Admixture of Holothurian Species in the Hellenic Seas (Eastern Mediterranean) as Revealed by RADseq
Abstract
1. Introduction
2. Materials and Method
2.1. Samplings
2.2. Molecular Procedures
2.2.1. DNA Extraction and Sequencing
2.2.2. Mapping, Variant Calling, and Filtering
2.2.3. Genomic Analysis
3. Results
3.1. SNPs Calling
3.2. Genetic Differentiation and Diversity
3.3. Coalescent Demographic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- de Bruyn, M.; Hall, B.L.; Chauke, L.F.; Baroni, C.; Koch, P.L.; Hoelzel, A.R. Rapid response of a marine mammal species to holocene climate and habitat change. PLoS Genet. 2009, 5, e1000554. [Google Scholar] [CrossRef]
- Gkafas, G.A.; Exadactylos, A.; Rogan, E.; Raga, J.A.; Reid, R.; Hoelzel, A.R. Biogeography and temporal progression during the evolution of striped dolphin population structure in European waters. J. Biogeogr. 2017, 44, 2681–3961. [Google Scholar] [CrossRef]
- Konstantinidis, I.; Gkafas, G.A.; Papathanasiou, V.; Orfanidis, S.; Küpper, F.C.; Arnaud-Haond, S.; Exadactylos, A. Biogeography pattern of the marine angiosperm Cymodocea nodosa in the eastern Mediterranean Sea related to the quaternary climatic changes. Ecol. Evol. 2022, 12, e8911. [Google Scholar] [CrossRef]
- Bargelloni, L.; Alarcon, J.A.; Alvarez, M.C.; Penzo, E.; Magoulas, A.; Palma, J.; Patarnello, T. The Atlantic-Mediterranean transition: Discordant genetic patterns in two seabream species Diplodus puntazzo (Cetti) and Diplodus sargus (L.). Mol. Phylogenetics Evol. 2005, 36, 523–535. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.X.; Wang, H.Y.; Zhang, T.; Liu, J.X. Population genetic structure and demographic history of Atrina pectinata based on mitochondrial DNA and microsatellite markers. PLoS ONE 2014, 9, e95436. [Google Scholar] [CrossRef]
- Vendrami, D.L.J.; De Noia, M.; Telesca, L.; Handal, W.; Charrier, G.; Boudry, P.; Eberhart-Phillips, L.; Hoffman, J.I. RAD sequencing sheds new light on the genetic structure and local adaptation of European scallops and resolves their demographic histories. Sci. Rep. 2019, 9, 7455. [Google Scholar] [CrossRef]
