Shall the Wild Boar Pass? A Genetically Assessed Ecological Corridor in the Geneva Region
Abstract
:1. Introduction
2. Materials and Methods
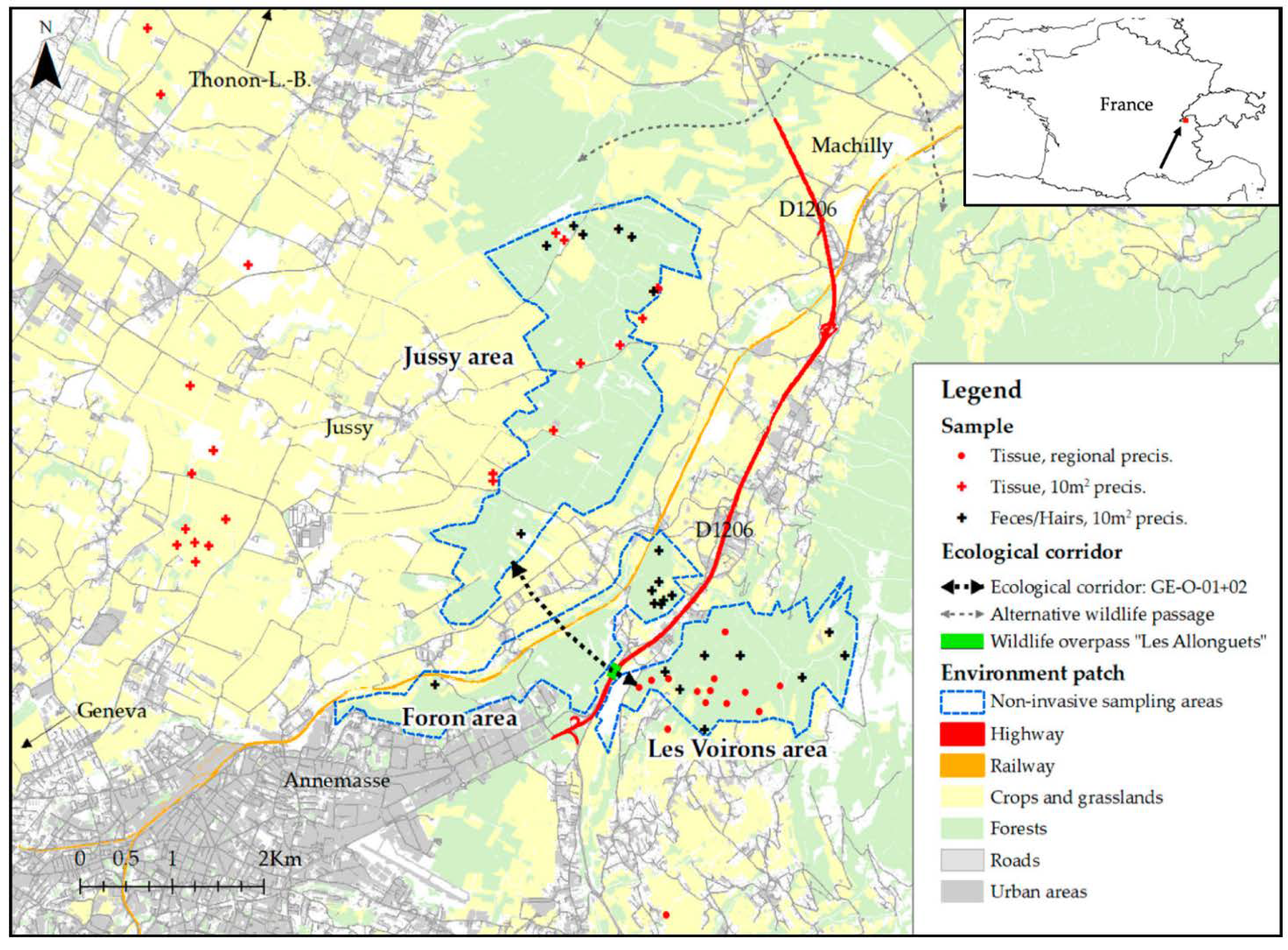
2.1. Studied Areas
2.2. Sample Collection
2.3. DNA Extraction
2.4. Amplification of Microsatellite Markers and Sex Gene
2.5. Statistical Analyses
2.5.1. Data Quality Check
2.5.2. Data Analysis
3. Results
3.1. Descriptive Statistics and General Genetic Characteristics
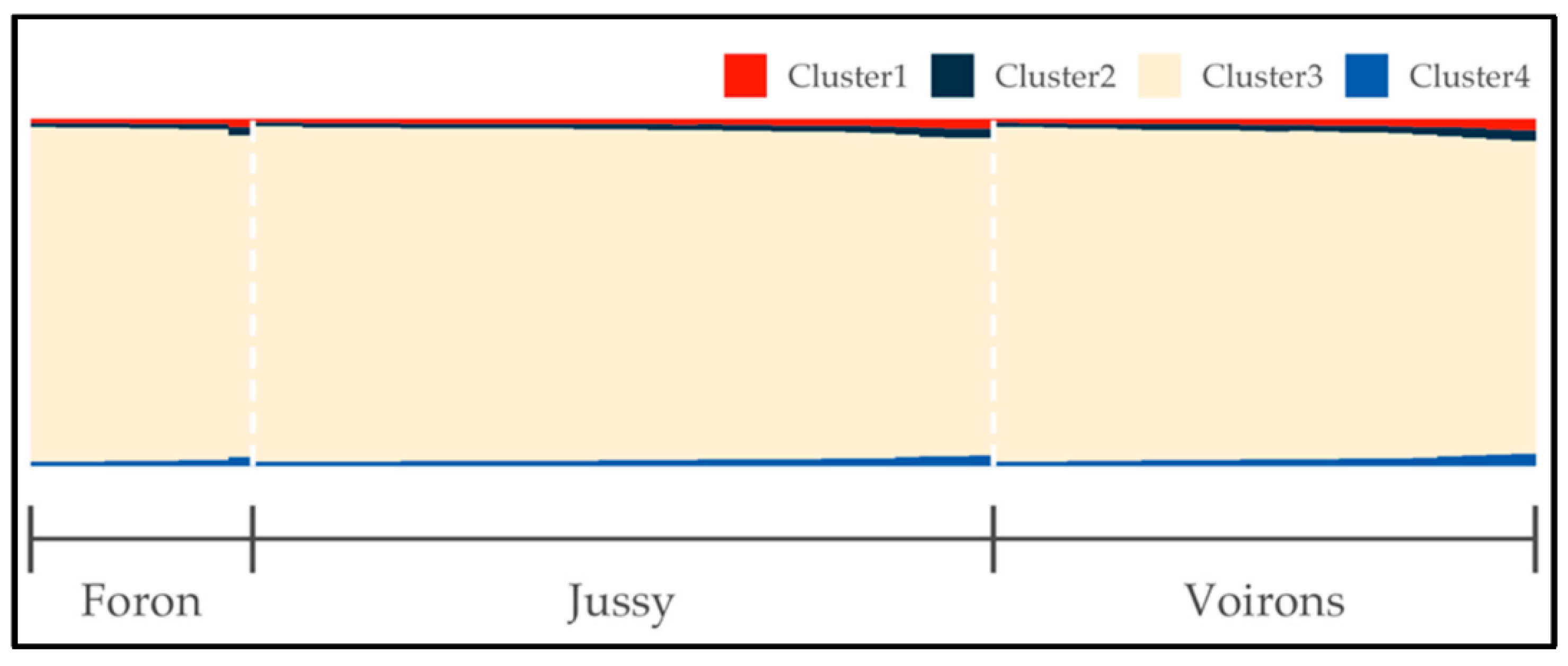
3.2. Population Genetic Structure Analysis
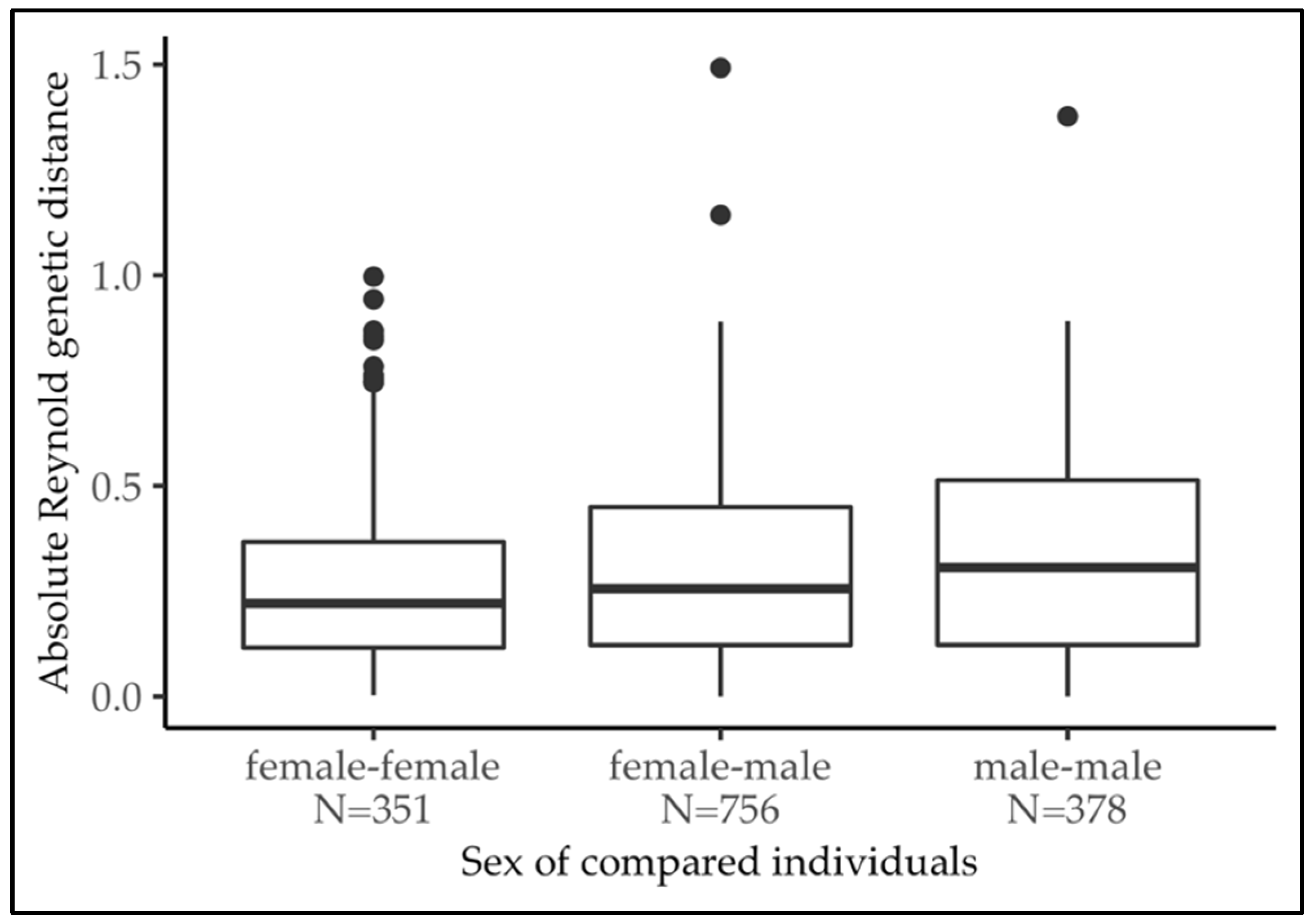
3.3. Sexual Genetic Structure Analysis
4. Discussion
4.1. Population Genetic Structure
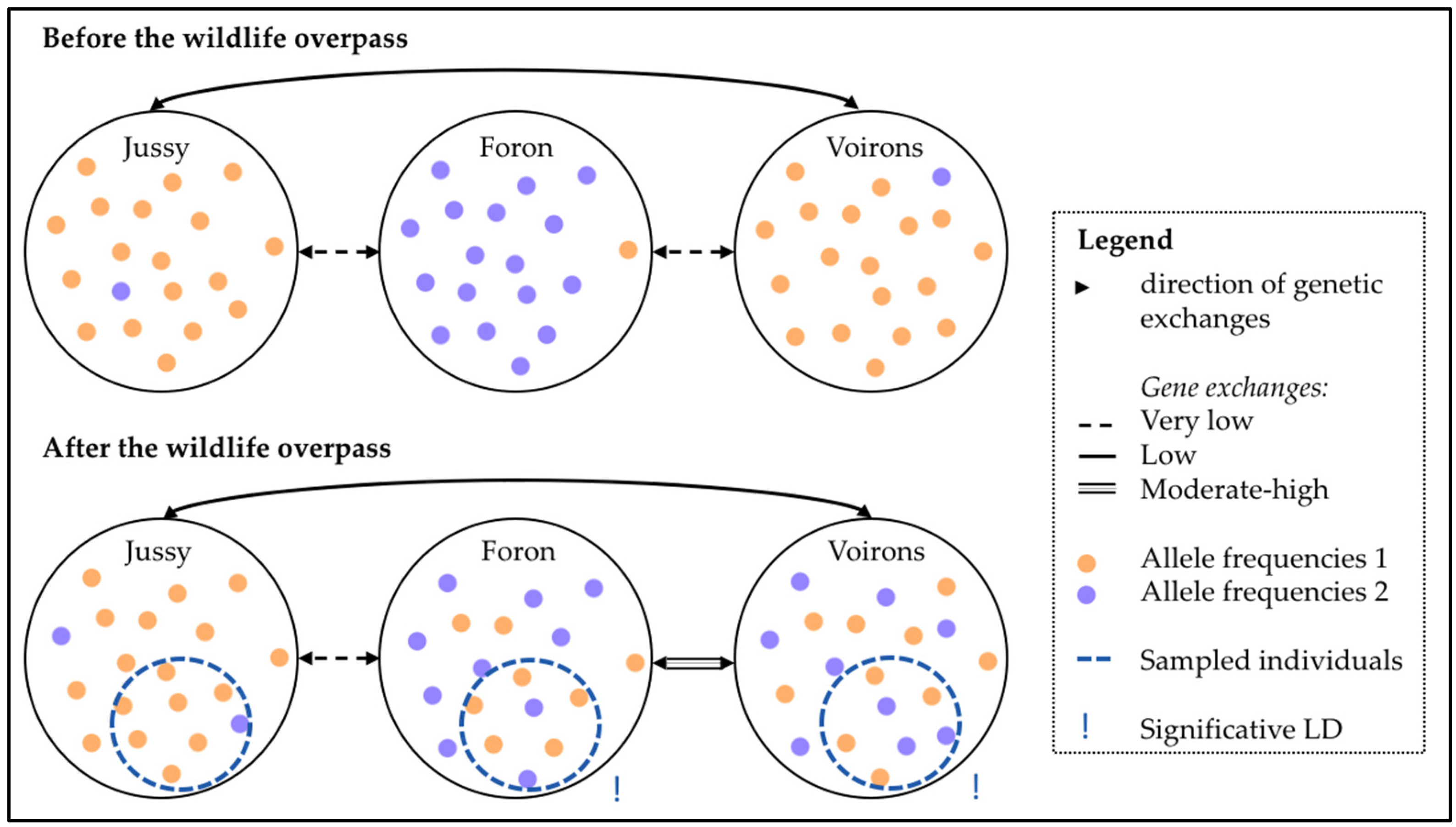
4.1.1. Population Differentiation
4.1.2. Clue of Genetic Reconnection
4.2. Sexual Genetic Structure
4.3. Perspective
4.4. Management Recommandations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- da Rocha, E.G.; Brigatti, E.; Niebuhr, B.B.; Ribeiro, M.C.; Vieira, M.V. Dispersal movement through fragmented landscapes: The role of stepping stones and perceptual range. Landsc. Ecol. 2021, 36, 3249–3267. [Google Scholar] [CrossRef]
- Coffin, A.W. From roadkill to road ecology: A review of the ecological effects of roads. J. Transp. Geogr. 2007, 15, 396–406. [Google Scholar] [CrossRef]
- Jackson, H.B.; Fahrig, L. Habitat loss and fragmentation. In Encyclopedia of Biodiversity, 2nd ed.; Elsevier Science; Levin, S.A., Ed.; Academic Press: Waltham, MA, USA, 2013; pp. 50–58. ISBN 978-0-12-384720-1. [Google Scholar]
