Abstract
Considerable attention has been paid to the establishment of an appropriate cropping patterns for the restoration of saline-alkali lands. This study’s goal was to explore changes in nitrogen-cycling (N-cycling) gene (nitrogen fixation: nifH; nitrification: AOA, AOB, and nxrB; denitrification: narG, norB, and nosZ) abundance of three cropping patterns at two soil depths in saline-alkali soils. Results showed that rotation and mixture promoted soil nutrients. N-cycling functional genes were significantly influenced by soil depths and cropping patterns. Compared with monoculture, rotation decreased the abundance of nifH, AOA, narG, and nosZ and increased the abundance of AOB; mixture decreased the abundance of AOA, narG, and nosZ and increased the abundance of AOB and nxrB in the 0–15 cm soil depth. Rotation increased all genes abundance; mixture increased nosZ abundance and decreased nxrB abundance in 15–30 cm soil depth. Soil protease, cellulase, nitrate reductase, pH, AK (available potassium), and AP (available phosphorus) were important factors influencing N-cycling gene abundance. In conclusion, rotation and mixture not only reduced soil salinity but also improved soil fertility and nitrogen cycling. These findings can provide some theories for the sustainable development of N-cycling during the restoration of saline-alkali soils.
1. Introduction
Saline-alkali soils contain high levels of soluble salts. Globally, about 932 million hectares (M ha), including 831 million hectares of agricultural land, are affected by salt damage [1]. Saline-alkaline soils have little productivity [2]. Several nations and regions pay significant attention to the improvement and extension of saline-alkali wastelands, especially considering the present global food crisis, and China has 99.13 × 106 ha of various forms of saline-alkali land [3]. Natural and anthropogenic increases in soil salinity are a key environmental issue that hinders plant development and production [4,5,6,7,8]. Due to their high salt concentration, saline-alkali soils affect the osmotic potential outside the root system, preventing plants from absorbing water and so affecting plant development [9,10,11,12,13]. The microbial populations responsible for the cycle of organic compounds are under a rising challenge from soil salinity [14]. In order to ensure national food and ecological security, the improvement and usage of salt-affected land are essential.
Planting alkali-tolerant plants has been observed to enhance microbial activity and restore soil fertility [15,16,17]. Oats, alfalfa, and tall wheatgrass are commonly used as species for the phytoremediation of saline lands. Alfalfa is a C3 plant and one of the world’s most important perennial leguminous forage grasses that improve soil fertility [18,19]. In addition, Li et al. [20] found that the introduction of oats mixed with other salinity-tolerant plants significantly contributed to the soil fungal communities in saline-alkali soils in Northeast China. Likewise, tall wheatgrass has a very high salinity tolerance and is a good choice when considering alkali-tolerant plants [21,22]. It can tolerate salt and sodium stress and has good production [23]. Phytoremediation does not only refer to the cropping of resistant plants but also includes field management practices, such as cropping patterns. Soil rotation has many advantageous uses, such as human food, bioenergy, and animal feed [24]. Crop rotation may exert beneficial effects on the physical, chemical, and biological properties of soils, help in the control of weeds and insect pests, and alleviate the adverse effects of climate and exogenous factors on soils [25]. In addition, mixed cropping systems can leverage resources, enhance biodiversity [26], provide insights for managing agricultural systems more sustainably, and improve ecosystem stability [27,28,29]. Nevertheless, information on the effects of saline land improvement under different cropping patterns is still insufficient.
Soil microorganisms are often considered indicators of soil health because they show great sensitivity to changes in agroecosystems [30], which is what allows researchers to infer the impact of a process or decision on the soil environment [31]. In addition, the critical contributions of soil biota to ecosystem services have now been universally recognized. It is an important cutting-edge research field of agroecosystems to fully improve the positive influences of soil microorganisms on sustainable agriculture cropping patterns [32]. Microorganisms play a crucial role in soil N-cycling and controlling the amount of soil N accessible to plants [33]. Nitrogen-cycle processes, such as assimilation, ammonification, nitrification, and denitrification, involve microorganisms [34]. Biological nitrogen fixation (BNF) is the primary route for nitrogen intake in the soil-nitrogen cycle, converting molecular nitrogen to ammonia or other nitrogen-containing compounds by the catalytic activity of nitrogenase in nitrogen-fixing bacteria [35]. The critical and rate-limiting phase in autotrophic nitrification is the oxidation of ammonium (NH4+) to nitrite (NO2−) by ammonia-oxidizing bacteria (AOB) and archaea (AOA) [36,37]. Nitrite is then oxidized to nitrate (NO3−), which is gradually reduced by denitrifying bacteria to gaseous products (NO, N2O and N2), removing nitrogen from the ecosystem [38]. However, information on how agricultural practices change the functional bacteria involved in the nitrogen cycle is still insufficient [39]. Numerous studies have demonstrated that quantitative marker genes are good indicators of key biogeochemical processes in the nitrogen cycle and have been used to quantify the quantity of bacteria in soils using genetic markers [40,41,42]. In the nitrogen cycle, the nifH gene (encoding nitrogenase) is related to nitrogen-fixation; amoA (ammonia monooxygenase) and nxrB (Nitrite oxidoreductase enzyme) are related to nitrification; and narG (nitric oxide reductase), norB (nitric oxide reductase), and nosZ (nitrous oxide reductase) are related to denitrification [43].
So far, the impact of different cropping patterns and soil depths on the abundance of functional genes for N-cycling in the saline-alkaline soils of Northeast China is unknown. Therefore, we used monoculture, rotation, and mixture in three experimental field distributions in Northeast China and evaluated the variations in N-cycling gene abundance by qPCR methods. The purpose of our study was to determine if the abundance of N-cycling genes in soil was responsive to changes in cropping patterns and soil depths. We hypothesized that (1) compared to monoculture, rotation and mixture will result in the restoration of soil nutrients, such as soil properties and enzymes; (2) rotation and mixture will cause changes in N-cycling gene abundance; and (3) variations in the abundance of N-cycling genes are associated with soil chemistry and enzymes.
2. Materials and Methods
2.1. Site Description
The Sifang Mountain Farm is located in Zhaodong City, Heilongjiang Province (125°45′–126°30′ E, 46°12′–46°22′ N), with flat terrain and a single type of landform. The area of soil is carbonate meadow alkaline soil and carbonate meadow soil. The average annual temperature is 2.4 °C, the annual evaporation is 1662 mm, the average annual rainfall is 396 mm, the maximum temperature is 39.0 °C, the minimum temperature is −37.5 °C, and the annual accumulated temperature is 2500–2700 °C. Before restoration, the soil pH in this location reached 11.00. According to the USDA soil taxonomy system, the soil was predominantly loamy [44] and saline-alkaline experimental, which is a soda saline soil dominated by sodium carbonate bicarbonate.
2.2. Experimental Design
This research was conducted in three experimental agricultural plots. These plots consisted of (a) a monoculture system in which alfalfa was cultivated annually; (b) a 13-year-old cycle of alfalfa–oats–tall wheatgrass, with annual alfalfa in the first year, followed by oats in the second year, and tall wheatgrass in the third year, in a three-year rotation; and (c) a mixture system plot in which alfalfa, oats, and tall wheatgrass were simultaneously planted, with each crop planted densely and virtually alternating in every other row. These three cropping patterns use a common approach to fertiliser management, applying nitrogen, phosphorus, and potash in the form of urea, calcium superphosphate, and potassium nitrate. During the experimental period, all plots had similar soil and climate conditions and were fertilized and managed in the same manner.
2.3. Soil Sampling
On the 17th of August 2017, soil samples were obtained from three alfalfa-planted plots. Before the alfalfa harvesting, samples were taken. A soil auger was used to collect soil samples from 0–15 cm and 15–30 cm depth in each repeated plot (8 cm diameter and 15 cm deep). Five samples were gathered in an “S” formation and then thoroughly combined to produce one composite sample for each replication, resulting in a total of 18 samples at the plot level. After removing roots and debris, each composite field soil sample was homogenized, packaged in sterile plastic bags, and delivered to the laboratory immediately. The soil was then passed through a 2 mm sieve and each soil sample was divided into two parts. One portion was frozen at −80 °C for DNA extraction, while the other was air-dried at ambient temperature for soil chemical testing.
2.4. Soil Chemical Analysis
The examined soil properties, including soil pH, exchange capacity (EC), soil organic matter (SOM), soil total nitrogen (TN), soil total phosphorus (TP) and potassium (TK), and soil available phosphorus (AP) and potassium (AK), were described by previous studies [45,46]. According to the description of Guan et al. [47], we determined the soil-enzyme activity. The urease activity was determined using urea as a substrate. The protease activity was determined using casein as a substrate. The nitrate reductase activity was determined using nitrate of potash as a substrate. The cellulase activity was determined using carboxymethylcellulose as a substrate. The β-glucosidase activity was determined using β-glucoso-saligenin (salicin) as a substrate.
2.5. Soil DNA Extraction and Quantitative PCR (qPCR)
The Power Soil DNA Isolation kit (MO BIO Laboratories Inc., Carlsbad, CA, USA) was used for DNA extraction from 0.25 g of soil following the manufacturer’s instructions. The quantity and quality of the extracted DNA were assessed using a NanoDrop ND2000c spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) and checked on 1% agarose gel. The DNA samples were used to quantify the functional N-cycling genes in the soil samples. Gene encoding the nitrogen fixation (nifH), nitrification (amoA bacteria, AOB; amoA archaea AOA; nxrB), and denitrification (narG, norB, and nosZ) processes (Table S1) were performed by quantitative real-time PCR (qPCR). ABI7500 Fluorescent Real-Time PCR Detection System (Applied Biosystems, Carlsbad, CA, USA) for qPCR. Primer sets and the PCR conditions of each gene are detailed in Table S2. The standard curves all had R2 values above 0.99, with slope values in the range of −3.28 and −3.59 and an estimated amplification efficiency in the range of 101.93 and 89.93%.
