Microbial and Physicochemical Status of Raw and Processed Sea Cucumbers from the Hellenic Seawaters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Provision, Processing and Sampling
2.2. Physicochemical Characteristics
2.2.1. Determination of pH
2.2.2. Determination of Water Activity
2.2.3. Determination of Moisture
2.2.4. Determination of Salt Content
2.2.5. Estimation of Water Phase Salt (WPS)
2.3. Microbiological Analysis
2.4. Metabarcoding Analysis of 16S rRNA Gene
2.4.1. Sample Preparation and DNA Extraction
2.4.2. Library Preparation, Next-Generation Sequencing and Bioinformatic Analysis
2.5. Statistical Analysis
3. Results
3.1. Physicochemical Characteristics
3.2. Microbiological Analysis
3.3. Metabarcoding Analysis of 16S rRNA Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zmemlia, N.; Bejaoui, S.; Khemiri, I.; Bouriga, N.; Louiz, I.; El-Bok, S.; Ben-Attia, M.; Souli, A. Biochemical composition and antioxidant potential of the edible Mediterranean sea cucumber Holothuria tubulosa. Grasas Y Aceites 2020, 71, e364. [Google Scholar] [CrossRef]
- Roggatz, C.C.; González-Wangüemert, M.; Pereira, H.; Rodrigues, M.J.; da Silva, M.M.; Barreira, L.; Varela, J.; Custódio, L. First report of the nutritional profile and antioxidant potential of Holothuria arguinensis, a new resource for aquaculture in Europe. Nat. Prod. Res. 2016, 30, 2034–2040. [Google Scholar] [CrossRef] [PubMed]
- Neofitou, N.; Lolas, A.; Ballios, I.; Skordas, K.; Tziantziou, L.; Vafidis, D. Contribution of sea cucumber Holothuria tubulosa on organic load reduction from fish farming operation. Aquaculture 2019, 501, 97–103. [Google Scholar] [CrossRef]
- Parra-Luna, M.; Martín-Pozo, L.; Hidalgo, F.; Zafra-Gómez, A. Common sea urchin (Paracentrotus lividus) and sea cucumber of the genus Holothuria as bioindicators of pollution in the study of chemical contaminants in aquatic media. A revision. Ecol. Indic. 2020, 113, 106185. [Google Scholar] [CrossRef]
- Ismail, H.; Lemriss, S.; Ben Aoun, Z.; Mhadhebi, L.; Dellai, A.; Kacem, Y.; Boiron, P.; Bouraoui, A. Antifungal activity of aqueous and methanolic extracts from the Mediterranean sea cucumber, Holothuria polii. J. Mycol. Med. 2008, 18, 23–26. [Google Scholar] [CrossRef]
- Bilgin, Ş.; Öztürk Tanrikulu, H. The changes in chemical composition of Holothuria tubulosa (Gmelin, 1788) with ambient-drying and oven-drying methods. Food Sci. Nutr. 2018, 6, 1456–1461. [Google Scholar] [CrossRef] [PubMed]
- Antoniadou, C.; Vafidis, D. Population structure of the traditionally exploited holothurian Holothuria tubulosa in the south Aegean Sea. Cah. Biol. Mar. 2011, 52, 171–175. [Google Scholar]
- González-Wangüemert, M.; Domínguez-Godino, J.; Cánovas, F. The fast development of sea cucumber fisheries in the Mediterranean and NE Atlantic waters: From a new marine resource to its over-exploitation. Ocean Coast Manag. 2018, 151, 165–177. [Google Scholar] [CrossRef]
- Kazanidis, G.; Lolas, A.; Vafidis, D. Reproductive cycle of the traditionally exploited sea cucumber Holothuria tubulosa (Holothuroidea: Aspidochirotida) in Pagasitikos Gulf, western Aegean Sea, Greece. Turk. J. Zool. 2014, 38, 306–315. [Google Scholar] [CrossRef]
