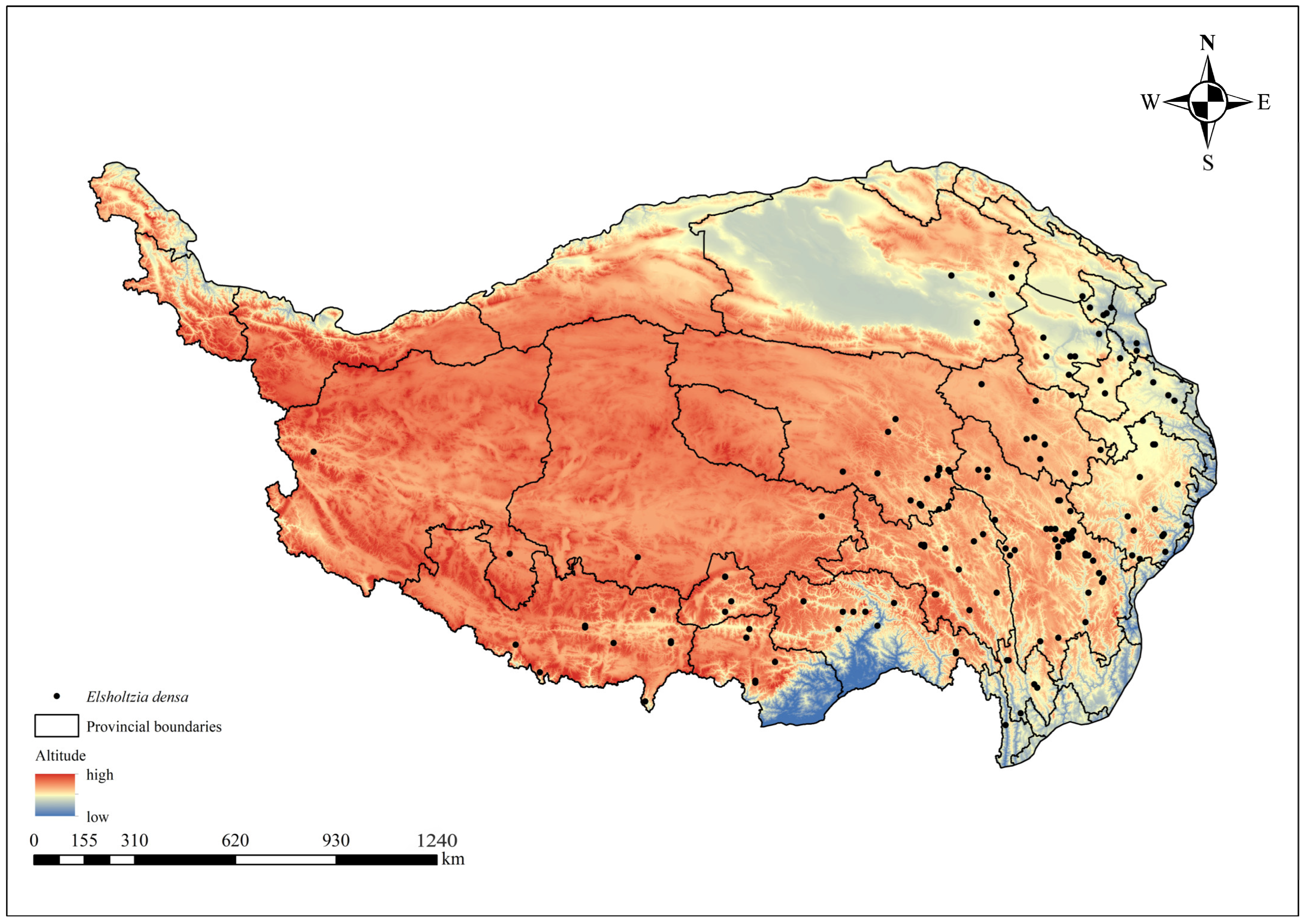



| Location | Altitude (m) | East Longitude | North Latitude |
|---|---|---|---|
| Zhongchuan Township, Minhe County | 1790 | 102°51′36.81″ | 35°52′12.09″ |
| Jishi Town, Xunhua County | 1871 | 102°28′50.64″ | 35°51′02.92″ |
| Gaomiao Town, Ledu County | 1936 | 102°32′29.58″ | 36°26′36.41″ |
| Donggou Township, Minhe County | 2208 | 102°42′58.71″ | 36°09′29.52″ |
| Gushan Township, Minhe County | 2219 | 102°48′31.61″ | 36°05′41.03″ |
| Hexi Township, Guide County | 2227 | 101°23′13.12″ | 36°02′03.90″ |
| Manping Township, Minhe County | 2286 | 102°46′35.74″ | 36°01′28.35″ |
| Shatangchuan Township, Huzhu County | 2332 | 101°53′49.92″ | 36°41′46.27″ |
| Sanhe Township, Ping ‘an County | 2391 | 101°56′56.67″ | 36°25′39.10″ |
| Zongzhai Township, Huangzhong County | 2422 | 101°41′19.08″ | 36°32′57.64″ |
| Shuangshu Township, Huzhu County | 2425 | 101°55′00.47″ | 36°46′26.25″ |
| Lijiashan Township, Ledu County | 2436 | 102°44′28.95″ | 36°07′21.44″ |
| Shagou Township, Ping ‘an County | 2508 | 102°02′43.71″ | 36°23′44.27″ |
| Wufeng Township, Huzhu County | 2530 | 101°51′21.42″ | 36°52′36.72″ |
| Tianjiazhai Township, Huangzhong County | 2584 | 101°48′53.74″ | 36°24′53.71″ |
| Machang Township, Ledu County | 2585 | 102°45′22.38″ | 36°26′37.17″ |
| Gangou Township, Minhe County | 2585 | 102°46′30.54″ | 37°24′14.95″ |
| Tumen Pass Township, Huangzhong County | 2597 | 101°43′41.09″ | 36°26′28.93″ |
| Dacai Township, Huangzhong County | 2610 | 101°28′56.18″ | 36°30′52.13″ |
| Lime Kiln Township, Ping ‘an County | 2619 | 101°53′37.16″ | 36°22′52.00″ |
| Shizhang Township, Huangzhong County | 2644 | 101°45′25.68″ | 36°24′08.99″ |
| Hualin Township, Datong County | 2646 | 101°43′57.96″ | 37°02′02.65″ |
| Luhua Township, Ledu County | 2653 | 102°43′11.30″ | 36°32′55.21″ |
| Angsi Duo, Hualong County | 2655 | 102°03′50.57″ | 36°10′15.45″ |
| Taizi Township, Huzhu County | 2661 | 101°56′15.04″ | 36°54′58.54″ |
| Chengguan Township, Huangyuan County | 2671 | 101°15′56.50″ | 36°41′19.23″ |
| Duolin Township, Datong County | 2672 | 101°27′39.63″ | 37°03′43.58″ |
| Zaba Township, Hualong County | 2702 | 101°59′41.22″ | 36°12′23.76″ |
| Baoku Township, Datong County | 2705 | 101°34′10.56″ | 37°06′53.28″ |
| Dahua Township, Huangyuan County | 2705 | 101°12′00.91″ | 36°42′55.99″ |
| Heping Township, Huangyuan County | 2709 | 101°15′25.85″ | 36°37′26.70″ |