- Harris, L.D.; Scheck, J. From implications to applications: The dispersal corridor principle applied to the conservation of biological diversity. In The Role of Corridors; Hobbs, R.J., Ed.; Surrey Beatty and Sons: Chipping Norton, Australia, 1991; Volume 2, pp. 189–220. ISBN 978-0-949324-35-1. [Google Scholar]
- Oddone Aquino, A.G.H.E.; Nkomo, S.L. Spatio-temporal patterns and consequences of road kills: A review. Animals 2021, 11, 799. [Google Scholar] [CrossRef] [PubMed]
- Corlatti, L.; Hackländer, K.; Frey-Roos, F. Ability of wildlife overpasses to provide connectivity and prevent genetic isolation. Conserv. Biol. 2009, 23, 548–556. [Google Scholar] [CrossRef]
- Frankham, R.; Ballou, J.D.; Briscoe, D.A.; McInnes, K. Section II-effects of population size reduction. In Introduction to Conservation Genetics; Cambridge University Press: Cambridge, MA, USA, 2012; pp. 225–360. ISBN 978-0-521-70271-3. [Google Scholar]
- Reed, D.H.; Frankham, R. Correlation between fitness and genetic diversity. Conserv. Biol. 2003, 17, 230–237. [Google Scholar] [CrossRef]
- Iuell, B.; Bekker, G.J.; Cuperus, R.; Dufek, J.; Fry, G.; Hicks, C.; Hlaváč, V.; Keller, V.B.; Rosell, C.; Sangwine, T.; et al. Widlife and Traffic: A European Handbook for Identifying Conflicts and Designing Solutions; European Co-operation in the Field of Scientific and Technical Research: Brussels, Belgium, 2003; ISBN 90-5011-186-6. [Google Scholar]