- Kazanidis, G.; Antoniadou, C.; Lolas, A.P.; Neofitou, N.; Vafidis, D.; Chintiroglou, C.; Neofitou, C. Population dynamics and reproduction of Holothuria tubulosa (Holothuroidea: Echinodermata) in the Aegean Sea. J. Mar. Biol. Assoc. U. K. 2010, 90, 895–901. [Google Scholar] [CrossRef]
- Coulon, P.; Jangoux, M. Feeding rate and sediment reworking by the holothuroid Holothuria tubulosa (Echinodermata) in a Mediterranean seagrass bed off Ischia Island, Italy. Mar. Ecol. Prog. Ser. 1993, 92, 201–204. [Google Scholar] [CrossRef]
- Özer, N.P.; Mol, S.; Varlık, C. Effect of the Handling Procedures on the Chemical Composition of Sea Cucumber. Turk. J. Fish. Aquat. Sci. 2005, 74, 71–74. [Google Scholar]
- Mamelona, J.; Pelletier, É.; Girard-Lalancette, K.; Legault, J.; Karboune, S.; Kermasha, S. Quantification of phenolic contents and antioxidant capacity of Atlantic sea cucumber, Cucumaria frondosa. Food Chem. 2007, 104, 1040–1047. [Google Scholar] [CrossRef]
- Aminin, D.L.; Agafonova, I.G.; Avilov, S.A.; Stonik, V.A.; Berdyshev, E.V.; Isachenko, E.G. Immunomodulatory Properties of Cucumariosides from the Edible Far-Eastern Holothurian Cucumaria japonica. J. Med. Food 2004, 4, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Abedin, M.Z.; Karim, A.A.; Ahmed, F.; Latiff, A.A.; Gan, C.Y.; Che Ghazali, F.; Islam Sarker, M.Z. Isolation and characterization of pepsin-solubilized collagen from the integument of sea cucumber (Stichopus vastus). J. Sci. Food Agric. 2013, 93, 1083–1088. [Google Scholar] [CrossRef] [PubMed]
- Gram, L.; Huss, H.H. Microbiological spoilage of fish and fish products. Int. J. Food Microbiol. 1996, 33, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Parlapani, F.F. Microbial diversity of seafood. Curr. Opin. Food Sci. 2021, 37, 45–51. [Google Scholar] [CrossRef]
- FAO. The State of World Fisheries and Aquaculture. In Contributing to Food Security and Nutrition for All; FAO: Rome, Italy, 2016; Available online: http://www.fao.org/3/a-i5555e.pdf (accessed on 29 June 2023).
- Syropoulou, F.; Parlapani, F.F.; Kakasis, S.; Nychas, G.J.E.; Boziaris, I.S. Primary Processing and Storage Affect the Dominant Microbiota of Fresh and Chill-Stored Sea Bass Products. Foods 2021, 10, 671. [Google Scholar] [CrossRef]
- Cocolin, L.; Mataragas, M.; Bourdichon, F.; Doulgeraki, A.; Pilet, M.F.; Jagadeesan, B.; Rantsiou, K.; Phister, T. Next generation microbiological risk assessment meta-omics: The next need for integration. Int. J. Food Microbiol. 2018, 287, 10–17. [Google Scholar] [CrossRef]
- Anagnostopoulos, D.A.; Parlapani, F.F.; Boziaris, I.S. The evolution of knowledge on seafood spoilage microbiota from the 20th to the 21st century: Have we finished or just begun? Trends Food Sci. Technol. 2022, 120, 236–247. [Google Scholar] [CrossRef]
- AOAC Official Method 937.09 Salt (Chlorine as Sodium Chloride) in Seafood-Methods of AOAC-International-Food Laws & Regulations-Documents-Global FoodMate. Available online: http://files.foodmate.com/2013/files_2962.html (accessed on 2 February 2023).