- Maggs, C.A.; Castilho, R.; Foltz, D.; Henzler, C.; Jolly, M.T.; Kelly, J.; Olsen, J.; Perez, K.E.; Stam, W.; Väinölä, R.; et al. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 2008, 89, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.; Godinho, R.; Randi, E.; Ferrand, N.; Alves, P.C. Molecular Analysis of Hybridisation between Wild and Domestic Cats (Felis silvestris) in Portugal: Implications for Conservation. Conserv. Genet. 2008, 9, 1–11. [Google Scholar] [CrossRef]
- Tea, Y.-K.; Hobbs, J.P.A.; Vitteli, F.; DiBattista, J.D.; Ho, S.Y.W.; Lo, N. Angels in disguise: Sympatric hybridization in the marine angelfishes is widespread and occurs between deeply divergent lineages. Proc. R. Soc. B 2020, 287, 1459. [Google Scholar] [CrossRef]
- Adavoudi, R.; Pilot, M. Consequences of Hybridization in Mammals. Genes 2022, 13, 50. [Google Scholar] [CrossRef]
- Bowen, B.W.; Rocha, L.A.; Toonen, R.J.; Karl, S.A.; Laboratory, T.T. Origins of tropical marine biodiversity. Trends Ecol. Evol. 2013, 28, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Allendorf, F.W.; Luikart, G.; Aitken, S.N. Conservation and the Genetics of Populations, 2nd ed.; USA Wiley-Blackwell: Hoboken, NJ, USA, 2013. [Google Scholar]
- Zinner, D.; Arnold, M.L.; Roos, C. The Strange Blood: Natural Hybridization in Primates. Evol. Anthropol. 2011, 20, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Jones, F.C.; Grabherr, M.G.; Chan, Y.F.; Russell, P.; Mauceli, E.; Johnson, J.; Swofford, R.; Pirun, M.; Zody, M.C.; White, S.; et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 2012, 484, 55–61. [Google Scholar] [CrossRef]
- Bernatchez, L.; Dodson, J.J. Allopatric origin of sympatric populations of lake whitefish (Coregonus clupeformis) as revealed by mitochondrial-DNA restriction analysis. Evolution 1990, 445, 1263–1271. [Google Scholar]
- Antoniou, A.; Frantzis, A.; Alexiadou, P.; Paschou, N.; Poulakakis, N. Evidence of introgressive hybridization between Stenella coeruleoalba and Delphinus delphis in th Greek Seas. Mol. Phylogenetics Evol. 2018, 129, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Simard, F.; Ayala, D.; Kamdem, G.C.; Pombi, M.; Etouna, J.; Ose, K.; Fotsing, J.M.; Fontenille, D.; Besansky, N.J.; Costantini, C. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: The ecological side of speciation. BMC Ecol. 2009, 91, 17. [Google Scholar] [CrossRef]