- Bänziger, R.; Bal, B.; Gilles, C. Etude de Base Pour L’élaboration D’un Contrat Corridors: Secteur Voirons-Jussy-Hermance; Cahier n° 13-53; Projet D’agglo Franco-Valdo-Genevois: Annemasse, France, 2010; Available online: https://www.grand-geneve.org/wp-content/uploads/cahier-13-53_contrat-corridors-secteur-voirons.pdf (accessed on 5 December 2020).
- Stratégie Biodiversité Genève 2030 (SBG-2030); Etat de Genève: Genève, Suisse, 2018; Available online: https://www.ge.ch/document/7302/telecharger (accessed on 6 December 2020).
- Montgelard, C.; Zenboudji, S.; Ferchaud, A.-L.; Arnal, V.; van Vuuren, B.J. Landscape genetics in mammals. Mammalia 2013, 78, 139–157. [Google Scholar] [CrossRef]
- Selkoe, K.A.; Toonen, R.J. Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol. Lett. 2006, 9, 615–629. [Google Scholar] [CrossRef]
- Frantzen, M.A.; Silk, J.B.; Ferguson, J.W.; Wayne, R.K.; Kohn, M.H. Empirical evaluation of preservation methods for faecal DNA. Mol. Ecol. 1998, 7, 1423–1428. [Google Scholar] [CrossRef] [Green Version]
- Haasl, R.J.; Payseur, B.A. Multi-locus inference of population structure: A comparison between single nucleotide polymorphisms and microsatellites. Heredity 2011, 106, 158–171. [Google Scholar] [CrossRef] [Green Version]
- Piggott, M.P.; Taylor, A.C. Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species. Wildl. Res. 2003, 30, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Beja-Pereira, A.; Oliveira, R.; Alves, P.C.; Schwartz, M.K.; Luikart, G. Advancing ecological understandings through technological transformations in noninvasive genetics. Mol. Ecol. Resour. 2009, 9, 1279–1301. [Google Scholar] [CrossRef] [PubMed]