2.6. Statistical Analysis
The Pearson correlation heat maps of soil properties and N-cycling genes were plotted with the “pheatmap” package [48]. In addition, the different N-cycling gene abundances were ordinated using non-metric multidimensional scaling (NMDS) with the dissimilarity matrices using the “metaMDS” function in the “vegan” package [49]. To analyse the soil chemical drivers of N-cycling gene abundance, we used the “rfPermute” package for random forest analysis [50]. The correlations between N-cycling gene abundance and soil important factors were analysed using Pearson linear regressions.
Using a two-way ANOVA, the impacts of cropping patterns and soil depths on soil chemical properties and enzyme activity were investigated. Subsequently, the effect of cropping patterns on soil properties, enzyme activity, N-cycle gene abundance, and their ratios at two soil depths was investigated using a one-way ANOVA. Before the ANOVA, all data were examined using Levene’s test for normality and homogeneity. Differences between groups were examined with Duncan’s post hoc test, p < 0.05. All the analyses above were carried out in R (v.4.2.2) [51].
3. Results
3.1. Soil Chemical Properties and Soil Enzymes
The presence of cropping patterns and soil depths can influence soil properties and soil enzymes differently, as shown in Table 1. Results from a two-way ANOVA indicate that pH and TN are significantly influenced by cropping patterns and soil depths, respectively (Table 2). The soil pH in all three cropping patterns ranged from 7.72 to 8.26, indicating that all samples are weakly basic and nearly neutral. Notably, the pH of monoculture soil was higher than that of rotation and mixture soils at two soil depths (p < 0.05) (Table 1). With increasing soil depths, the TN content of monoculture, rotation, and mixture soils decreased by 22.99%, 10.79%, and 44.15%, respectively (p < 0.05) (Table 1). Cropping patterns and soil depths also significantly influenced EC and AK content (p < 0.05) (Table 2). Specifically, at 0-15 cm soil depth, the EC of monoculture soil was significantly higher than that of rotation and mixture soils (p < 0.05) (Table 1). In addition, the EC of rotation soil was significantly higher than that of mixture soils (p < 0.05) (Table 1). At 15–30 cm soil depth, there were no significant differences in EC among the three cropping patterns (p > 0.05) (Table 1). The AK content of mixture soil was significantly higher than that of rotation (+73.22%) and monoculture (+43.67%) soils at 0–15 cm soil depth (p < 0.05) (Table 1). At 15–30 cm soil depth, the AK content of rotation soil was significantly higher than that of mixture and monoculture soils (p < 0.05) (Table 1).

Table 1.
The soil chemical properties and soil enzymes in the three cropping patterns.

Table 2.
Two-way ANOVAs for the impact of cropping patterns (CP), soil depths (SD) and their interaction (CP × SD) on soil chemical properties and soil enzymes.
Table 2 shows that soil enzyme activity is influenced by cropping patterns and soil depths. When comparing different cultivation methods, the activity of urease, cellulase, and nitrate reductase was significantly higher in mixed soils at a soil depth of 0–15 cm, compared to rotated and monoculture soils (p < 0.05) (Table 1). Furthermore, the activity of urease and cellulase was significantly higher in rotation soils than in monoculture soils (p < 0.05) (Table 1). The activity of β-glucosidase was significantly lower in monoculture soils than in mixed and rotation soils (p < 0.05) (Table 1), while the activity of protease was significantly lower in monoculture soils compared to mixed soils (p < 0.05) (Table 1). At a soil depth of 15–30 cm, the activity of urease was significantly lower in monoculture soils than in mixed soils (p < 0.05) (Table 1). Moreover, the activity of nitrate reductase, β-glucosidase, and cellulase was significantly higher in mixed soils than in rotated and monoculture soils (p < 0.05) (Table 1). The nitrate reductase activity was significantly higher in monoculture soils than in rotation soils, while cellulase activity was higher in rotation soils compared to monoculture soils (p < 0.05) (Table 1).
3.2. N-Cycling Gene Abundance
Cropping patterns and soil depths had a significant impact on all N-cycling functional genes (Table 3). At a soil depth of 0–15 cm, the abundance of nifH was significantly higher in monoculture than in rotation (p < 0.05) (Figure 1). Similarly, the abundance of AOA, narG, and nosZ was significantly higher in monoculture than in rotation and mixture soils (p < 0.05), while the AOB abundance was significantly lower in monoculture than in rotation and mixture soils (p < 0.05). The AOA abundance in mixture soils was significantly lower than in rotation (p < 0.05), and the nxrB abundance in monoculture and rotation was significantly lower than in mixture soils (p < 0.05). There was no significant difference in norB abundance among the three cropping patterns (p > 0.05), as shown in Figure 1.

Table 3.
Two-way ANOVAs for the impact of cropping patterns (CP), soil depths (SD) and their interaction (CP × SD) on N-cycling gene abundance.
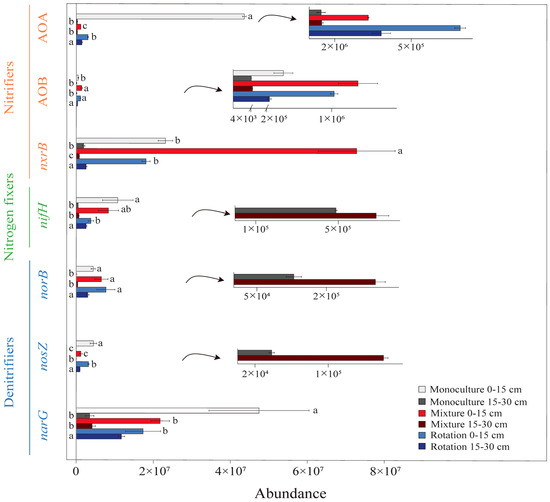
Figure 1.
The abundance of N-cycling functional genes in different cropping patterns at two soil depths. Different small letters indicate significant differences between treatments (p < 0.05). The letters on the right side of the bar chart are for 0–15 cm soil depth, while the letters on the left side are for 15–30 cm soil depth.
At a soil depth of 15–30 cm, all N-cycling abundances in rotation were significantly higher than in mixture and monoculture soils (p < 0.05) (Figure 1). In addition, the nosZ abundance in mixture soils was significantly higher than in monoculture soils (p < 0.05), while the nxrB abundance in monoculture was significantly higher than in mixture soils (p < 0.05).
3.3. N-Cycling Gene Abundance Ratio
To investigate how soil depths and cropping patterns impact N-cycling processes associated with gene abundance, we calculated the ratios for different N-cycling abundances. At surface soil, we found no significant difference in the nifH/(AOA +AOB + nxrB) ratio among all cropping patterns. However, at deep soil, the nifH/(AOA + AOB + nxrB) ratio in monoculture was significantly lower than in rotation and mixture (p < 0.05) (Figure 2a). The nifH/(AOA + AOB + nxrB) ratio in rotation and mixture at deep soil was significantly higher than that at surface soil (p < 0.05) (Figure 2a). Furthermore, the AOA/AOB ratio in mixture and rotation was significantly lower than in monoculture at both soil depths (p < 0.05) (Figure 2b). The AOA/AOB ratio in monoculture at surface soil was significantly higher than that at deep soil (P<0.05) (Figure 2b). We found no significant difference in the nifH/(narG + norB +nosZ) ratio among the three cropping patterns at both soil depths (p < 0.05) (Figure 2c). The norB/nosZ ratio in mixtures was significantly higher than in monoculture at surface soil and lower than in monoculture and rotation at deep soil (p < 0.05) (Figure 2d).
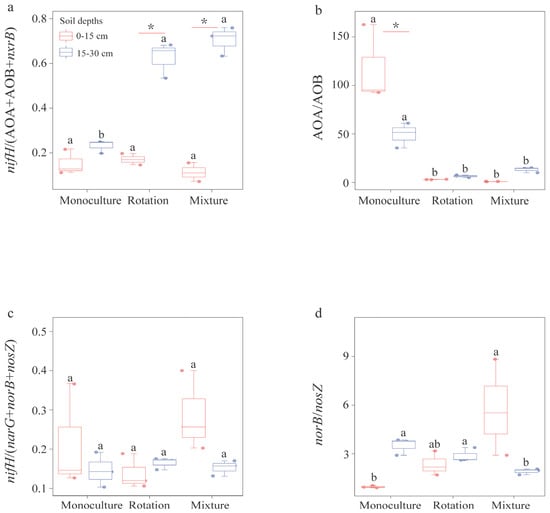
Figure 2.
The efficiency of N-cycling processes were inferred from the calculation of different N-cycling gene abundance ratios. (a–d) represent the nitrogen sequestration process, ammonia oxidation process, nitrogen loss process, and N2O production process, respectively. Different small letters indicate significant differences among the three cropping patterns (p < 0.05). A significant difference between soil depths and statistical significance is indicated as follows. * p < 0.05.