- Lamichhaney, S.; Han, F.; Webster, M.T.; Andersson, L.; Grant, B.R.; Grant, P.R. Rapid hybrid speciation in Darwin’s finches. Science 2018, 359, 224–227. [Google Scholar] [CrossRef]
- Nosil, P. Ecological Speciation; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- Hendry, A.P. Ecological speciation! Or the lack thereof? Can. J. Fish. Aquat. Sci. 2009, 66, 1383–1398. [Google Scholar] [CrossRef]
- Zhang, L.; He, J.; Tan, P.; Gong, Z.; Qian, S.; Mia, Y.; Zhang, H.Y.; Tu, G.; Chen, Q.; Zhong, Q.; et al. The genome of an apodid holothuroid (Chiridota heheva) provides insights into its adaptation to a deep-sea reducing environment. Commun. Biol. 2022, 5, 224. [Google Scholar] [CrossRef]
- Borrero-Pérez, G.H.; Gómez-Zurita, J.; González-Wangüemert, M.; Marcos, C.; Pérez-Ruzafa, A. Molecular systematics of the genus Holothuria in the Mediterranean and Northeastern Atlantic and a molecular clock for the diversification of the Holothuriidae (Echinodermata: Holothuroidea). Mol. Phylogenetics Evol. 2010, 57, 899–906. [Google Scholar] [CrossRef]
- Belbachir, N.E.; Lepoint, G.; Mezali, K. Comparison of isotopic niches of four sea cucumbers species (Holothuroidea: Echinodermata) inhabiting two seagrass meadows in the southwestern Mediterranean Sea (Mostaganem Algeria). Belg. J. Zool. 2019, 149, 95–106. [Google Scholar] [CrossRef]
- Boncagni, P.; Rakaj, A.; Fianchin, A.; Vizzini, S. Preferential assimilation of seagrass detritus by two coexisting Mediterranean sea cucumbers: Holothuria polii and Holothuria tubulosa. Estuar. Coast. Shelf Sci. 2019, 231, 106464. [Google Scholar] [CrossRef]
- Antoniadou, C.; Vafidis, D. Population structure of the traditionally exploited holothurian Holothuria tubulosa in the south Aegean Sea. Cah. Biol. Mar. 2011, 52, 171–175. [Google Scholar]
- Gonzalez-Wangüemert, M.; Domínguez-Godino, J.; Cánovas, F. The fast development of sea cucumber fisheries in the Mediterranean and NE Atlantic waters: From a new marine resource to its over-exploitation. Ocean. Coast. Manag. 2018, 151, 165–177. [Google Scholar] [CrossRef]