- Piggott, M.P.; Taylor, A.C. Extensive evaluation of faecal preservation and DNA extraction methods in Australian native and introduced species. Aust. J. Zool. 2003, 51, 341–355. [Google Scholar] [CrossRef]
- Bonin, A.; Bellemain, E.; Eidesen, P.B.; Pompanon, F.; Brochmann, C.; Taberlet, P. How to track and assess genotyping errors in population genetics studies. Mol. Ecol. 2004, 13, 3261–3273. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.M.; Sabino-Marques, H.; Barbosa, S.; Costa, P.; Encarnação, C.; Alpizar-Jara, R.; Pita, R.; Beja, P.; Mira, A.; Searle, J.B.; et al. Genetic non-invasive sampling (gNIS) as a cost-effective tool for monitoring elusive small mammals. Eur. J. Wildl. Res. 2018, 64, 46:1–46:15. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.L.; Duarte, J.M.B. Amplifiability of mitochondrial, microsatellite and amelogenin DNA loci from fecal samples of red brocket deer Mazama Americana (Cetartiodactyla, Cervidae). Genet. Mol. Res. 2013, 12, 44–52. [Google Scholar] [CrossRef]
- Henry, P.; Henry, A.; Russello, M.A. A noninvasive hair sampling technique to obtain high quality DNA from elusive small mammals. J. Vis. Exp. 2011, ep2791, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Fattebert, J.; Baubet, E.; Slotow, R.; Fischer, C. Landscape effects on wild boar home range size under contrasting harvest regimes in a human-dominated agro-ecosystem. Eur. J. Wildl. Res. 2017, 63, 32:1–32:9. [Google Scholar] [CrossRef]