- Dowd, S.E.; Callaway, T.R.; Wolcott, R.D.; Sun, Y.; McKeehan, T.; Hagevoort, R.G.; Edrington, T.S. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 2008, 8, 125. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Bateman, A. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Eren, A.M.; Zozaya, M.; Taylor, C.M.; Dowd, S.E.; Martin, D.H.; Ferris, M.J. Exploring the Diversity of Gardnerella vaginalis in the Genitourinary Tract Microbiota of Monogamous Couples through Subtle Nucleotide Variation. PLoS ONE 2011, 6, e26732. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and gene-centric metagenomics of the canine intestinal microbiome reveals similarities with humans and mice. ISME J. 2010, 5, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Nunes, M.L.; Batista, I.; De Campos, R.M. Physical, chemical and sensory analysis of sardine (Sardina pilchardus) stored in ice. J. Sci. Food Agric. 1992, 59, 37–43. [Google Scholar] [CrossRef]
- El Marrakchi, A.; Bennour, M.; Bouchriti, N.; Hamama, A.; Tagafait, H. Sensory, Chemical, and Microbiological Assessments of Moroccan Sardines (Sardina pilchardus) Stored in Ice. J. Food Prot. 1990, 53, 600–605. [Google Scholar] [CrossRef]
- Zhang, Y.; Hou, H.; Fan, Y.; Zhang, F.; Li, B.; Xue, C. Effect of moisture status on the stability of thermal gels from the body wall of sea cucumbers (Apostichopus japonicus). LWT 2016, 74, 294–302. [Google Scholar] [CrossRef]
- Tapía, M.S.; Alzamora, S.M.; Chirife, J. Effects of Water Activity (aw) on Microbial Stability as a Hurdle in Food Preservation. In Water Activity in Foods: Fundamentals and Applications, 2nd ed.; Wiley: Hoboken, NJ, USA, 2020; pp. 323–355. [Google Scholar] [CrossRef]
- Rahmati, T.; Labbe, R. Levels and toxigenicity of Bacillus cereus and Clostridium perfringens from retail seafood. J. Food Prot. 2008, 71, 1178–1185. [Google Scholar] [CrossRef]
- Karyantina, M.; Anggrahini, S.; Utami, T.; Rahayu, E.S. Moderate Halophilic Lactic Acid Bacteria from Jambal roti: A Traditional Fermented Fish of Central Java, Indonesia. J. Aquat. Food Prod. Technol. 2020, 29, 990–1000. [Google Scholar] [CrossRef]