- Bulteel, P.; Jangoux, M.; Coulon, P. Biometry bathymetric distribution and reproductive cycle of the holothuroid Holothuria tubulosa (Echinodermata) in Mediterranean seagrass beds. Mar. Ecol. 1992, 13, 53–62. [Google Scholar] [CrossRef]
- Simunovic, A.; Piccinetti, C.; Bartulovic, M.; Grubelic, I. Distribution and abundance of the species Holothuria tubulosa Gmelin 1788 and Holothuria forskali Delle Chiaje 1823 (Holothuria Echinodermata) in the Adriatic Sea. Acta Adriat. 2000, 41, 3–16. [Google Scholar]
- Kazanidis, G.; Lolas, A.; Vafidis, D. Reproductive cycle of the traditionally exploited sea cucumber Holothuria tubulosa (Holothuroidea: Aspidochirotida) in Pagasitikos Gulf western Aegean Sea Greece. Turk. Zool. Derg. 2014, 38, 306–315. [Google Scholar] [CrossRef]
- Rakaj, A.; Fianchini, A.; Boncagni, P.; Lovatelli, A.; Scardi, M.; Cataudella, S. Spawning and rearing of Holothuria tubulosa: A new candidate for aquaculture in the Mediterranean region. Aquac. Res. 2018, 49, 557–568. [Google Scholar] [CrossRef]
- Uthicke, S.; Purcell, S.; Blockmans, B. Natural hybridization does not dissolve species boundaries in commercially important sea cucumbers. Biol. J. Linn. Soc. 2005, 85, 261–270. [Google Scholar] [CrossRef]
- Feidantsis, K.; Gkafas, G.A.; Exadactylos, A.; Michaelidis, B.; Staikou, A.; Hatziioannou, M.; Apostologamvrou, C.; Sarantopoulou, J.; Vafidis, D. Different Interspecies Demographic Histories within the Same Locality: A Case Study of Sea Cucumbers Cuttlefish and Clams in Greek Waters. Sustainability 2022, 14, 14380. [Google Scholar] [CrossRef]
- Peterson, B.K.; Weber, J.N.; Kay, E.H.; Fisher, H.S.; Hoekstra, H.E. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLoS ONE 2012, 7, e37135. [Google Scholar] [CrossRef] [PubMed]
- Lepais, O.; Weir, J.T. SimRAD: An R package for simulation–based prediction of the number of loci expected in RADseq and similar genotyping by sequencing approaches. Mol. Ecol. Resour. 2014, 14, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Catchen, J.; Hohenlohe, P.A.; Bassham, S.; Amores, A.; Cresko, W.A. Stacks: An analysis tool set for population genomics. Mol. Ecol. 2013, 22, 3124–3140. [Google Scholar] [CrossRef] [PubMed]