3.4. Association of Soil Properties and Enzyme Activity with Gene Abundance
Based on the NMDS results, we observed that both soil depths and cropping patterns had a significant impact on the abundance of N-cycling genes (Figure 3a). We then proceeded to investigate the individual effects of surface soil and deep soil cropping patterns on the abundance of N-cycling genes (Figure 3b,c). Furthermore, we carried out a correlation analysis between soil properties, enzymes, and N-cycling gene abundance (Figure 3d,e). Specifically, we found a significant positive correlation between pH and nosZ abundance, and a negative correlation between urease and nosZ abundance, within a soil depth range of 0–15 cm (Figure 3d). AOB abundance was positively correlated with protease and β-glucosidase, and negatively correlated with soil EC (Figure 3d). Additionally, the nifH/(AOA + AOB + nxrB) ratio was negatively associated with cellulase, while the nifH/(narG + norB + nosZ) ratio was positively correlated with SOM and TP (Figure 3d). Furthermore, the abundance of nxrB was positively correlated with TP in deep soil (Figure 3e), and the norB/nosZ ratio was positively correlated with EC and negatively correlated with protease (Figure 3e).
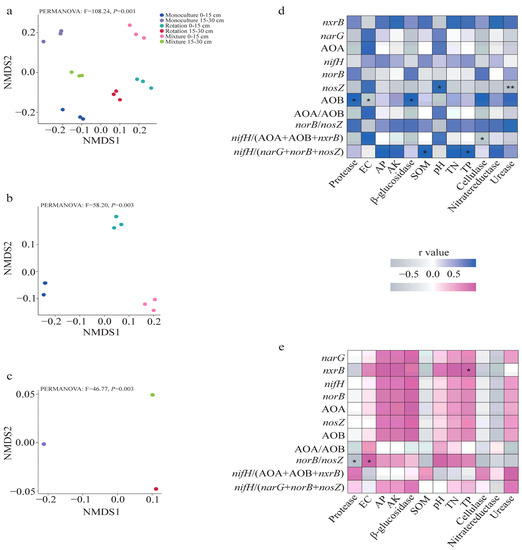
Figure 3.
The NMDS analysis of cropping patterns and soil depths on functional genes for N-cycling (a) and separate analysis of cropping patterns at 0–15 cm (b) and 15-30 cm (c). Correlation analysis of soil properties and enzyme activities with N-cycling gene abundance at 0–15 cm (d) and 15–30 cm (e) soil depths. A significant difference between soil depths and statistical significance is indicated as follows. * p < 0.05, ** p < 0.01.
3.5. Random Forest Analysis
We utilized a random forest model (Figure 4) to predict the impact of soil chemical properties and enzymes on the abundance of different N-cycling genes. At a soil depth of 0–15 cm, the abundance of nifH and norB was primarily driven by the activity of protease, while cellulase was the main driver for the abundance of AOA and nosZ. Additionally, pH was the primary driver for the abundance of AOB and narG, while nitrate reductase drove the abundance of nxrB (Figure 4a). At a soil depth of 15–30 cm, nitrate reductase was the primary driver for the abundance of AOA and nifH, AK was the main driver for the abundance of nxrB, and AP was the primary driver for the abundance of AOB, narG, norB, and nosZ (Figure 4b). We further investigated the relationship between important soil factors and N-cycling gene abundance through linear regression (Figure 5).
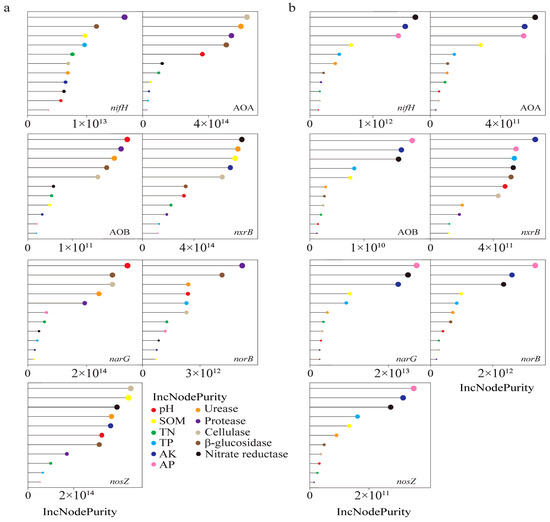
Figure 4.
Random forest analysis of soil chemistry and soil enzyme activity on functional genes abundance of N-cycling at 0–15 cm (a) and 15–30 cm soil depths (b).
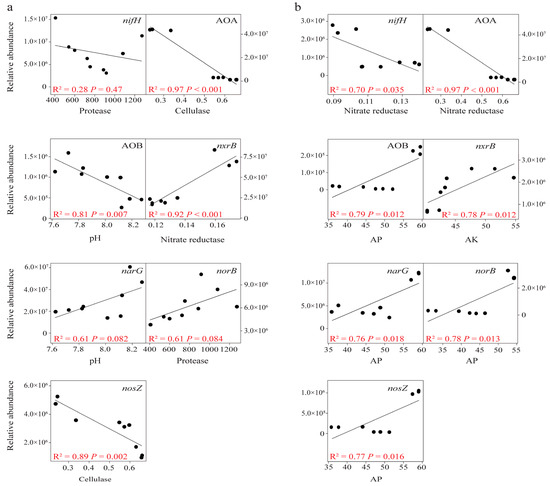
Figure 5.
Linear regression analysis of important soil properties and soil enzyme on functional genes abundance of N-cycling at 0–15 cm (a) and 15–30 cm soil depths (b).
A significant linear pattern was observed between soil factors and the abundance of nitrification functional genes (AOA, AOB, and nxrB) and nosZ at 0–15 cm soil depth (Figure 5a). At a soil depth of 15–30 cm, all models of N-cycling gene abundance were significantly correlated with important soil factors (Figure 5b).
4. Discussion
The security and sustainability of food production is one of the key challenges of the 21st century, and mixture and rotation are essential for soil fertility maintenance as conservation tillage practices. Our study found that complex cropping patterns promoted soil nutrients and prevented the further development of soil salinization (Table 1). This seems to be due to the effective increase of root secretions containing organic acids with the use of mixture and rotation, which in turn neutralized the CO32− and HCO3− content on soil colloids [52]. In addition, plants could absorb soil salts because of their ability to absorb salt ions directly [53,54].
4.1. Complex Cropping Patterns Alter nifH Gene Expression
Soil nitrogen is critical and the lack of nitrogen will severely limit agricultural production [55]. In the biosphere, nitrogen can only be fixed naturally by nitrogen-fixing microorganisms [56]. Zhou et al. found that crop rotation significantly affected the community structure of nifH diazotrophs, especially the community composition, by comparing soil samples from uncultivated and rotation fields [57]. Our study showed that soil depth may be an important factor affecting nifH expression in rotation soils, specifically by decreasing in topsoil and increasing in deeper soils. Previous studies have shown that high soil NH4+ amounts can suppress the abundance of the nifH gene [58,59,60]. Unfortunately, we did not test the NH4+ content of the three cropping patterns. Therefore, we speculate that it may be caused by high plant N uptake and N loss from different plants in the rotation soils triggered by heavy summer rainfall [61]. Hao et al. [62] also showed by a meta-analysis that long-term crop diversification appears to lead to a decrease in the abundance of the nifH gene. Our study found that the lowest AP content was detected in surface rotation soils (Table 1). The decrease in the abundance of nifH in the soil also seems to be closely related to the reduction of AP content [63]. Furthermore, we noted that proteases were the most important factors driving the abundance of the nifH gene (Figure 4). Soil microorganisms are capable of secreting extracellular proteins to influence soil nutrient cycling, so changes in proteases that are sensitive to the external environment may affect soil microbial-mediated nutrient cycling [64,65]. Proteases could suppress the abundance of the nifH gene from rotation soil in 0-15 cm soil depth, even though the two do not show significant correlation (Figure 3d and Figure 5a). This may be due to the fact that proteases provide bioavailable nitrogen by breaking down organic matter in the soil [66] and smaller organic matter molecules such as oligopeptides and amino acids [67]. In addition, Wang et al. also showed that the abundance of nifH may also be negatively correlated with bioavailable nitrogen in soil due to changes in the ecological niche, which further confirms our speculations [68]. The abundance of nifH increases with increasing soil depth. It appears to exhibit higher abundance for rotation soils in the 15–30 cm soil depth [69]. By comparing continuous and rotational cropping treatments, Zou et al. [70] showed that rotational cropping systems could significantly increase the abundance of the nifH gene at 0–20 cm soil depth in the black soils of Northeast China. This may be due to the perturbation of the multi-year crop rotation promoting an increase in the abundance of the nifH gene that was not as stable as in monoculture [71]. In addition, the abundance of nifH seems to be associated with soil N2 fixation [72]. It has been shown that the abundance of nifH genes detected in Australian soils during the disturbance experiment was significantly and positively correlated with the rate of nitrogen-fixation processes, indicating the vigorous activity of bacterial populations [73]. By comparing the abundance of functional genes mediating different nitrogen-cycling processes, we found that the nitrogen-fixation efficiency of rotation and mixture soils may be higher than that of monoculture in deep soil (Figure 2a). The increase in soluble organic carbon promoted by the litter left in the soil by rotation and mixture can provide a sufficient carbon source for the enrichment of diazotrophic bacteria [72], which promotes the nitrogen-fixation efficiency of the rotation and mixture in the deep soil (Figure 2a). In addition, we noted that soil depth significantly influenced the nitrogen-fixation efficiency in complex cropping systems (Figure 2a). Deeper soils have higher soil temperatures than topsoil, which promotes the decomposition of soil organic matter and thus increases the efficiency of N fixation. Soil quality improvement can improve greenhouse-gas (N2O) emissions, and the significant correlation between nitrate reduction and the abundance of nifH may be due to the relationship established by N2O emissions [74]. It has been established that N2O emissions appear to be positively correlated with nitrate-reductase activity, while a reduction in soil N2O emissions is able to increase the abundance of N-fixing microorganisms [74]. Soil nitrate reductase may have influenced N2O emissions and, in turn, the abundance of nifH. The increased abundance of nifH is supported by the possible higher availability of oxygen in the soil, as low oxygen conditions are also considered to be one of the controlling factors for microbial nitrogen fixation [75,76]. In addition, deeper soils have higher soil temperatures than topsoil, which promotes the decomposition of soil organic matter and thus increases the efficiency of N fixation.