- Hebeisen, C.; Fattebert, J.; Baubet, E.; Fischer, C. Estimating wild boar (Sus scrofa) abundance and density using capture–resights in Canton of Geneva, Switzerland. Eur. J. Wildl. Res. 2008, 54, 391–401. [Google Scholar] [CrossRef]
- Gilliéron, J.; Morel, J. Atlas des Mammifères Terrestres du Bassin Genevois; Editions Faune Genève: Genève, Swizterland, 2018; ISBN 978-2-8399-2405-4. [Google Scholar]
- Casas-Díaz, E.; Closa-Sebastià, F.; Peris, A.; Miño, A.; Torrentó, J.; Casanovas, R.; Marco, I.; Lavín, S.; Fernández-Llario, P.; Serrano, E. Recorded dispersal of wild boar (Sus scrofa) in northeast Spain: Implications for disease-monitoring programs. Wildl. Biol. Pract. 2013, 9, 19–26. [Google Scholar] [CrossRef] [Green Version]
- Valente, A.M.; Acevedo, P.; Figueiredo, A.M.; Fonseca, C.; Torres, R.T. Overabundant wild ungulate populations in Europe: Management with consideration of socio-ecological consequences. Mam. Rev. 2020, 50, 353–366. [Google Scholar] [CrossRef]
- Frantz, A.C.; Bertouille, S.; Eloy, M.C.; Licoppe, A.; Chaumont, F.; Flamand, M.C. Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa). Mol. Ecol. 2012, 21, 3445–3457. [Google Scholar] [CrossRef] [PubMed]
- Griciuvienė, L.; Janeliūnas, Ž.; Jurgelevičius, V.; Paulauskas, A. The effects of habitat fragmentation on the genetic structure of wild boar (Sus scrofa) population in Lithuania. BMC Genom. Data 2021, 22, 53:1–53:8. [Google Scholar] [CrossRef] [PubMed]