- Medina-Feliciano, J.G.; Pirro, S.; García-Arrarás, J.E.; Mashanov, V.; Ryan, J.F. Draft Genome of the Sea Cucumber Holothuria glaberrima a Model for the Study of Regeneration. Front. Mar. Sci. 2021, 8, 603410. [Google Scholar] [CrossRef]
- Li, H. Exploring single–sample SNP and INDEL calling with whole–genome de novo assembly. Bioinformatics 2012, 28, 1838–1844. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Durbin, R. Genome Project Data Processing Subgroup Genome Project Data Processing Subgroup. 2009 The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A toolset for whole-genome association and population based linkage analysis. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- De Jong, M.J.; de Jong, J.F.; Hoelzel, A.R.; Janke, A. SambaR: An R package for fast easy and reproducible population-genetic analyses of biallelic SNP datasets. Mol. Ecol. Resour. 2021, 21, 1369–1379. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing R Foundation for Statistical Computing: Vienna Austria 2023. Available online: https://www.R-project.org/ (accessed on 15 March 2023).
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar]
- Raj, A.; Stephens, M.; Pritchard, J.K. fastSTRUCTURE: Variational inference of population structure in large SNP data sets. Genetics 2014, 197, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; von Holdt, B.M. Structure Harvester: A website and program for visualizing Structure output and implementing the Evanno method. Conserv. Genet. Res. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef]
- Anderson, E.C.; Thompson, E.A. A Model-Based Method for Identifying Species Hybrids Using Multilocus Genetic Data. Genetics 2002, 160, 1217–1229. [Google Scholar] [CrossRef]
- Santiago, E.; Novo, I.; Pardinas, F.; Saura, M.; Wang, J.; Caballero, A. Recent Demographic History Inferred by High-Resolution Analysis of Linkage Disequilibrium. Mol. Biol. Evol. 2020, 37, 3642–3653. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hansen, M.M. PSMC (pairwise sequentially Markovian coalescent) analysis of RAD (restriction site associated DNA) sequencing data. Mol. Ecol. Resour. 2017, 17, 631–641. [Google Scholar] [CrossRef]
- Excoffier, L.; Marchi, N.; Marques, D.A.; Matthey-Doret, R.; Gouy, A.; Sousa, V.C. fastsimcoal2: Demographic inference under complex evolutionary scenarios. Bioinformatics 2021, 37, 4882–4885. [Google Scholar] [CrossRef]