4.2. Cropping Patterns Ecological Niches Affect Nitrifying Bacteria Abundance
Nitrification can be driven by AOB or AOA and is a central process in the N-cycling process [77]. Our study found that the abundance of AOA of monoculture was significantly higher than in rotation and mixture soils in surface soil, which seemed to be caused by differences in soil oxygen levels (Figure 1). The multi-year rotation of soils introduces different vegetation types of apoplastic material, which can act as cover crops that can play a role in increasing soil oxygen [78]. In addition, mixture soils have greater planting density, and more plant roots and an excess root system ensure sufficient soil pore space to store oxygen [79]. Interestingly, we noted a significant negative correlation between cellulase activity and AOA gene abundance (Figure 5a). Zhang et al. [80] reported that elevated CO2 can lead to a decrease in soil-cellulase activity. This indicates that the link between AOA abundance and the presence of cellulose seems to be caused by changes in CO2 due to changes in oxygen content. In contrast to the abundance of AOA, more complex cropping patterns appear to have led to an increase in the abundance of AOB in 0–15 cm soil depth (Figure 1). Although a study reported that crop rotation seems to have little effect on AOB abundance [81], for saline low-nutrient soils, the restoration of soil nutrients seems to provide more ecological niches for increased AOB abundance. Rotation and mixture will inevitably introduce more nutrient inputs than monoculture soils, and this will inevitably increase the abundance of AOB [82,83]. Many studies have confirmed that pH is a driving factor affecting the abundance of AOB [84,85,86]. Our study found that soil pH was significantly and negatively correlated with the abundance of AOB (Figure 5a). It was reported that AOA seems to prefer an acidic environment while AOB prefers an alkaline environment [87]. AOB is more sensitive to environmental changes and more susceptible to soil-salt stress than AOA [88]. Saline and alkaline stress reduced the number of AOB and reduced the preference of AOB for saline and alkaline environments [89]. Furthermore, in addition to soil acidity, the ecological niche specialization between AOA and AOB depends on the availability of nutrients from soil resources [90]. Soil proteases and β-glucosidases have been shown to be significantly correlated with soil-nutrient content [91], suggesting that AOB seems to be able to benefit soil nutrients better than AOA to ensure bacterial population growth. In addition, a significant negative correlation between EC and AOB abundance has also been reported, and EC may have an effect on AOB abundance [92]. Luo et al. [93] also reported a significant positive correlation between soil AP content and AOB abundance. In deeper soils, we observed significantly higher abundances of both AOA and AOB in rotation than in monoculture and mixture soils (Figure 1). The changes in AOA and AOB abundance were consistent, showing that AOA also plays an important role in soil nitrification in deep soils [94]. Crop rotation can better increase the carbon and nitrogen content of the soil and maintain soil quality as well as nutrient balance [95,96]. It has been confirmed that nitrate reductase is closely related to the AOA community [97].
We noted that the mixture possessed higher nxrB abundance than monoculture and rotation at a soil depth of 0–15 cm (Figure 1). It is reported that nxrB is a functional gene of the Nitrospira bacterial populations [98] and mixture soils are richer in root secretions and thus recruit Nitrospira [99]. We also found that the abundance of nxrB appeared to be significantly higher in monoculture than in a mixture at a soil depth of 15–30 cm (Figure 1). Zhang et al. [100] found that a conventional monocrop pattern could increase the abundance of Nitrospira, which may explain the higher abundance of nxrB in monoculture soils. Nitrospira is a diverse group of nitrite-oxidizing bacteria (NOB) and among the environmentally most widespread nitrifiers [101] and it responded significantly to the gradient of environmental nutrients, which included soil TP content [102]. In addition, Yu et al. [103] tested nine classes of Nitrospira and three of them showed a significant positive correlation with soil TP, which would suggest that a significant positive correlation between nxrB and TP is possible. Hu et al. found that the AOA/AOB ratio decreased with increasing pH by gaining insight into the ecological characteristics of AOB in 65 soils collected from various soil and ecosystem types [104]. This contrasts with our findings that AOA microorganisms seem to remain active in saline soils and that AOA and AOB do not compete. In addition, the higher soil nutrient content of rotation and mixture promoted the increase in AOB abundance, resulting in a lower AOA/AOB ratio [105]. Szukics et al. [106] found that AOA abundance can also be favored by high pH through the study of eight mountain grasslands in Austria, France, and the UK. In addition to this, we observed a significant effect of soil depth on AOA/AOB (Figure 2). AOA has the advantage of being more numerous in the amoA gene pool relative to AOB, and as AOA decreases with depth leading to the same trend in the AOA/AOB ratio [107].
4.3. Mixture and Rotation Reduce N Losses by Reducing narG and nosZ Abundance
In this study, we observed that the abundance of narG and nosZ was significantly higher in monoculture than in rotation and mixture soils in a soil depth of 0–15 cm (Figure 1). The decrease in the abundance of narG may be related to the shallow groundwater of the soil. Zhang et al. found that the abundance of narG genes decreases as the water table decreases and the soil profile continues to dry [108]. Crop rotation soils have more plant litter in the surface soil to play a certain role in water absorption [109]. In addition, more roots in mixture soils also led to increased water demand [110]. Soil pH appears to be an important factor affecting nosZ abundance, and the abundance of nosZ was inhibited by low pH and increased with increasing pH [111]. The present study confirms this idea, with lower pH values in rotation and mixture than in monoculture soils (Figure 3d and Table 1). In addition, an increase in the denitrifying bacteria abundance and a significant decrease in urease activity have been reported, which is consistent with the results of the study (Figure 3d). The abundance of narG, norB and nosZ genes were significantly higher in rotation than in monoculture and mixture soils in 15–30 cm soil depth (Figure 1) Crop rotation can increase the rate of denitrification, which also increases when a mixture of crop residues is added to the soil [112]. It seems that the difference in oxygen levels due to soil depth provides a better environment for denitrifying bacterial populations to survive, and residual apoplast is less likely to decompose and meet nutrient requirements. In addition, we also observed that the abundance of nosZ in the mixture was significantly higher than in monoculture in deep soils (Figure 1). Castellano et al. demonstrated that mixed crops of both legumes and non-legumes significantly increased nosZ abundance and significantly reduced N2O emissions by mixing two types of plants, legumes, and non-legumes [113]. This may be due to the influence of soil moisture. It has been reported that nosZ gene abundance is increased by reducing soil moisture [114]. More roots in the mixture reduce soil water retention compared to monoculture soils in the deep soil. Notably, we found that the norB/nosZ ratio in the mixture soils was significantly higher than the monoculture soils in surface soil (Figure 2d). This suggests that N2O emissions in monoculture may be greater than in mixture soils. A tighter link between N2O-reducers and mixture-composition compared to microbes involved in N2O production, which may be of agronomic relevance as microbes containing the nosZ gene are the only known biological sink for N2O emissions [115]. This indicates that the abundance of nosZ is closely related to N2O emissions, and the abundance of nosZ is significantly higher in monoculture than in mixture soils in 0–15 cm soil depth (Figure 1). Unlike the topsoil, the norB/nosZ ratio was significantly lower in the mixture than in the rotation and monoculture soils in 15–30 cm (Figure 2d). This may be due to changes in microbial community structure as well as changes in nosZ community diversity. Different mixed-crop species affect the abundance of nosZ and nosZ communities [116]. This is due to differences in the composition of plant root exudates and root morphology, which determine the degree of competition between plant and microbial communities for available soil N [117,118]. In addition, our study found a significant correlation of the norB/nosZ ratio with soil EC and protease activity (Figure 3e). Adviento et al. found that the maximum N2O loss occurred at the highest EC level by exploring the effect of electrical conductivity (EC) on N2O production in five soils under intensive cultivation [119]. It has also been confirmed that soil N2O emissions are significantly correlated with soil protease activity [120], which remains consistent with the results of this study (Figure 3e).
5. Conclusions
The introduction of a mixture of oats and tall wheatgrass, along with rotation, has been found to promote soil desalination and nutrient restoration, particularly soil enzyme activity, in saline soils. The abundance of N-cycling is strongly influenced by cropping patterns and soil depths, with rotation decreasing nifH abundance in surface soils and increasing nifH abundance, AOA, and AOB abundance in deep soils. Crop diversity is crucial, resulting in the highest nxrB abundance in mixture surface soils. However, complex cropping patterns decrease AOA abundance and increase AOB abundance in surface soils. They also reduce narG and nosZ abundance in surface soils while increasing denitrification in deep soils. Mixtures may promote N2O emissions from surface soils and suppress N2O emissions in deep soils. Changes in soil chemistry and enzyme activity play a vital role in the genetic variation of N-cycling functions in saline soils. Further investigation into changes in microbial communities of different N-cycling genes is necessary to better understand the mechanisms of genetic variation in nitrogen-cycling processes in saline soils.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/su15086592/s1, Table S1: Summary of genes investigated, the enzymes they encode, and their function in the nitrogen cycle; Table S2: PCR primers used for quantitative PCR and reaction conditions. References [121,122,123,124,125,126] are cited in the Supplementary Materials.