- Papadopoulou, C.; Economou, E.; Zakas, G.; Salamoura, C.; Dontorou, C.; Apostolou, J. Microbiological and pathogenic contaminants of seafood in Greece. J. Food Qual. 2007, 30, 28–42. [Google Scholar] [CrossRef]
- Boziaris, I.S.; Stamatiou, A.P.; Nychas, G.J.E. Microbiological aspects and shelf life of processed seafood products. J. Sci. Food Agric. 2013, 93, 1184–1190. [Google Scholar] [CrossRef] [PubMed]
- Erkan, N.; Tosun, Ş.Y.; Alakavuk, D.Ü.; Ulusoy, Ş. Keeping quality of different packaged salted atlantic bonito “lakerda”. J. Food Biochem. 2009, 33, 728–744. [Google Scholar] [CrossRef]
- Ben Braïek, O.; Ghomrassi, H.; Cremonesi, P.; Morandi, S.; Fleury, Y.; Le Chevalier, P.; Hani, K.; Bel Hadj, O.; Ghrairi, T. Isolation and characterisation of an enterocin P-producing Enterococcus lactis strain from a fresh shrimp (Penaeus vannamei). Antonie Leeuwenhoek/Int. J. Gen. Mol. Microbiol. 2017, 110, 771–786. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.H.; Kang, S.J.; Oh, T.K. Marinomonas dokdonensis sp. nov., isolated from sea water. Int. J. Syst. Evol. Microbiol. 2005, 55, 2303–2307. [Google Scholar] [CrossRef] [PubMed]
- Jung, Y.T.; Oh, T.K.; Yoon, J.H. Marinomonas hwangdonensis sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 2012, 62, 2062–2067. [Google Scholar] [CrossRef]
- Broekaert, K.; Noseda, B.; Heyndrickx, M.; Vlaemynck, G.; Devlieghere, F. Volatile compounds associated with Psychrobacter spp. and Pseudoalteromonas spp., the dominant microbiota of brown shrimp (Crangon crangon) during aerobic storage. Int. J. Food Microbiol. 2013, 166, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Broekaert, K.; Heyndrickx, M.; Herman, L.; Devlieghere, F.; Vlaemynck, G. Molecular identification of the microbiota of peeled and unpeeled brown shrimp (Crangon crangon) during storage on ice and at 7.5 °C. Food Microbiol. 2013, 36, 123–134. [Google Scholar] [CrossRef]
- Møretrø, T.; Moen, B.; Heir, E.; Hansen, A.; Langsrud, S. Contamination of salmon fillets and processing plants with spoilage bacteria. Int. J. Food Microbiol. 2016, 237, 98–108. [Google Scholar] [CrossRef]