- Gutenkunst, R.N.; Hernandez, R.D.; Williamson, S.H.; Bustamante, C.D. Inferring the joint demographic history of multiple populations from multidimensional SNP data. PLoS Genet. 2009, 5, e1000695. [Google Scholar] [CrossRef]
- Crossman, C.A.; Taylor, E.B.; Barrett-Lennard, L.G. Hybridization in the Cetacea: Widespread occurrence and associated morphological behavioral and ecological factors. Ecol. Evol. 2016, 6, 1293–1303. [Google Scholar] [CrossRef] [PubMed]
- Mezali, K.; Zupo, V.; Francour, P. Population dynamics of Holothuria (Holothuria) tubulosa and Holothuria (Lessonothuria) poli of an Algerian Posidonia oceanica meadow. Biol. Mar. Mediterr. 2006, 13, 158–161. [Google Scholar]
- Aydin, M. Some biological characteristics of the sea cucumber Holothuria (Platyperona) sanctori Delle Chiaje, 1823 (Echinodermata: Holothuroidea). Biol. Div. Conserv. 2015, 6, 153–159. [Google Scholar]
- Kazanidis, G.; Antoniadou, C.; Lolas, A.; Neofitou, N.; Vafidis, D.; Chintiroglou, C.; Neofitou, C. Population dynamics and reproduction of Holothuria tubulosa (Holothuroidea: Echinodermata) in the Aegean Sea. J. Mar. Biol. Assoc. UK 2010, 90, 895–901. [Google Scholar] [CrossRef]
- Lee, J.; Byrne, M.; Uthicke, S. The influence of population density on fission and growth of Holothuria atra in natural mesocosms. J. Exp. Mar. Biol. Ecol. 2008, 365, 126–135. [Google Scholar] [CrossRef]
- Kamarudin, K.R.; Rehan, M.M. Morphological and Molecular Identification of Holothuria (Merthensiothuria) leucospilota and Stichopus horrens from Pangkor Island Malaysia. Trop. Life Sci. Res. 2015, 26, 87–99. [Google Scholar]
- Moussa, R.; Wirawati, I. Observations on some biological characteristics of Holothuria polii and Holothuria sanctori from Mediterranean Egypt. Int. J. Fish. Aquat. Sci. 2018, 6, 351–357. [Google Scholar]
- Utzeri, V.J.; Ribani, A.; Bovo, S.; Taurisano, V.; Calassanzio, M.; Baldo, D.; Fontanesi, L. Microscopic ossicle analyses and the complete mitochondrial genome sequence of Holothuria (Roweothuria) polii (Echinodermata Holothuroidea) provide new information to support the phylogenetic positioning of this sea cucumber species. Mar. Genom. 2022, 51, 100735. [Google Scholar] [CrossRef]