- Mihalik, B.; Frank, K.; Astuti, P.K.; Szemethy, D.; Szendrei, L.; Szemethy, L.; Kusza, S.; Stéger, V. Population genetic structure of the wild boar (Sus scrofa) in the Carpathian basin. Genes 2020, 11, 1194. [Google Scholar] [CrossRef]
- Rutten, A.; Cox, K.; Scheppers, T.; Broecke, B.V.; Leirs, H.; Casaer, J. Analysing the recolonisation of a highly fragmented landscape by wild boar using a landscape genetic approach. Wildl. Biol. 2019, 1, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lecis, R.; Dondina, O.; Orioli, V.; Biosa, D.; Canu, A.; Fabbri, G.; Iacolina, L.; Cossu, A.; Bani, L.; Apollonio, M.; et al. Main roads and land cover shaped the genetic structure of a Mediterranean island wild boar population. Ecol. Evol. 2022, 12, e8804:1–e8804:13. [Google Scholar] [CrossRef] [PubMed]
- Holzgang, O.; Pfister, P.H.; Heynen, D.; Blant, M.; Righetti, A.; Berthoud, G.; Marchesi, P.; Maddalena, T.; Müri, H.; Wendelspiess, M.; et al. Les Corridors Faunistiques en Suisse; Cahier de l’environnement n° 326; Office Fédéral de l’Environnement, des Forêts et du Paysage (OFEFP): Bern, Switzerland; Société Suisse de Biologie de la Faune (SSBF): Bern, Switzerland; Station Ornithologique Suisse de Sempach: Bern, Switzerland, 2001; Available online: https://www.bafu.admin.ch/dam/bafu/fr/dokumente/biodiversitaet/uw-umwelt-wissen/korridore_fuer_wildtiereinderschweizgrundlagenzurueberregionalen.pdf.download.pdf/les_corridors_faunistiquesensuissebasespourlamiseenreseausuprare.pdf (accessed on 12 December 2020).