Author Contributions
Conceptualization, J.D., M.S. and X.L.; methodology, J.D. and B.L.; data processing, J.D. and M.S.; literature review, B.L. and J.D.; writing—original draft preparation, B.L. and J.D.; writing—review and editing, X.L.; supervision, M.S. and X.L.; funding acquisition, J.D. All authors have read and agreed to the published version of the manuscript.
Funding
This study received financial support from the Natural Science Foundation of Heilongjiang Province (Grant No. LH2021D014).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fagodiya, R.K.; Malyan, S.K.; Singh, D.; Kumar, A.; Yadav, R.K.; Sharma, P.C.; Pathak, H. Greenhouse gas emissions from salt-affected soils: Mechanistic understanding of interplay factors and reclamation approaches. Sustainability 2022, 14, 11876. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.-L.; Wang, S. Brief analysis on the trend of improve saline alkali soil in China. Hubei Agric. Sci. 2021, 59, 302–306. [Google Scholar]
- Ma, Y.; Tashpolat, N. Current Status and Development Trend of Soil Salinity Monitoring Research in China. Sustainability 2023, 15, 5874. [Google Scholar] [CrossRef]
- Shao, Q.; Han, N.; Ding, T.; Zhou, F.; Wang, B. SsHKT1; 1 is a potassium transporter of the C3 halophyte Suaeda salsa that is involved in salt tolerance. Funct. Plant Biol. 2014, 41, 790–802. [Google Scholar] [CrossRef]
- Song, J.; Wang, B. Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann. Bot. 2015, 115, 541–553. [Google Scholar] [CrossRef]
- Yuan, F.; Lyu, M.J.A.; Leng, B.Y.; Zhu, X.G.; Wang, B.S. The transcriptome of NaCl-treated Limonium bicolor leaves reveals the genes controlling salt secretion of salt gland. Plant Mol. Biol. 2016, 91, 241–256. [Google Scholar] [CrossRef]
- Yuan, F.; Liang, X.; Li, Y.; Yin, S.; Wang, B. Methyl jasmonate improves tolerance to high salt stress in the recretohalophyte Limonium bicolor. Funct. Plant Biol. 2019, 46, 82–92. [Google Scholar] [CrossRef]
- AbdElgawad, H.; Zinta, G.; Hegab, M.M.; Pandey, R.; Asard, H.; Abuelsoud, W. High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 2016, 7, 276. [Google Scholar] [CrossRef]
- Ding, T.; Yang, Z.; Wei, X.; Yuan, F.; Yin, S.; Wang, B. Evaluation of salt-tolerant germplasm and screening of the salt-tolerance traits of sweet sorghum in the germination stage. Funct. Plant Biol. 2018, 45, 1073–1081. [Google Scholar] [CrossRef]
- Wang, F.X.; Yin, C.H.; Song, Y.P.; Li, Q.; Tian, C.Y.; Song, J. Reproductive allocation and fruit-set pattern in the euhalophyte Suaeda salsa in controlled and field conditions. Plant Biosyst. 2018, 152, 749–758. [Google Scholar] [CrossRef]
- Liu, J.; Li, L.; Yuan, F.; Chen, M. Exogenous salicylic acid improves the germination of Limonium bicolor seeds under salt stress. Plant Signal. Behav. 2019, 14, e1644595. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, Y.; Wei, X.; Zhao, X.; Wang, B.; Sui, N. Transcription profiles of genes related to hormonal regulations under salt stress in sweet sorghum. Plant Mol. Biol. Rep. 2017, 35, 586–599. [Google Scholar] [CrossRef]
- Song, Y.; Ma, L.; Zhang, H.; Fu, R.; Liang, X.; Li, J.; Li, J.; Li, M.; Shan, Y.; Cheng, J.; et al. The diversity and structure of diazotrophic communities in the rhizosphere of coastal saline plants is mainly affected by soil physicochemical factors but not host plant species. Front. Microbiol. 2022, 9, 2634. [Google Scholar] [CrossRef]
- Boyrahmadi, M.; Raiesi, F. Plant roots and species moderate the salinity effect on microbial respiration, biomass, and enzyme activities in a sandy clay soil. Biol. Fert. Soils. 2018, 54, 509–521. [Google Scholar] [CrossRef]
- Choudhary, M.; Chandra, P.; Dixit, B.; Nehra, V.; Choudhary, U.; Choudhary, S. Plant growth-promoting microbes: Role and prospective in amelioration of silk stress. Commun. Soil Sci. Plan. 2022, 53, 1692–1711. [Google Scholar] [CrossRef]
- Luo, S.S.; Tian, L.; Chang, C.L.; Wang, S.J.; Zhang, J.F.; Zhou, X.; Li, X.J.; Tran, L.S.P.; Tian, C.J. Grass and maize vegetation systems restore saline-sodic soils in the Songnen Plain of northeast China. Land Degrad. Dev. 2018, 29, 1107–1119. [Google Scholar] [CrossRef]
- Guo, Z.G.; Liu, H.X.; Wang, S.M.; Tian, F.P.; Cheng, G.D. Biomass, persistence and drought resistance of nine lucerne varieties in the dry environment of west China. Aust. J. Exp. Agr. 2005, 45, 59–64. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, W.; Xi, L. Alfalfa cover crops influence the soil fungal community and function in apple orchards in arid desert oases in northwest China. Sustainability 2022, 14, 11816. [Google Scholar] [CrossRef]
- Li, B.; Liu, X.; Zhu, D.; Su, H.; Guo, K.; Sun, G.; Li, X.; Sun, L. Crop diversity promotes the recovery of fungal communities in saline-alkali areas of the Western Songnen Plain. Front. Microbiol. 2023, 14, 52. [Google Scholar] [CrossRef]
- Singh, A.; Quinn, N.W.T.; Benes, S.E.; Cassel, F. Policy-driven sustainable saline drainage disposal and forage production in the western San Joaquin Valley of California. Sustainability 2020, 12, 6362. [Google Scholar] [CrossRef]
- Jensen, K.B.; Pearse, G.; Larson, S.R.; Robins, J.G. ‘AlkarXL’, a new tall wheatgrass cultivar for use on saline semiarid lands. J. Plant Regist. 2020, 14, 298–305. [Google Scholar] [CrossRef]
- Suyama, H.; Benes, S.E.; Robinson, P.H.; Getachew, G.; Grattan, S.R.; Grieve, C.M. Biomass yield and nutritional quality of forage species under long-term irrigation with saline-sodic drainage water: Field evaluation. Anim. Feed Sci. Technol. 2007, 135, 329–345. [Google Scholar] [CrossRef]
- Balota, E.L.; Colozzi, A.; Andrade, D.S.; Dick, R.P. Microbial biomass in soils under different tillage and crop rotation systems. Biol. Fert. Soils 2003, 38, 15–20. [Google Scholar] [CrossRef]
- Zeng, L.; Wang, F.; Zhou, H.; Lyu, S.; Liu, W.; Cui, G.; Fan, Z.; Liu, L.; Zhang, K. Preliminary Screening of Vegetable Varieties Suitable for Rotation to Prevent and Control Banana Fusarium Wilt. Chin. J. Trop. Crops 2021, 42, 1678–1684. [Google Scholar]
- Liu, C.L.C.; Kuchma, O.; Krutovsky, K.V. Mixed-species versus monocultures in plantation forestry: Development, benefits, ecosystem services and perspectives for the future. Glob. Ecol. Conserv. 2018, 15, e00419. [Google Scholar] [CrossRef]
- Bainard, L.D.; Evans, B.; Malis, E.; Yang, T.; Bainard, J.D. Influence of Annual Plant Diversity on Forage Productivity and Nutrition, Soil Chemistry, and Soil Microbial Communities. Front. Microbiol. 2020, 4, 560479. [Google Scholar] [CrossRef]
- Brooker, R.W.; Bennett, A.E.; Cong, W.F.; Daniell, T.J.; George, T.S.; Hallett, P.D.; Hawes, C.; Iannetta, P.P.; Jones, H.G.; Karley, A.J.J.N.P. Improving intercropping: A synthesis of research in agronomy, plant physiology and ecology. New Phytol. 2015, 206, 107–117. [Google Scholar] [CrossRef]
- Gaba, S.; Lescourret, F.; Boudsocq, S.; Enjalbert, J.; Hinsinger, P.; Journet, E.P.; Navas, M.L.; Wery, J.; Louarn, G.; Malézieux, E.; et al. Multiple cropping systems as drivers for providing multiple ecosystem services: From concepts to design. Agron. Sustain. Dev. 2015, 35, 607–623. [Google Scholar] [CrossRef]
- Rosa, S.M.; Kraemer, F.B.; Soria, M.A.; Guerrero, L.D.; Morrás, H.J.; Figuerola, E.L.; Erijman, L.J.A.S.E. The influence of soil properties on denitrifying bacterial communities and denitrification potential in no-till production farms under contrasting management in the Argentinean Pampas. Appl. Soil Ecol. 2014, 75, 172–180. [Google Scholar] [CrossRef]
- Martins, S.R.; Casalinho, H.; Silva, J.J.C.A.S. Technology, Qualidade do solo como indicador de sustentabilidade de agroecossistemas. Curr. Agr. Sci. Technol. 2007, 13, 195–203. [Google Scholar]