- Fitzpatrick, S.W.; Reid, B.N. Does gene flow aggravate or alleviate maladaptation to environmental stress in small populations? Evol. Appl. 2019, 12, 1402–1416. [Google Scholar] [CrossRef]
- Massin, C.; Mercier, A.; Hamel, J.F. Ossicle change in Holothuria scabra with a discussion of ossicle evolution within the Holothuriidae (Echinodermata). Acta Zool. 2000, 81, 77–91. [Google Scholar] [CrossRef]
- Fraïsse, C.; Roux, C.; Welch, J.J.; Bierne, N. Gene-flow in a mosaic hybrid zone: Is local introgression adaptive? Genetics 2014, 197, 939–951. [Google Scholar] [CrossRef]
- Quilodrán, C.S.; Tsoupas, A.; Currat, M. The spatial signature of introgression after a biological invasion with hybridization. Front. Ecol. Evol. 2020, 8, 321. [Google Scholar] [CrossRef]
- Soliman, T.; Fernandez-Silva, I.; Reimer, J.D. Genetic population structure and low genetic diversity in the over-exploited sea cucumber Holothuria edulis Lesson 1830 (Echinodermata: Holothuroidea) in Okinawa Island. Conserv. Genet. 2016, 17, 811–821. [Google Scholar] [CrossRef]
- Willing, E.M.; Dreyer, C.; Oosterhout, C.V. Estimates of genetic differentiation measured by FST do not necessarily require large sample sizes when using many SNP markers. PLoS ONE 2012, 7, e42649. [Google Scholar] [CrossRef] [PubMed]
- Nazareno, A.G.; Bemmels, J.B.; Dick, C.W.; Lohmann, L.G. Minimum sample sizes for population genomics: An empirical study from an Amazonian plant species. Mol. Ecol. Resour. 2017, 17, 1136–1147. [Google Scholar] [CrossRef]
- Yin, B.; Yang, S.; Shang, G.; Wei, W. Effects of predation risk on behavior hormone levels and reproductive success of plateau pikas. Ecosphere 2017, 8, e01643. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Multimodel Inference: Understanding AIC and BIC in Model Selection. Sociol. Methods Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
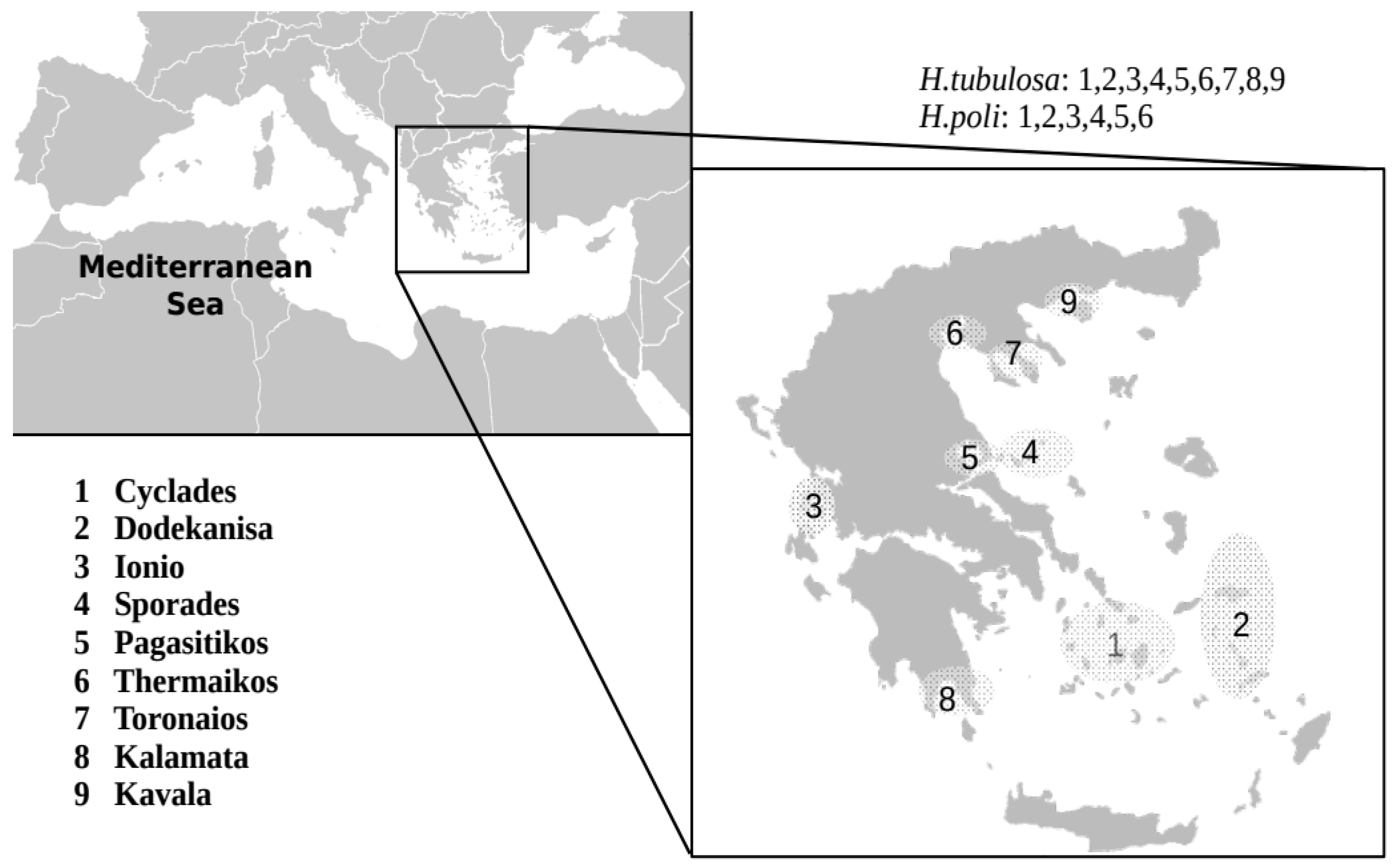
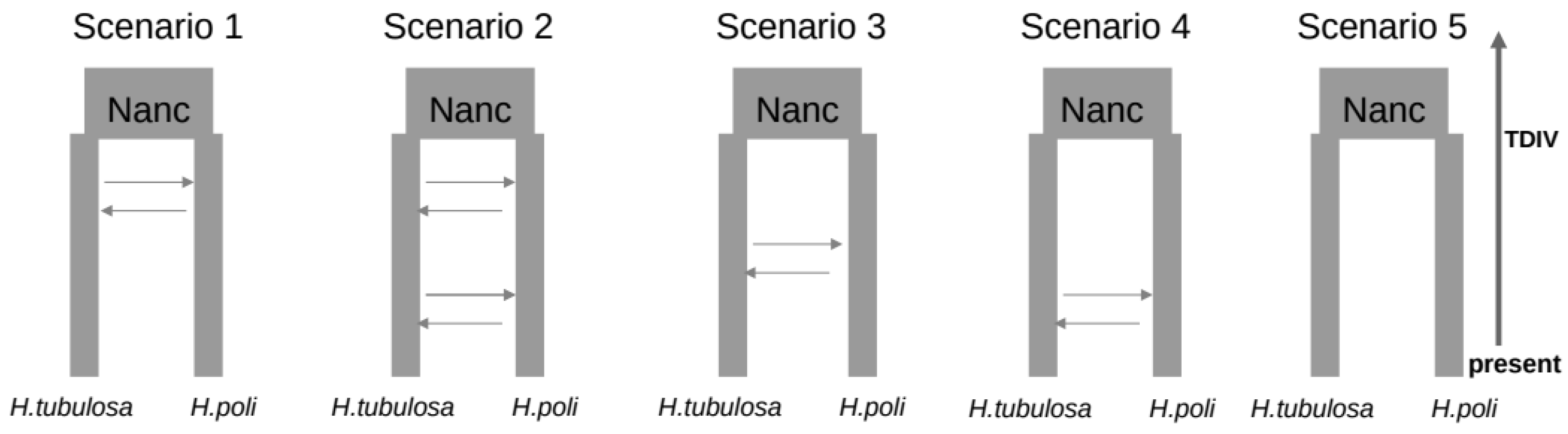
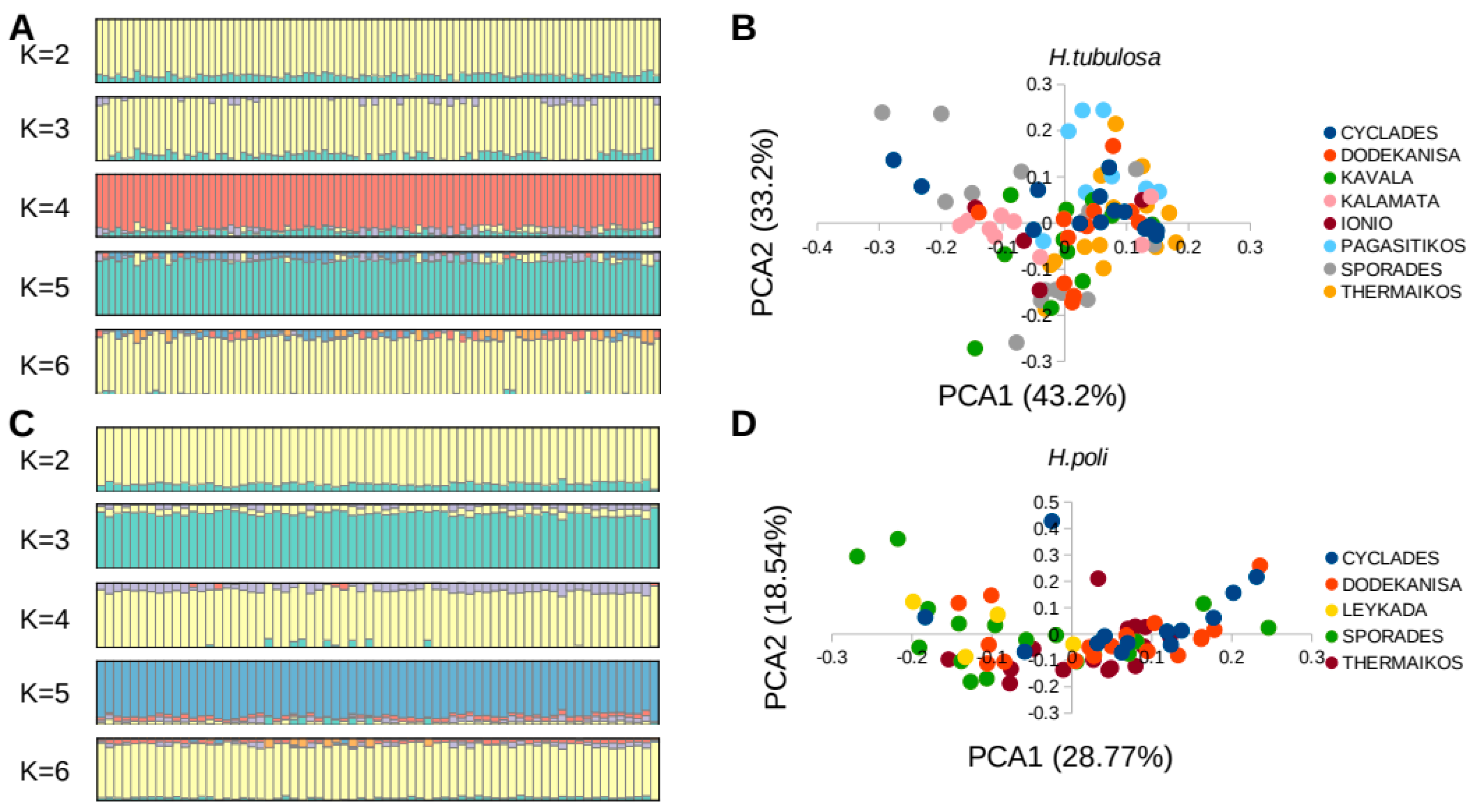
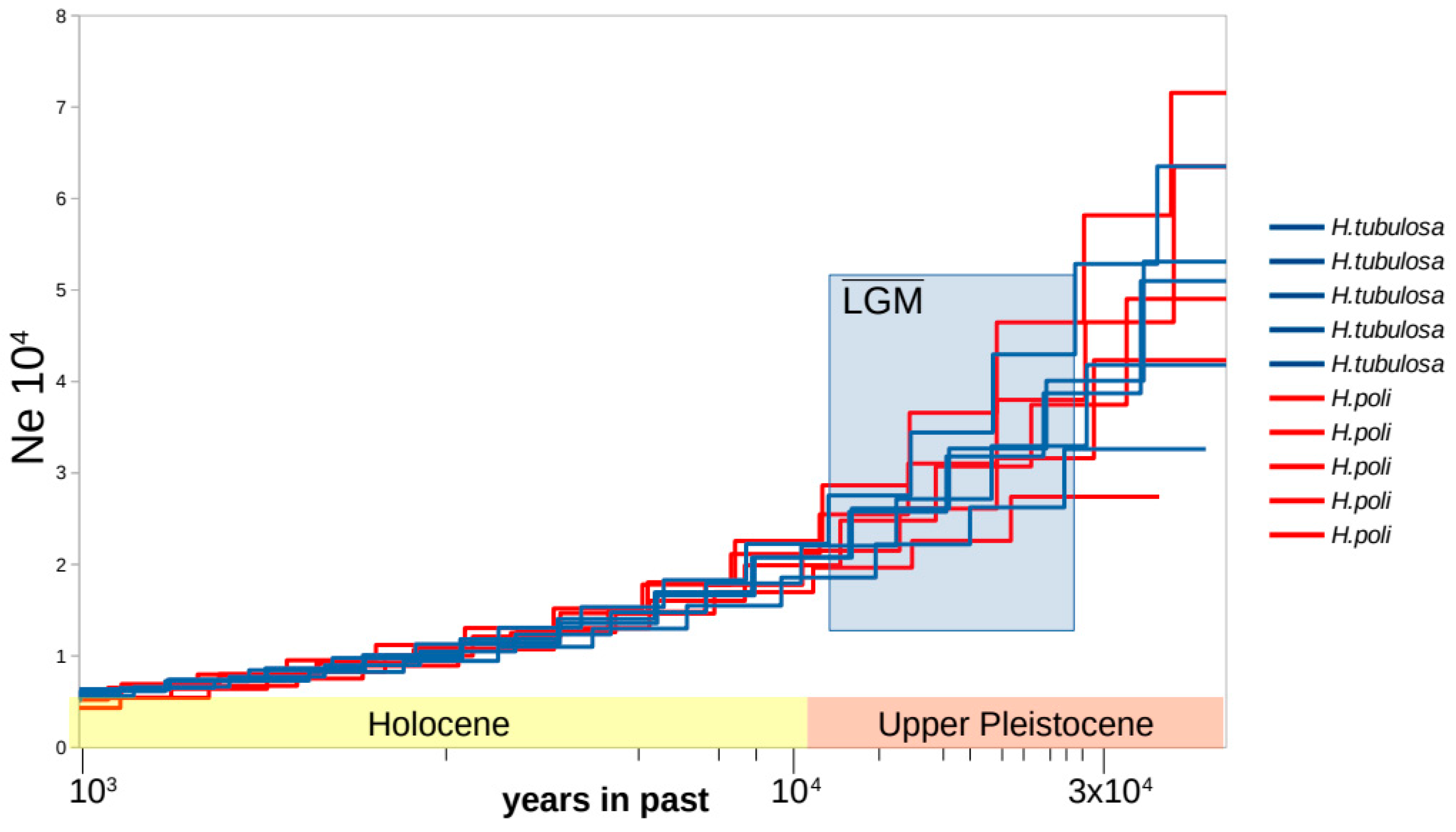
| Species | CYCL | DOD | ION | KAL | KAV | PAG | SPO | |
|---|---|---|---|---|---|---|---|---|