- Comité de Pilotage du Contrat Corridors du Secteur Arve-Lac. Contrat Corridors du Secteur Arve-Lac; Cahier n° 13-61; Grand Genève: Geneva, Suisse, 2012; Available online: https://www.grand-geneve.org/wp-content/uploads/cahier-13-61_contrat-corridor_arve-lac_nov2012.pdf (accessed on 5 December 2020).
- Boschung, O. Gestion des Écoponts du Bassin Genevois. Bachelor’s Thesis, HEPIA, Haute Ecole du Paysage, D’ingénierie et D’architecture de Genève, Jussy-Geneva, Switzerland, 2020. [Google Scholar]
- Berthoud, G.; Lebeau, R.P.; Righetti, A. Réseau Ecologique National REN, Rapport Final; Cahier de l’environnement n° 373; Office fédéral de L’environnement, des Forêts et du Paysage: Bern, Switzerland, 2004; Available online: https://www.bafu.admin.ch/dam/bafu/fr/dokumente/biodiversitaet/uw-umwelt-wissen/nationales_oekologischesnetzwerkrenschlussbericht.pdf.download.pdf/reseau_ecologiquenationalrenrapportfinal.pdf (accessed on 10 December 2020).
- Comité Régional Franco-Genevois. Synthèse 2019, Ain-Haute-Savoie-Genève-Vaud; Observatoire Statistique Transfrontalier: Carouge, Suisse, 2019; Available online: https://www.ge.ch/statistique/tel/publications/2019/hors_collection/ost/Synthese_2019.pdf (accessed on 4 January 2022).
- Les services de l’Etat en Haute-Savoie. Available online: https://www.haute-savoie.gouv.fr/Politiques-publiques/Votre-departement/Deplacements/Circulation-routiere/Carte-interactive-du-trafic (accessed on 10 December 2021).
- DREAL Auvergne-Rhône-Alpes. Available online: http://www.auvergne-rhone-alpes.developpement-durable.gouv.fr/presentation-de-la-section-chasseurs-machilly-a10184.html (accessed on 24 December 2020).
- Evesque, F.; Macian, F.; Menu, C. Inauguration de la Mise à 2x2 Voies de la RD 1206 Entre le Carrefour des Chasseurs et Machilly; Préfet de la région Rhône-Alpes: Clermont-Ferrand, France, 2014; Available online: http://www.auvergne-rhone-alpes.developpement-durable.gouv.fr/IMG/pdf/20141120-DP-InaugurationChasseursMachilly-V5_cle5e44a1.pdf (accessed on 10 January 2022).