- Zhang, W.; Shen, Z.; Shao, Y.; Shi, L.; Liu, S.; Shi, N.; Fu, S. Soil biota and sustainable agriculture: A review. Acta Ecol. Sin. 2020, 40, 3183–3206. [Google Scholar]
- Lee, T.K.; Han, I.; Kim, M.S.; Seong, H.J.; Kim, J.S.; Sul, W.J. Characterization of a nifH-Harboring Bacterial Community in the Soil-Limited Gotjawal Forest. Front. Microbiol. 2019, 10, 1858. [Google Scholar] [CrossRef]
- Dai, S.; Liu, Q.; Zhao, J.; Zhang, J. Ecological niche differentiation of ammonia-oxidising archaea and bacteria in acidic soils due to land use change. Soil Res. 2017, 56, 71–79. [Google Scholar] [CrossRef]
- Gross, C.; Hossen, S.; Hartmann, H.; Noll, M.; Borken, W. Biological nitrogen fixation and nifH gene abundance in dead wood of 13 different tree species. Biogeochemistry 2022, 161, 353–371. [Google Scholar] [CrossRef]
- Prosser, J.I.; Nicol, G.W. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ. Microbiol. 2008, 10, 2931–2941. [Google Scholar] [CrossRef] [PubMed]
- Prosser, J.I.; Nicol, G.W. Archaeal and bacterial ammonia-oxidisers in soil: The quest for niche specialisation and differentiation. Trends Microbiol. 2012, 20, 523–531. [Google Scholar] [CrossRef]
- Dell, E.A.; Bowman, D.; Rufty, T.; Shi, W. The community composition of soil-denitrifying bacteria from a turfgrass environment. Res. Microbiol. 2010, 161, 315–325. [Google Scholar] [CrossRef]
- Rosa Portilho, I.I.; Savin, M.C.; Borges, C.D.; Tsai, S.M.; Mercante, F.M.; Roscoe, R.; de Carvalho, L.A. Maintenance of N cycling gene communities with crop-livestock integration management in tropical agriculture systems. Agr. Ecosyst. Environ. 2018, 267, 52–62. [Google Scholar] [CrossRef]
- Dai, Z.M.; Li, Y.; Zhang, X.J.; Wu, J.J.; Luo, Y.; Kuzyakov, Y.; Brookes, P.C.; Xu, J.M. Easily mineralizable carbon in manure-based biochar added to a soil influences N2O emissions and microbial-N cycling genes. Land Degrad. Dev. 2019, 30, 406–416. [Google Scholar] [CrossRef]
- Liu, H.; Wu, X.; Wang, Q.; Wang, S.; Liu, D.; Liu, G. Responses of soil ammonia oxidation and ammonia-oxidizing communities to land-use conversion and fertilization in an acidic red soil of southern China. Eur. J. Soil Biol. 2017, 80, 110–120. [Google Scholar] [CrossRef]
- Ma, B.; Wang, Y.; Ye, S.; Liu, S.; Stirling, E.; Gilbert, J.A.; Faust, K.; Knight, R.; Jansson, J.K.; Cardona, C.; et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 2020, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Levy-Booth, D.J.; Prescott, C.E.; Grayston, S.J. Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 2014, 75, 11–25. [Google Scholar] [CrossRef]
- Soil Survey Staff. Keys to Soil Taxonomy, 11th ed.; United States Department of Agriculture (USDA) & Natural Resources Conservation Service (NRCS): Washington, DC, USA, 2010. [Google Scholar]
- Chen, K.; Peng, J.; Li, J.; Yang, Q.; Zhan, X.M.; Liu, N.; Han, X.R. Stabilization of soil aggregate and organic matter under the application of three organic resources and biochar-based compound fertilizer. J. Soil Sediment. 2020, 20, 3633–3643. [Google Scholar] [CrossRef]
- Hu, X.J.; Liu, J.J.; Liang, A.Z.; Li, L.J.; Yao, Q.; Yu, Z.H.; Li, Y.S.; Jin, J.; Liu, X.B.; Wang, G.H. Conventional and conservation tillage practices affect soil microbial co-occurrence patterns and are associated with crop yields. Agric. Ecosyst. Environ. 2021, 319, 107534. [Google Scholar] [CrossRef]
- Guan, S.Y. Soil Enzymes and Their Research Method; Higher Education Press: Beijing, China, 1986; pp. 275–312. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. R Package, Version 1.0.12.; R Project: Tartu, Estonia, 2019.
- Oksanen, J.J. Vegan: Community Ecology Package; R Project: Helsinki, Finland, 2010. [Google Scholar]
- Archer, E. rfPermute: Estimate Permutation p-Values for Random Forest Importance Metrics; R Project: Indianapolis, IN, USA, 2016. [Google Scholar]
- Team, R.J.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2009; Volume 14, pp. 12–21. [Google Scholar]
- Guo, Y.D.; Cheng, M.; Zhao, X.F.; Hao, B.P.; Zhang, Y.F.; Cao, W.D.; Zheng, P.S. Effects of green manure rotation on soil properties and yield and quality of silage maize in saline-alkali soils. Chin. J. Eco-Agric. 2018, 26, 856–864. [Google Scholar]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Bhowmik, P.C.; Hossain, M.A.; Rahman, M.M.; Prasad, M.N.V.; Ozturk, M.; Fujita, M. Potential use of halophytes to remediate saline soils. Biomed. Res. Int. 2014, 2014, 589341. [Google Scholar] [CrossRef]
- Yuan, F.; Leng, B.; Wang, B. Progress in studying salt secretion from the salt glands in recretohalophytes: How do plants secrete salt? Front. Plant Sci. 2016, 7, 977. [Google Scholar] [CrossRef]
- Zhu, Y.; Shao, T.; Zhou, Y.; Zhang, X.; Gao, X.; Long, X.; Rengel, Z.J.L.D. Periphyton improves soil conditions and offers a suitable environment for rice growth in coastal saline alkali soil. Land Degrad. Dev. 2021, 32, 2775–2788. [Google Scholar] [CrossRef]
- Alaylar, B.; Gulluce, M.; Karadayi, M. Detection of the nifH gene in nitrogen fixing bacteria from agricultural areas in erzurum. Fresen Environ. Bull. 2020, 29, 809–814. [Google Scholar]
- Zhou, L.T.; Li, J.J.; Pokhrel, G.R.; Chen, J.; Zhao, Y.L.; Bai, Y.; Zhang, C.; Lin, W.X.; Wu, Z.Y.; Wu, C.Z. nifH Gene sequencing reveals the effects of successive monoculture on the soil diazotrophic microbial community in Casuarina equisetifolia plantations. Front. Plant Sci. 2021, 11, 578812. [Google Scholar] [CrossRef] [PubMed]
- Lei, Z.; Li, Q.Q.; Tang, Y.N.; Zhang, H.; Han, C.; Wang, X.; Zhao, X.H.; Shi, G.Y. Selenium enhanced nitrogen accumulation in legumes in soil with rhizobia bacteria. J. Clean. Prod. 2022, 380, 134960. [Google Scholar] [CrossRef]
- Dai, X.L.; Song, D.L.; Guo, Q.K.; Zhou, W.; Liu, G.R.; Ma, R.P.; Liang, G.Q.; He, P.; Sun, G.; Yuan, F.S.; et al. Predicting the influence of fertilization regimes on potential N fixation through their effect on free-living diazotrophic community structure in double rice cropping systems. Soil Boil. Biochem. 2021, 156, 108220. [Google Scholar] [CrossRef]
- Wang, L.N.; Yu, Z.; Yang, J.; Zhou, J. Diazotrophic bacterial community variability in a subtropical deep reservoir is correlated with seasonal changes in nitrogen. Environ. Sci. Pollut. Res. 2015, 22, 24. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.Z.; Hong, H.S.; Zhang, Y.Z.; Chen, N.W.; Zeng, Y.; Wang, W.P. Health, Anthropogenic nitrogen sources and exports in a village-scale catchment in Southeast China. Environ. Geochem. Health 2006, 28, 45. [Google Scholar] [CrossRef]
- Hao, J.Q.; Feng, Y.Z.; Wang, X.; Yu, Q.; Zhang, F.; Yang, G.H.; Ren, G.X.; Han, X.H.; Wang, X.J.; Ren, C.J. Soil microbial nitrogen-cycling gene abundances in response to crop diversification: A meta-analysis. Sci. Total Environ. 2022, 838, 156621. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Wang, J.; Wang, J.D.; Zhang, Y.C. Long-term fertilization with high nitrogen rates decreased diversity and stability of diazotroph communities in soils of sweet potato. Appl. Soil Ecol. 2022, 170, 104266. [Google Scholar] [CrossRef]
- Xu, H.; Wu, X.; Zhao, L.; Zhao, Y.; Wu, T.; Hu, G.; Li, W.; Ding, Y. Changes in soil enzyme activities under different vegetation types of the northern fringe of the permafrost regions in the Qinghai-Tibetan Plateau. Fresen. Environ. Bull. 2015, 24, 4720–4728. [Google Scholar]
- Graham, E.B.; Yang, F.; Bell, S.; Hofmockel, K.S. High Genetic Potential for Proteolytic Decomposition in Northern Peatland Ecosystems. Appl. Environ. Microb. 2019, 85, e02851-18. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R. Regulation of extracellular protease activity in soil in response to different sources and concentrations of nitrogen and carbon. Soil Boil. Biochem. 2008, 40, 3040–3048. [Google Scholar] [CrossRef]