| Shidacang Township, Hualong County | 2725 | 102°21′17.03″ | 36°05′55.13″ |
| Tangnaihai Township, Xinghai County | 2729 | 100°08′30.88″ | 35°30′39.82″ |
| Zama Shi Township, Qilian County | 2741 | 100°03′40.17″ | 38°12′59.43″ |
| Xiejiatan Township, Hualong County | 2749 | 102°16′32.00″ | 36°04′55.03″ |
| Chuankou Town, Menyuan County | 2751 | 101°51′14.50″ | 37°19′11.10″ |
| Babao Township, Qilian County | 2754 | 100°15′00.68″ | 38°10′23.07″ |
| Bohang Township, Huangyuan County | 2765 | 101°12′33.87″ | 36°39′48.37″ |
| Qushi ‘an Township, Xinghai County | 2767 | 100°14′01.28″ | 35°19′31.49″ |
| Maying Township, Ledu County | 2768 | 102°39′34.88″ | 36°33′14.39″ |
| Gucheng Town, Ping ‘an County | 2789 | 101°59′25.11″ | 36°19′13.97″ |
| Nuomuhong Township, Dulan County | 2791 | 96°27′26.91″ | 36°26′07.85″ |
| Bayan Township, Huangyuan County | 2806 | 101°08′03.19″ | 36°46′00.25″ |
| Hedong Township, Golmud City | 2810 | 94°54′43.96″ | 36°24′53.97″ |
| Qingshan Township, Datong County | 2815 | 101°23′46.29″ | 37°05′29.93″ |
| Dayuan Township, Huangzhong County | 2824 | 101°31′18.56″ | 36°27′51.40″ |
| Chabuqia Town, Gonghe County | 2840 | 100°36′55.75″ | 36°16′40.85″ |
| Bayan Township, Hualong County | 2841 | 102°14′58.05″ | 36°06′58.00″ |
| Huaitoutala Township, Delingha City | 2872 | 96°44′51.06″ | 37°21′01.87″ |
| Shazhuyu Township, Gonghe County | 2877 | 100°15′42.99″ | 36°15′39.80″ |
| Hongshuiquan Township, Ping ‘an County | 2879 | 101°53′25.42″ | 36°27′45.39″ |
| Nanmenxia Township, Huzhu County | 2880 | 101°54′27.02″ | 37°00′17.55″ |
| Donghe Township, Huzhu County | 2887 | 102°03′39.37″ | 36°55′13.51″ |
| Bagou Township, Tongde County | 2890 | 100°20′11.30″ | 35°17′46.66″ |
| Qingshizui Town, Menyuan County | 2970 | 101°23′38.84″ | 37°28′29.15″ |
| Gannan Village, Delingha City | 2977 | 97°22′13.52″ | 37°21′21.66″ |
| Ertang Township, Hualong County | 2979 | 102°11′56.13″ | 36°08′05.85″ |
| Riyue Township, Huangyuan County | 3018 | 101°09′22.72″ | 36°31′07.00″ |
| Wulan County Town | 3052 | 98°31′51.02″ | 36°56′02.44″ |
| Tangge Mu, Gonghe County | 3057 | 99°57′50.57″ | 36°12′07.24″ |
| Tongpu Township, Wulan County | 3081 | 98°32′09.14″ | 36°59′48.01″ |
| Xiangride Township, Dulan County | 3084 | 97°51′30.92″ | 35°59′13.34″ |
| Tiegai Township, Gonghe County | 3103 | 100°12′32.47″ | 35°59′47.78″ |