- Chen, H.; Wang, M.; Yang, C.; Wan, X.; Ding, H.H.; Shi, Y.; Zhao, C. Bacterial spoilage profiles in the gills of Pacific oysters (Crassostrea gigas) and Eastern oysters (C. virginica) during refrigerated storage. Food Microbiol. 2019, 82, 209–217. [Google Scholar] [CrossRef]
- Parlapani, F.F.; Michailidou, S.; Anagnostopoulos, D.A.; Koromilas, S.; Kios, K.; Pasentsis, K.; Psomopoulos, F.; Argiriou, A.; Haroutounian, S.A.; Boziaris, I.S. Bacterial communities and potential spoilage markers of whole blue crab (Callinectes sapidus) stored under commercial simulated conditions. Food Microbiol. 2019, 82, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.P.; Xie, J.; Qian, Y.F. Determination of Spoilage Microbiota of Pacific White Shrimp during Ambient and Cold Storage Using Next-Generation Sequencing and Culture-Dependent Method. J. Food Sci. 2017, 82, 1178–1183. [Google Scholar] [CrossRef] [PubMed]
- DePaola, A.; Nordstrom, J.L.; Bowers, J.C.; Wells, J.G.; Cook, D.W. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl. Environ. Microbiol. 2003, 69, 1521–1526. [Google Scholar] [CrossRef]
- Su, Y.C.; Liu, C. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007, 24, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Reed, E.; Ottesen, A. Exploring the microbiome of Callinectes sapidus (Maryland blue crab). Genome Announc. 2018, 6, e00466-18. [Google Scholar] [CrossRef] [PubMed]
- Antunes-rohling, A.; Calero, S.; Halaihel, N.; Marquina, P.; Raso, J.; Calanche, J.; Beltr, A.; Ignacio, Á.; Cebri, G. Characterization of the Spoilage Microbiota of Hake Different Temperatures. Foods 2019, 2, 489. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xie, J. Characterization of the volatiles and quality of hybrid grouper and their relationship to changes of microbial community during storage at 4 °C. Molecules 2020, 25, 818. [Google Scholar] [CrossRef]
- Zotta, T.; Parente, E.; Ianniello, R.G.; De Filippis, F.; Ricciardi, A. Dynamics of bacterial communities and interaction networks in thawed fish fillets during chilled storage in air. Int. J. Food Microbiol. 2019, 293, 102–113. [Google Scholar] [CrossRef]