- Ladd, J.; Butler, J. Short-term assays of soil proteolytic enzyme activities using proteins and dipeptide derivatives as substrates. Soil Biol. Biochem. 1972, 4, 19–30. [Google Scholar] [CrossRef]
- Wang, C.; Zheng, M.M.; Song, W.F.; Wen, S.L.; Wang, B.R.; Zhu, C.Q.; Shen, R.F. Impact of 25 years of inorganic fertilization on diazotrophic abundance and community structure in an acidic soil in southern China. Soil Biol. Biochem. 2017, 113, 240–249. [Google Scholar] [CrossRef]
- Wang, J.C.; Zhang, D.; Zhang, L.; Li, J.; Raza, W.; Huang, Q.W.; Shen, Q.R. Temporal variation of diazotrophic community abundance and structure in surface and subsoil under four fertilization regimes during a wheat growing season. Agric. Ecosyst. Environ. 2016, 216, 116–124. [Google Scholar] [CrossRef]
- Zou, J.X.; Yao, Q.; Liu, J.J.; Li, Y.S.; Song, F.Q.; Liu, X.B.; Wang, G.H. Changes of diazotrophic communities in response to cropping systems in a Mollisol of Northeast China. PeerJ 2020, 8, e9550. [Google Scholar] [CrossRef] [PubMed]
- Collavino, M.M.; Tripp, H.J.; Frank, I.E.; Vidoz, M.L.; Calderoli, P.A.; Donato, M.; Zehr, J.P.; Aguilar, O.M. nifH pyrosequencing reveals the potential for location-specific soil chemistry to influence N2-fixing community dynamics. Environ. Microbiol. 2014, 16, 3211–3223. [Google Scholar] [CrossRef]
- Xie, Z.; Yu, Z.; Li, Y.; Wang, G.; Tang, C.; Mathesius, U.; Liu, X.; Liu, J.; Liu, J.; Herbert, S.J.; et al. Linking rhizospheric diazotrophs to the stimulation of soybean N2 fixation in a Mollisol amended with maize straw. Plant Soil 2021, 463, 279–289. [Google Scholar] [CrossRef]
- Colloff, M.J.; Wakelin, S.A.; Gomez, D.; Rogers, S.L. Detection of nitrogen cycle genes in soils for measuring the effects of changes in land use and management. Soil Biol. Biochem. 2008, 40, 1637–1645. [Google Scholar] [CrossRef]
- Harter, J.; Krause, H.M.; Schuettler, S.; Ruser, R.; Fromme, M.; Scholten, T.; Kappler, A.; Behrens, S. Linking N2O emissions from biochar-amended soil to the structure and function of the N-cycling microbial community. ISME J. 2014, 8, 660–674. [Google Scholar] [CrossRef]
- Ventouses, P.M.; Cassman, K.; Cleveland, C.; Crews, T.; Field, C.B.; Grimm, N.B.; Howarth, R.W.; Marino, R.; Martinelli, L.; Rastetter, E.B.; et al. Towards an Ecological Understanding of Biological Nitrogen Fixation. In The Nitrogen Cycle at Regional to Global Scales; Springer: Dordrecht, The Netherlands, 2022. [Google Scholar] [CrossRef]
- Reed, S.C.; Cleveland, C.C.; Townsend, A.R. Functional ecology of free-living nitrogen fixation: A contemporary perspective. Annu. Rev. Ecol. Evol. Syst. 2011, 42, 489–512. [Google Scholar] [CrossRef]
- Liu, H.Y.; Qin, S.Y.; Liu, H.E. Comammox Nitrospira and AOB communities are more sensitive than AOA community to different fertilization strategies in a fluvo-aquic soil. Agr. Ecosyst. Environ. 2023, 342. [Google Scholar] [CrossRef]
- Sias, C.; Wolters, B.R.; Reiter, M.S.; Flessner, M.L. Cover crops as a weed seed bank management tool: A soil down review. Ital. J. Agron. 2021, 16, 1852. [Google Scholar] [CrossRef]
- Ile, O.J.; Aguilos, M.; Morkoc, S.; Heitman, J.; King, J.S. Root biomass distribution and soil physical properties of short-rotation coppice American sycamore (Platanus occidentalis L.) grown at different planting densities. Forests 2021, 12, 1806. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, Y.; Chen, L.; Wu, Z. Response of soil saccharidase activities to free-air carbon dioxide enrichment (FACE) under rice-wheat rotation. Ying Yong Sheng Tai Xue Bao = J. Appl. Ecol. 2004, 15, 1019–1024. [Google Scholar]
- Munroe, J.W.; McCormick, I.; Deen, W.; Dunfield, K.E. Effects of 30 years of crop rotation and tillage on bacterial and archaeal ammonia oxidizers. J. Environ. Qual. 2016, 45, 940–948. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Bai, N.L.; Sun, H.F.; Zhou, S.; Zheng, X.Q.; Li, S.X.; Zhang, J.Q.; Zhang, H.Y.; Lv, W.G. Spatial and temporal responses of ammonia-oxidizing bacteria and archaea to organic amendments in rice-wheat rotation system. Appl. Soil Ecol. 2019, 139, 94–99. [Google Scholar] [CrossRef]
- Zhang, H.L.; Sun, H.F.; Zhou, S.; Bai, N.L.; Zheng, X.Q.; Li, S.X.; Zhang, J.Q.; Lv, W.G. Effect of straw and straw biochar on the community structure and diversity of ammonia-oxidizing bacteria and archaea in rice-wheat rotation ecosystems. Sci. Rep. 2019, 9, 9367. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.J.; Tang, C.Y.; Cao, Y.J.; Li, X.; Huang, P.Y. Contribution of ammonia-oxidizing archaea and bacteria to nitrification under different biogeochemical factors in acidic soils. Environ. Sci. Pollut. R. 2022, 29, 17209–17222. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Dong, Z.X.; Wang, Z.; Xiao, L.W.; Zhu, B. The contributions of ammonia oxidizing bacteria and archaea to nitrification-dependent N2O emission in alkaline and neutral purple soils. Sci. Rep. 2022, 12, 19928. [Google Scholar] [CrossRef]
- Ying, J.Y.; Li, X.X.; Wang, N.N.; Lan, Z.C.; He, J.Z.; Bai, Y.F. Contrasting effects of nitrogen forms and soil pH on ammonia oxidizing microorganisms and their responses to long-term nitrogen fertilization in a typical steppe ecosystem. Soil Biol. Biochem. 2017, 107, 10–18. [Google Scholar] [CrossRef]
- Robinson, A.; Di, H.J.; Cameron, K.C.; Podolyan, A.; He, J. The effect of soil pH and dicyandiamide (DCD) on N2O emissions and ammonia oxidiser abundance in a stimulated grazed pasture soil. J. Soil. Sediment. 2014, 14, 1434–1444. [Google Scholar] [CrossRef]
- Zhang, Q.; Liang, G.; Myrold, D.D.; Zhou, W. Variable responses of ammonia oxidizers across soil particle-size fractions affect nitrification in a long-term fertilizer experiment. Soil Biol. Biochem. 2017, 105, 25–36. [Google Scholar] [CrossRef]
- Guo, J.X.; Zhou, Y.X.; Guo, H.J.; Min, W. Saline and alkaline stresses alter soil properties and composition and structure of gene-based nitrifier and denitrifier communities in a calcareous desert soil. BMC Microbiol. 2021, 21, 246. [Google Scholar] [CrossRef] [PubMed]
- Balume, I.; Agumas, B.; Musyoki, M.; Marhan, S.; Cadisch, G.; Rasche, F. Potential proteolytic enzyme activities modulate archaeal and bacterial nitrifier abundance in soils differing in acidity and organic residue treatment. Appl. Soil Ecol. 2022, 169, 104188. [Google Scholar] [CrossRef]
- Adetunji, A.T.; Lewu, F.B.; Mulidzi, R.; Ncube, B. The biological activities of beta-glucosidase, phosphatase and urease as soil quality indicators: A review. J. Soil Sci. Plant Nut. 2017, 17, 794–807. [Google Scholar] [CrossRef]
- He, Y.; Hu, W.; Ma, D.; Yang, Y.; Lan, H.; Gao, Y. Diversity and abundance of ammonia-oxidizing microorganisms in relation to soil environment in rhizosphere soil of Halocnemum strobilaceum in Ebinur Lake wetland. Acta Sci. Circumstantiae 2017, 37, 1967–1975. [Google Scholar]
- Luo, P.; Fan, Y.; Yang, J.; Ge, Y.; Cai, F.; Han, X. Influence of long-term fertilization on abundance of ammonia oxidizing bacteria and archaea in brown soil. J. Plant Nutr. Fert. 2017, 23, 678–685. [Google Scholar]
- Li, S.; Jiang, X.; Wang, X.; Wright, A.L. Tillage effects on soil nitrification and the dynamic changes in nitrifying microorganisms in a subtropical rice-based ecosystem: A long-term field study. Soil. Till. Res. 2015, 150, 132–138. [Google Scholar] [CrossRef]
- Tiemann, L.K.; Grandy, A.S.; Atkinson, E.E.; Marin-Spiotta, E.; McDaniel, M.D. Crop rotational diversity enhances belowground communities and functions in an agroecosystem. Ecol. Lett. 2015, 18, 761–771. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Tiemann, L.K.; Grandy, A.S. Does agricultural crop diversity enhance soil microbial biomass and organic matter dynamics? A meta-analysis. Ecol. Appl. 2014, 24, 560–570. [Google Scholar] [CrossRef]