| Guomaying Township, Guinan County | 3106 | 101°06′49.74″ | 35°48′38.11″ |
| Mangqu Township, Guinan County | 3122 | 100°46′50.88″ | 35°34′11.97″ |
| Laiyihai Township, Guinan County | 3164 | 100°46′31.61″ | 35°34′09.45″ |
| Sizhai Township, Huangyuan County | 3172 | 101°02′13.26″ | 36°45′11.25″ |
| Chasu Town, Dulan County | 3190 | 98°05′17.39″ | 36°17′50.82″ |
| Tanggu Town, Tongde County | 3316 | 100°32′08.30″ | 35°11′26.50″ |
| Climate Factors | Name | Unit |
|---|---|---|
| bio1 | Annual mean temperature | °C |
| bio2 | Mean diurnal temperature range | °C |
| bio3 | Isothermality | - |
| bio4 | Temperature seasonality (standard deviation × 100) | - |
| bio5 | Maximum temperature of warmest month | °C |
| bio6 | Minimum temperature of coldest month | °C |
| bio7 | Temperature annual range | °C |
| bio8 | Mean temperature of wettest quarter | °C |
| bio9 | Mean temperature of driest quarter | °C |
| bio10 | Mean temperature of warmest quarter | °C |
| bio11 | Mean temperature of coldest quarter | °C |
| bio12 | Annual precipitation | mm |
| bio13 | Precipitation of wettest month | mm |
| bio14 | Precipitation of driest month | mm |
| bio15 | Precipitation seasonality | mm |
| bio16 | Precipitation of wettest quarter | mm |
| bio17 | Precipitation of driest quarter | mm |
| bio18 | Precipitation of warmest quarter | mm |
| bio19 | Precipitation of coldest quarter | mm |
| Model | Mean_AUC_ratio | pval_pROC | Omission_rate_at_5% | AICc | delta_AICc | W_AICc | num_parameters |
|---|---|---|---|---|---|---|---|
| M_2.5_F_LQTH_Set2 | 1.661067 | 0 | 0.055556 | 2999.589 | 0 | 1 | 28 |
| M_3.1_F_LQTH_Set2 | 1.656331 | 0 | 0.055556 | 3001.345 | 1.756348 | 1 | 24 |
| M_2.7_F_LQTH_Set2 | 1.671476 | 0 | 0.055556 | 3001.437 | 1.847758 | 1 | 27 |
| Environmental Variables | Free Extrapolation | Extrapolation with Clamping | No Extrapolation |
|---|---|---|---|
| bio1 (Annual mean temperature) | 2.5–8.6 °C | 2.6–9.0 °C | 2.6–9.0 °C |
| bio2 (Mean diurnal temperature range) | 13.7–16.5 °C | 13.4–15.4 °C | 13.5–15.7 °C |
| bio7 (Temperature annual range) | 25.4–35.6 °C | 24.6–34.5 °C | 26.9–34.3 °C |
| bio12 (Annual precipitation) | 490.2–715.5 mm | 469.1–738.5 mm | 451.4–815.3 mm |
| Extrapolation Model | Periods | Low Suitability Area (×104 km2) | Trends (×104 km2) | Moderate Suitability Area (×104 km2) | Trends (×104 km2) | High Suitability Area (×104 km2) | Trends (×104 km2) |