- Kuuliala, L.; Al Hage, Y.; Ioannidis, A.G.; Sader, M.; Kerckhof, F.M.; Vanderroost, M.; Boon, N.; De Baets, B.; De Meulenaer, B.; Ragaert, P.; et al. Microbiological, chemical and sensory spoilage analysis of raw Atlantic cod (Gadus morhua) stored under modified atmospheres. Food Microbiol. 2018, 70, 232–244. [Google Scholar] [CrossRef]
- Jääskeläinen, E.; Jakobsen, L.M.A.; Hultman, J.; Eggers, N.; Bertram, H.C.; Björkroth, J. Metabolomics and bacterial diversity of packaged yellowfin tuna (Thunnus albacares) and salmon (Salmo salar) show fish species-specific spoilage development during chilled storage. Int. J. Food Microbiol. 2019, 293, 44–52. [Google Scholar] [CrossRef]
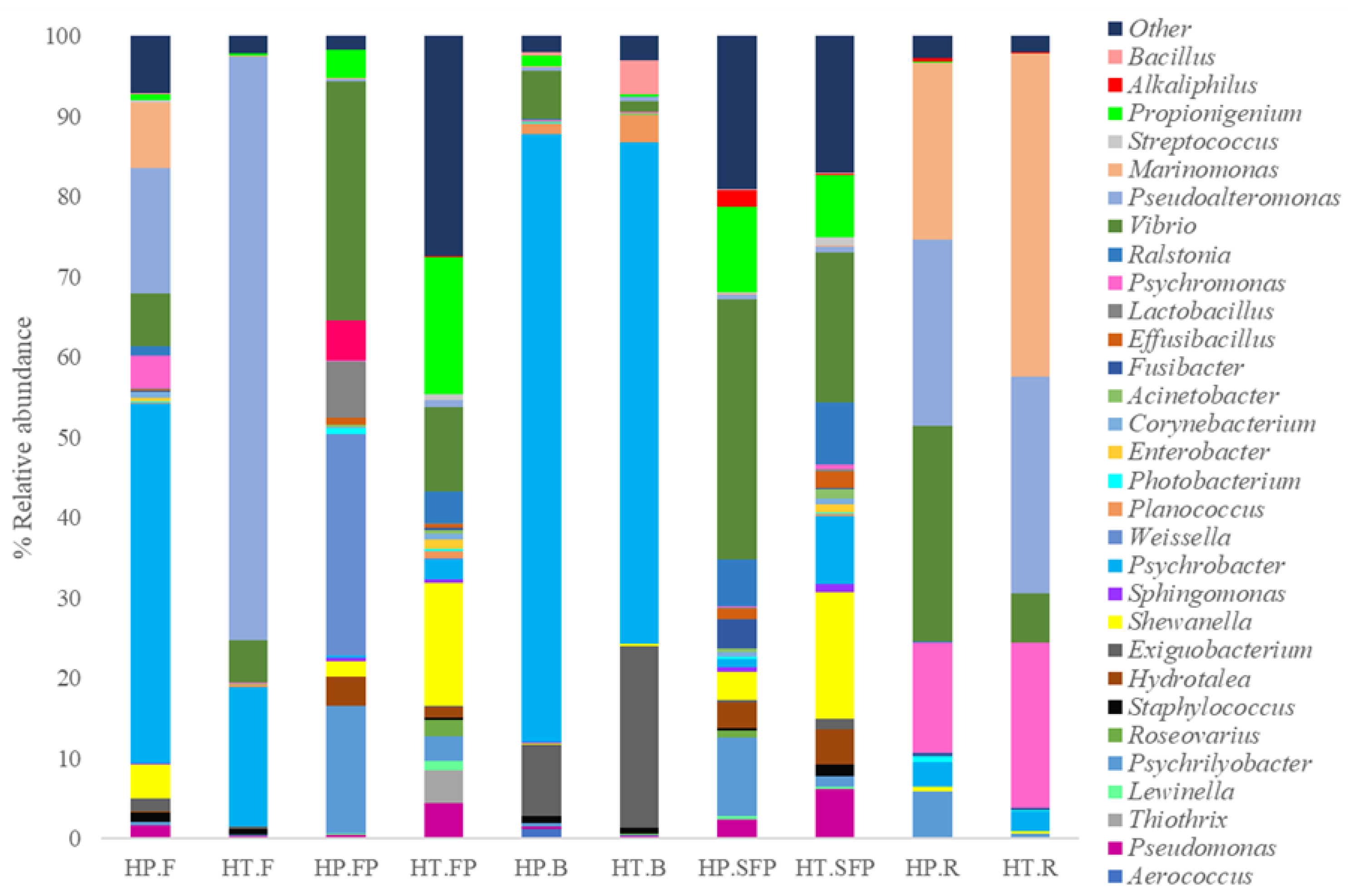
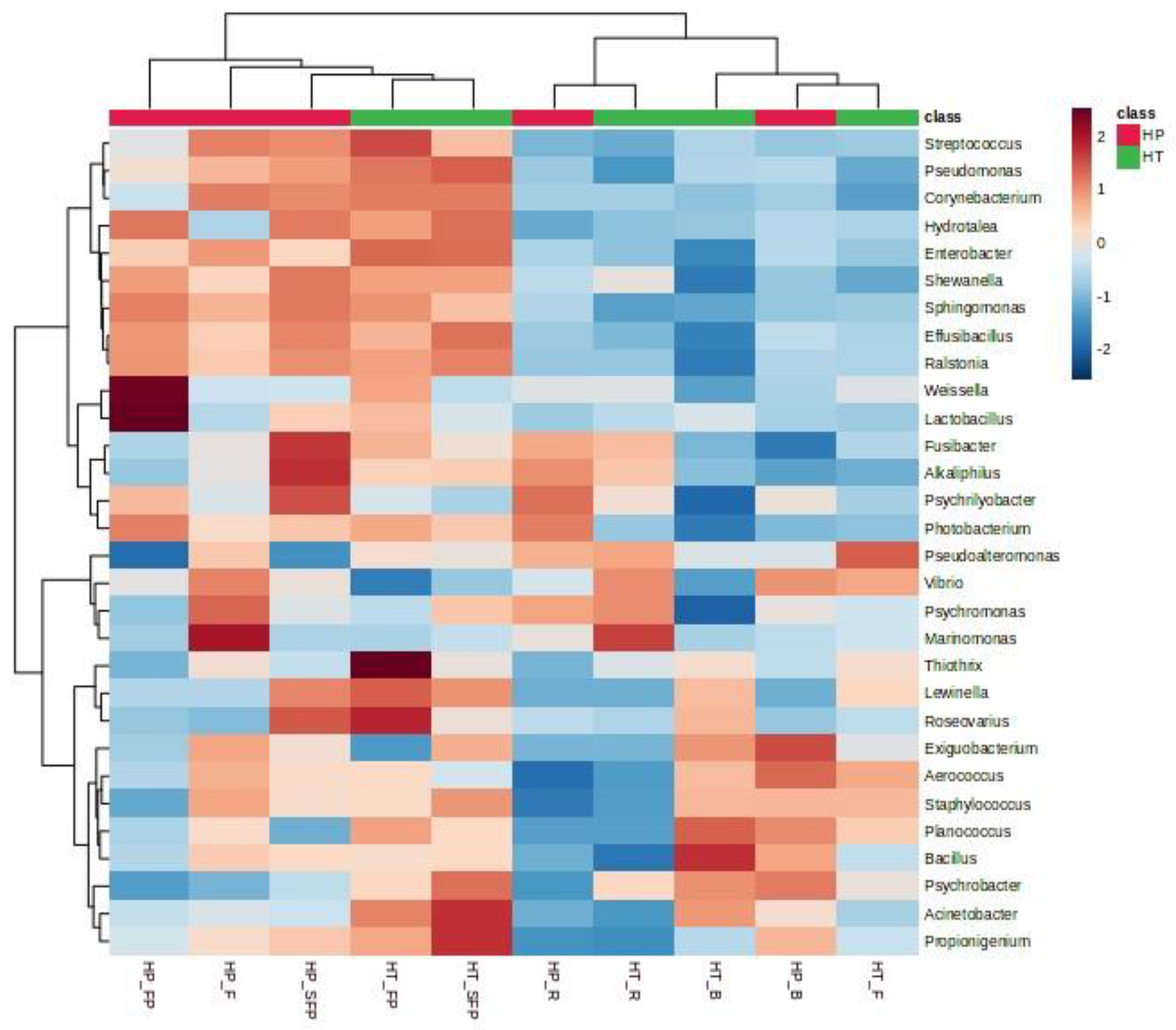
| Species | Type of Product | Product Code |
|---|---|---|
| H. polii | Raw | HP.R |
| H. polii | Boiled | HP.B |
| H. polii | Frozen | HP.F |
| H. polii | Dehydrated | HP.FP |
| H. polii | Salted | HP.SFP |
| H. tubulosa | Raw | HT.R |
| H. tubulosa | Boiled | HT.B |
| H. tubulosa | Frozen | HT.F |
| H. tubulosa | Dehydrated | HT.FP |
| H. tubulosa | Salted | HT.SFP |
| Sample Code | pH | aw | Salt (%) | Moisture (%) | WPS% (Calculated) |
|---|---|---|---|---|---|
| HP.R | 7.32 ± 0.01 b | 0.961 ± 0.002 a | 2.64 ± 0.31 b | 80.30 ± 2.22 a | 3.18 |
| HT.R | 7.38 ± 0.15 b | 0.953 ± 0.009 a | 2.75 ± 0.26 b | 83.53 ± 1.87 a | 3.19 |
| HP.B | 7.72 ± 0.09 a | 0.932 ± 0.030 b | 2.33 ± 0.06 bc | 68.36 ± 2.61 b | 3.29 |
| HT.B | 7.69 ± 0.15 a | 0.956 ± 0.018 b | 2.06 ± 0.12 bc | 69.45 ± 0.94 b | 2.88 |
| HP.F | 7.40 ± 0.14 b | 0.909 ± 0.023 c | 2.47 ± 0.14 bc | 69.82 ± 1.15 b | 3.42 |
| HT.F | 7.43 ± 0.03 b | 0.917 ± 0.015 c | 2.46 ± 0.14 bc | 68.62 ± 1.79 b | 3.46 |
| HP.FP | 7.00 ± 0.10 c | 0.847 ± 0.068 d | 3.43 ± 0.06 a | 15.35 ± 3.59 c | 18.26 |
| HT.FP | 6.99 ± 0.38 c | 0.853 ± 0.060 d | 3.22 ± 0.35 a | 13.87 ± 2.75 cd | 18.84 |
| HP.SFP | 6.87 ± 0.10 c | 0.565 ± 0.007 e | 3.43 ± 0.06 a | 11.03 ± 2.19 cd | 23.72 |
| HT.SFP | 6.15 ± 0.33 d | 0.447 ± 0.011 f | 3.47 ± 0.18 a | 9.97 ± 2.71 d | 25.81 |
| Sample | TVC 1 | LAB 2 | Yeasts | Osmophilic Yeasts | H2S Producing Bacteria | Enterobacteriaceae | Coliforms | E. coli |
|---|---|---|---|---|---|---|---|---|
| logcfu/g | ||||||||