- Wei, D.; Zeng, S.; Hou, D.; Zhou, R.; Xing, C.; Deng, X.; Yu, L.; Wang, H.; Deng, Z.; Weng, S.; et al. Community diversity and abundance of ammonia-oxidizing archaea and bacteria in shrimp pond sediment at different culture stages. J. Appl. Microbiol. 2021, 130, 1442–1455. [Google Scholar] [CrossRef]
- Pester, M.; Maixner, F.; Berry, D.; Rattei, T.; Koch, H.; Luecker, S.; Nowka, B.; Richter, A.; Spieck, E.; Lebedeva, E.; et al. NxrB encoding the beta subunit of nitrite oxidoreductase as functional and phylogenetic marker for nitrite-oxidizing Nitrospira. Environ. Microbiol. 2014, 16, 3055–3071. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.L.; Mei, L.L.; Wei, Q.H.; Li, B.; Zhang, P.; Sun, S.X.; Cui, G.W. Leymus chinensis resists degraded soil stress by modulating root exudate components to attract beneficial microorganisms. Front. Microbiol. 2022, 13, 951838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.Y.; Shi, Y.C.; Dong, Y.X.; Lapen, D.R.; Liu, J.H.; Chen, W. Subsoiling and conversion to conservation tillage enriched nitrogen cycling bacterial communities in sandy soils under long-term maize monoculture. Soil Till. Res. 2022, 215, 105197. [Google Scholar] [CrossRef]
- Koch, H.; Lucker, S.; Albertsen, M.; Kitzinger, K.; Herbold, C.; Spieck, E.; Nielsen, P.H.; Wagner, M.; Daims, H. Expanded metabolic versatility of ubiquitous nitriteoxidiz-ing bacteria from the genus Nitrospira. Proc. Natl. Acad. Sci. USA 2015, 112, 11371–11376. [Google Scholar] [CrossRef]
- Liu, S.F.; Wang, H.Y.; Chen, L.M.; Wang, J.W.; Zheng, M.S.; Liu, S.T.; Chen, Q.; Ni, J.R. Comammox Nitrospira within the Yangtze River continuum: Community, biogeography, and ecological drivers. ISME J. 2020, 14, 2488–2504. [Google Scholar] [CrossRef]
- Yu, J.-L.; Xia, J.-J.; Li, C.-H.; Zhang, S.-H.; Li, X.; Lu, Y.; Xininigen. Niche differentiation of Nitrospira and associated environmental driving forces in Xilin river basin. Microbiol. China 2020, 47, 1418–1429. [Google Scholar]
- Hu, H.W.; Zhang, L.M.; Dai, Y.; Di, H.J.; He, J.Z. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J. Soil Sediment. 2013, 13, 1439–1449. [Google Scholar] [CrossRef]
- Behnke, G.D.; Zabaloy, M.C.; Riggins, C.W.; Rodriguez-Zas, S.; Huang, L.; Villamil, M.B. Acidification in corn monocultures favor fungi, ammonia oxidizing bacteria, and nirK-denitrifier groups. Sci. Total Environ. 2020, 720, 137514. [Google Scholar] [CrossRef]
- Szukics, U.; Grigulis, K.; Legay, N.; Kastl, E.M.; Baxendale, C.; Bardgett, R.D.; Clement, J.C.; Lavorel, S.; Schloter, M.; Bahn, M. Management versus site effects on the abundance of nitrifiers and denitrifiers in European mountain grasslands. Sci. Total Environ. 2019, 648, 745–753. [Google Scholar] [CrossRef]
- Banning, N.C.; Maccarone, L.D.; Fisk, L.M.; Murphy, D.V. Ammonia-oxidising bacteria not archaea dominate nitrification activity in semi-arid agricultural soil. Sci. Rep. 2015, 5, e11146. [Google Scholar] [CrossRef]
- Zhang, D.; Cui, R.Y.; Fu, B.; Yang, Y.X.; Wang, P.L.; Mao, Y.T.; Chen, A.Q.; Lei, B.K. Shallow groundwater table fluctuations affect bacterial communities and nitrogen functional genes along the soil profile in a vegetable field. Appl. Soil Ecol. 2020, 146, 103368. [Google Scholar] [CrossRef]
- Luan, L.; Zhang, G.; Sun, L.; Geng, R.; Wang, H. Spatial variation in water-holding properties of typical plant litters in the loess hilly region. J. Soil Water Conserv. 2015, 29, 225–230. [Google Scholar]
- Lobet, G.; Couvreur, V.; Meunier, F.; Javaux, M.; Draye, X. Plant water uptake in drying soils. Plant Physiol. 2014, 164, 1619–1627. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Z.; Guo, B.L.; Liu, H.S.; Wu, Y.Q.; Yu, J.Q.; Ding, H.; Jiang, X.H.; Luo, Q.D.; Zhang, Y.S. Low pH inhibits soil nosZ without affecting N2O uptake. J. Soil. Sediment. 2023, 23, 422–430. [Google Scholar] [CrossRef]
- Zhao, W.; Li, S.; Dong, L.; Wang, P.; Lu, X.; Zhang, X.; Su, Z.; Guo, Q.; Ma, P. Effects of different crop rotations on the incidence of cotton Verticillium wilt and structure and function of the rhizospheric microbial community. Plant Soil 2023. [Google Scholar] [CrossRef]
- Castellano-Hinojosa, A.; Martens-Habbena, W.; Strauss, S.L. Cover crop composition drives changes in the abundance and diversity of nitrifiers and denitrifiers in citrus orchards with critical effects on N2O emissions. Geoderma 2022, 422, 115952. [Google Scholar] [CrossRef]
- Pereira, E.I.P.; Chung, H.; Scow, K.; Six, J. Microbial communities and soil structure are affected by reduced precipitation, but not by elevated carbon dioxide. Soil Sci. Soc. Am. J. 2013, 77, 482–488. [Google Scholar] [CrossRef]
- Jones, C.M.; Spor, A.; Brennan, F.P.; Breuil, M.-C.; Bru, D.; Lemanceau, P.; Griffiths, B.; Hallin, S.; Philippot, L. Recently identified microbial guild mediates soil N2O sink capacity. Nat. Clim. Chang. 2014, 4, 801–805. [Google Scholar] [CrossRef]
- Graf, D.R.; Saghaï, A.; Zhao, M.; Carlsson, G.; Jones, C.M.; Hallin, S.J.S.B. Lucerne (Medicago sativa) alters N2O-reducing communities associated with cocksfoot (Dactylis glomerata) roots and promotes N2O production in intercropping in a greenhouse experiment. Soil Biol. Biochem. 2019, 137, 107547. [Google Scholar] [CrossRef]
- Cantarel, A.A.; Pommier, T.; Desclos-Theveniau, M.; Diquélou, S.; Dumont, M.; Grassein, F.; Kastl, E.-M.; Grigulis, K.; Laîné, P.; Lavorel, S.J.E. Using plant traits to explain plant–microbe relationships involved in nitrogen acquisition. Ecology 2015, 96, 788–799. [Google Scholar] [CrossRef]
- Moreau, D.; Pivato, B.; Bru, D.; Busset, H.; Deau, F.; Faivre, C.; Matejicek, A.; Strbik, F.; Philippot, L.; Mougel, C.J.E. Plant traits related to nitrogen uptake influence plant-microbe competition. Ecology 2015, 96, 2300–2310. [Google Scholar] [CrossRef]
- Adviento-Borbe, M.A.A.; Doran, J.W.; Drijber, R.A.; Dobermann, A. Soil electrical conductivity and water content affect nitrous oxide and carbon dioxide emissions in intensively managed soils. J. Environ. Qual. 2006, 35, 1999–2010. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.S.; Qu, T.H.; Li, Y.F.; Van Zwieten, L.; Wang, H.L.; Chen, J.H.; Song, X.Z.; Lin, Z.; Zhang, X.P.; Luo, Y.; et al. Biochar-based fertilizer decreased while chemical fertilizer increased soil N2O emissions in a subtropical Moso bamboo plantation. Catena 2021, 202, 105257. [Google Scholar] [CrossRef]
- Fan, K.; Delgado-Baquerizo, M.; Guo, X.; Wang, D.; Wu, Y.; Zhu, M.; Yu, W.; Yao, H.; Zhu, Y.; Chu, H. Suppressed N fixation and diazotrophs after four decades of fertilization. Microbiome 2019, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Meinhardt, K.A.; Bertagnolli, A.; Pannu, M.W.; Strand, S.E.; Brown, S.L.; Stahl, D.A. Evaluation of revised polymerase chain reaction primers for more inclusive quantification of ammonia-oxidizing archaea and bacteria. Environ. Microbiol. Rep. 2015, 7, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, L.; Lu, Q.; Raza, W.; Huang, Q.; Shen, Q. Ammonia oxidizer abundance in paddy soil profile with different fertilizer regimes. Appl. Soil Ecol. 2014, 84, 38–44. [Google Scholar] [CrossRef]
- Cassman, N.A.; Soares, J.R.; Pijl, A.; Lourenço, K.S.; van Veen, J.A.; Cantarella, H.; Kuramae, E.E. Nitrification inhibitors effectively target N2O-producing Nitrosospira spp. in tropical soil. Environ. Microbiol. 2019, 21, 1241–1254. [Google Scholar] [CrossRef] [PubMed]
- Bru, D.; Sarr, A.; Philippot, L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl. Environ. Microbiol. 2007, 73, 5971–5974. [Google Scholar] [CrossRef]
- Wang, J.P.; Meng, F.G.; Li, W. The short- and long-term effects of formic acid on rapid nitritation start-up. Environ. Int. 2020, 135. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).