|---|---|---|---|---|---|---|---|
| E | Current | 32.05 | - | 16.32 | - | 7.88 | - |
| RCP2.6 | 40.35 | ↑ 8.3 | 22.97 | ↑ 6.65 | 20.21 | ↑ 12.33 | |
| RCP4.5 | 42.39 | ↑ 10.34 | 27.04 | ↑ 10.72 | 29.17 | ↑ 21.29 | |
| RCP8.5 | 43.41 | ↑ 11.36 | 33.01 | ↑ 16.69 | 41.93 | ↑ 34.05 | |
| EC | Current | 34.39 | - | 17.48 | - | 7.73 | - |
| RCP2.6 | 43.21 | ↑ 8.82 | 24.82 | ↑ 7.34 | 19.93 | ↑ 12.20 | |
| RCP4.5 | 45.00 | ↑ 10.62 | 29.60 | ↑ 12.13 | 29.01 | ↑ 21.28 | |
| RCP8.5 | 45.80 | ↑ 11.41 | 36.26 | ↑ 18.78 | 40.87 | ↑ 33.14 | |
| NE | Current | 33.77 | - | 17.16 | - | 8.15 | - |
| RCP2.6 | 42.42 | ↑ 8.64 | 25.55 | ↑ 8.38 | 21.13 | ↑ 12.99 | |
| RCP4.5 | 44.52 | ↑ 10.75 | 31.05 | ↑ 13.88 | 31.11 | ↑ 22.97 | |
| RCP8.5 | 45.41 | ↑ 11.64 | 35.89 | ↑ 18.73 | 45.21 | ↑ 37.07 |
| Extrapolation Model | Climate Scenarios | Area (×104 Km2) | |||
|---|---|---|---|---|---|
| Persistently Unsuitable | Suitability Gain | Suitability Loss | Persistently Suitable | ||
| RCP2.6 | 146.52 | 27.43 | 0.07 | 81.07 | |
| E | RCP4.5 | 130.19 | 43.76 | 0.10 | 81.03 |
| RCP8.5 | 112.25 | 61.70 | 0.03 | 81.10 | |
| RCP2.6 | 151.16 | 28.04 | 0.18 | 75.70 | |
| EC | RCP4.5 | 134.84 | 44.37 | 0.20 | 75.68 |
| RCP8.5 | 117.48 | 61.72 | 0.15 | 75.73 | |
| RCP2.6 | 151.34 | 30.11 | 0.80 | 72.84 | |
| NE | RCP4.5 | 133.36 | 48.09 | 0.94 | 72.69 |
| RCP8.5 | 114.57 | 66.88 | 1.71 | 71.93 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Wei, Y.; Cheng, L.; Chen, H.; Weng, H. Optimized Extrapolation Methods Enhance Prediction of Elsholtzia densa Distribution on the Tibetan Plateau. Sustainability 2025, 17, 8206. https://doi.org/10.3390/su17188206
Liu Z, Wei Y, Cheng L, Chen H, Weng H. Optimized Extrapolation Methods Enhance Prediction of Elsholtzia densa Distribution on the Tibetan Plateau. Sustainability. 2025; 17(18):8206. https://doi.org/10.3390/su17188206
Chicago/Turabian StyleLiu, Zeyuan, Youhai Wei, Liang Cheng, Hongyu Chen, and Hua Weng. 2025. "Optimized Extrapolation Methods Enhance Prediction of Elsholtzia densa Distribution on the Tibetan Plateau" Sustainability 17, no. 18: 8206. https://doi.org/10.3390/su17188206
APA StyleLiu, Z., Wei, Y., Cheng, L., Chen, H., & Weng, H. (2025). Optimized Extrapolation Methods Enhance Prediction of Elsholtzia densa Distribution on the Tibetan Plateau. Sustainability, 17(18), 8206. https://doi.org/10.3390/su17188206