| H. tubulosa | DOD | 0.002 | ||||||
| ION | 0.0005 | 0.008 | ||||||
| KAL | 0.004 | 0.005 | 0.006 | |||||
| KAV | 0.002 | 0.003 | −0.002 | 0.005 | ||||
| PAG | 0.004 | 0.008 | −0.001 | 0.014 | 0.008 | |||
| SPO | 0.003 | 0.003 | −0.003 | 0.003 | −0.001 | 0.001 | ||
| THE | 0.011 | 0.011 | 0.0006 | 0.025 | 0.006 | 0.015 | 0.007 | |
| CYCL | SPO | DOD | ION | |||||
| H. poli | SPO | 0.011 | ||||||
| DOD | 0.004 | 0.004 | ||||||
| ION | 0.006 | −0.003 | 0.003 | |||||
| THE | 0.005T | 0.006 | 0.002 | 0.003 |
| Priors | Median | 95% CI (Low–High) | GONE | |
|---|---|---|---|---|
| NH.tubulosa | 100–107 | 5.6 × 106 | 3.59 × 104 –9.32 × 106 | 4.43 × 106 |
| NH.poli | 100–107 | 6.0 × 105 | 5.62 × 104 –6.26 × 105 | 4.74 × 106 |
| NANC | 100–107 | 5.3 × 104 | 7.26 × 103–9.32 × 104 | |
| TDIV | 100–107 | 4.5 × 103 (13.5 × 103) | 3.32 × 103–9.92 × 103 | |
| mig1 | 10−6–10−3 | 0.002 | 9.7 × 106–4 × 10−3 | |
| mig2 | 10−6–10−3 | 8.5 × 10−6 | 1.5 × 106–4 × 10−4 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gkafas, G.A.; Sarantopoulou, J.; Apostologamvrou, C.; Antoniadou, C.; Exadactylos, A.; Fleris, G.; Vafidis, D. Admixture of Holothurian Species in the Hellenic Seas (Eastern Mediterranean) as Revealed by RADseq. Sustainability 2023, 15, 11493. https://doi.org/10.3390/su151511493
Gkafas GA, Sarantopoulou J, Apostologamvrou C, Antoniadou C, Exadactylos A, Fleris G, Vafidis D. Admixture of Holothurian Species in the Hellenic Seas (Eastern Mediterranean) as Revealed by RADseq. Sustainability. 2023; 15(15):11493. https://doi.org/10.3390/su151511493
Chicago/Turabian StyleGkafas, Georgios A., Joanne Sarantopoulou, Chrysoula Apostologamvrou, Chryssanthi Antoniadou, Athanasios Exadactylos, Georgios Fleris, and Dimitris Vafidis. 2023. "Admixture of Holothurian Species in the Hellenic Seas (Eastern Mediterranean) as Revealed by RADseq" Sustainability 15, no. 15: 11493. https://doi.org/10.3390/su151511493
APA StyleGkafas, G. A., Sarantopoulou, J., Apostologamvrou, C., Antoniadou, C., Exadactylos, A., Fleris, G., & Vafidis, D. (2023). Admixture of Holothurian Species in the Hellenic Seas (Eastern Mediterranean) as Revealed by RADseq. Sustainability, 15(15), 11493. https://doi.org/10.3390/su151511493