| HP.R | 5.31 ± 0.41 b | <1 | 5.20 ± 0.10 b | <2 | 4.87 ± 0.21 a | <1 | <1 | <1 |
| HT.R | 4.35 ± 0.08 c | <1 | 4.97 ± 0.26 cd | <2 | 2.26 ± 0.24 c | <1 | <1 | <1 |
| HP.B | 4.51 ± 0.42 c | 4.51 ± 0.19 b | 5.97 ± 0.04 a | <2 | 4.41 ± 0.51 a | <1 | <1 | <1 |
| HT.B | 4.91 ± 0.21 bc | 4.69 ± 0.13 ab | 6.00 ± 0.00 a | <2 | 3.23 ± 0.53 b | <1 | <1 | <1 |
| HP.F | 3.93 ± 0.20 d | <1 | 4.36 ± 0.13 d | <2 | 3.82 ± 0.54 b | <1 | <1 | <1 |
| HT.F | 5.00 ± 0.00 bc | 4.86 ± 0.13 a | 5.00 ± 0 c | <2 | 4.95 ± 0.41 a | 1.67 ± 0.03 | <1 | <1 |
| HP.FP | 2.67 ± 0.57 e | <1 | <2 | <2 | <1 | <1 | <1 | <1 |
| HT.FP | 2 ± 0 e | <1 | <2 | <2 | <1 | <1 | <1 | <1 |
| HP.SFP | 6.23 ± 0.04 a | <1 | 5.30 ± 0.44 b | <2 | <1 | <1 | <1 | <1 |
| HT.SFP | 6.24 ± 0.02 a | 3.26 ± 0.24 c | 5.17 ± 0.33 b | <2 | <1 | <1 | <1 | <1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boziaris, I.S.; Anagnostopoulos, D.A.; Parlapani, F.F.; Syropoulou, F.; Martsikalis, P.V.; Apostologamvrou, C.; Kokioumi, D.; Vafidis, D. Microbial and Physicochemical Status of Raw and Processed Sea Cucumbers from the Hellenic Seawaters. Sustainability 2023, 15, 13467. https://doi.org/10.3390/su151813467
Boziaris IS, Anagnostopoulos DA, Parlapani FF, Syropoulou F, Martsikalis PV, Apostologamvrou C, Kokioumi D, Vafidis D. Microbial and Physicochemical Status of Raw and Processed Sea Cucumbers from the Hellenic Seawaters. Sustainability. 2023; 15(18):13467. https://doi.org/10.3390/su151813467
Chicago/Turabian StyleBoziaris, Ioannis S., Dimitrios A. Anagnostopoulos, Foteini F. Parlapani, Faidra Syropoulou, Petros V. Martsikalis, Chrysoula Apostologamvrou, Despoina Kokioumi, and Dimitris Vafidis. 2023. "Microbial and Physicochemical Status of Raw and Processed Sea Cucumbers from the Hellenic Seawaters" Sustainability 15, no. 18: 13467. https://doi.org/10.3390/su151813467
APA StyleBoziaris, I. S., Anagnostopoulos, D. A., Parlapani, F. F., Syropoulou, F., Martsikalis, P. V., Apostologamvrou, C., Kokioumi, D., & Vafidis, D. (2023). Microbial and Physicochemical Status of Raw and Processed Sea Cucumbers from the Hellenic Seawaters. Sustainability, 15(18), 13467. https://doi.org/10.3390/su151813467