- Hunting Federation of Haute-Savoie. Suivi du Monitoring des Corridors Ecologiques de Machilly à Gaillard; Route du Pont-Rouge: Cranves-Sales, France, 2021. [Google Scholar]
- Ripoll, J.; Bon, M.-C.; Jones, W. Optimalisation de l’extraction d’ADN génomique de la morelle jaune (Solanum elaeagnifolium Cav.), une plante invasive des milieux cultivés en région méditerranéenne. Biotechnol. Agron. Soc. Environ. 2011, 15, 95–100. [Google Scholar]
- Jeffery, K.J.; Abernethy, K.A.; Tutin, C.E.G.; Bruford, M.W. Biological and environmental degradation of gorilla hair and microsatellite amplification success. Biol J. Linn. Soc. Lond. 2007, 91, 281–294. [Google Scholar] [CrossRef]
- Wehausen, J.D.; Ramey, R.R.; Epps, C.W. Experiments in DNA extraction and PCR amplification from bighorn sheep feces: The importance of DNA extraction method. J. Hered. 2004, 95, 503–509. [Google Scholar] [CrossRef] [Green Version]
- Conyers, C.M.; Allnutt, T.R.; Hird, H.J.; Kaye, J.; Chisholm, J. Development of a microsatellite-based method for the differentiation of european wild boar (Sus scrofa scrofa) from domestic pig breeds (Sus scrofa domestica) in food. J. Agric. Food Chem. 2012, 60, 3341–3347. [Google Scholar] [CrossRef]
- ISAG/FAO Standing Committee. Secondary Guidelines for Development of National Farm Animal Genetic Resources Management Plans, Measurement of Domestic Animal Diversity (MoDAD): Recommended Microsatellite Markers; Food and Agriculture Organization of the United Nations: Rome, Italy, 2004; Available online: https://www.fao.org/3/aq569e/aq569e.pdf (accessed on 23 February 2020).
- Robic, A.; Dalens, M.; Woloszyn, N.; Milan, D.; Riquet, J.; Gellin, J. Isolation of 28 new porcine microsatellites revealing polymorphism. Mamm. Genome 1994, 5, 580–583. [Google Scholar] [CrossRef]
- Fontanesi, L.; Scotti, E.; Russo, V. Differences of the porcine amelogenin X and Y chromosome genes (AMELX and AMELY) and their application for sex determination in pigs. Mol. Reprod. Dev. 2008, 75, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.I.; Amos, W. Microsatellite genotyping errors: Detection approaches, common sources and consequences for paternal exclusion. Mol. Ecol. 2005, 14, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, R.; Zhong, Y.; Jin, L.; Budowle, B. Nondetectability of restriction fragments and independence of DNA fragment sizes within and between loci in RFLP typing of DNA. Am. J. Hum. Genet. 1994, 55, 391–401. [Google Scholar] [PubMed]
- Jombart, T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef] [Green Version]
- Adamack, A.T.; Gruber, B. PopGenReport: Simplifying basic population genetic analyses in R. Methods Ecol. Evol. 2014, 5, 384–387. [Google Scholar] [CrossRef]
- Cutter, A.D. Recombination and linkage disequilibrium in evolutionary signature. In A Primer of Molecular Population Genetics; CPI Group (UK) Ltd.: Oxford, UK, 2019; pp. 113–128. ISBN 978-0-19-883894-4. [Google Scholar]
- Foll, M.; Gaggiotti, O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A bayesian perspective. Genetics 2008, 180, 977–993. [Google Scholar] [CrossRef] [Green Version]
- El Mousadik, A.; Petit, R.J. High level of genetic differentiation for allelic richness among populations of the argan tree [Argania spinosa (L.) Skeels] endemic to Morocco. Theor. Appl. Genet. 1996, 92, 832–839. [Google Scholar] [CrossRef]
- Goudet, J. Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 2005, 5, 184–186. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987; ISBN 978-0-231-88671-0. [Google Scholar]
- Weir, B.S.; Goudet, J. A unified characterization of population structure and relatedness. Genetics 2017, 206, 2085–2103. [Google Scholar] [CrossRef] [Green Version]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Lischer, H.E.L.; Excoffier, L. PGDSpider: An automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 2012, 28, 298–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pritchard, J.K.; Wen, X.; Falush, D. Documentation for Structure Software: Version 2.3; Department of Human Genetics, University of Chicago: Chicago, IL, USA; Department of Statistics, University of Oxford: Oxford, UK, 2009; Available online: https://burfordreiskind.com/wp-content/uploads/Structure_Manual_doc.pdf (accessed on 2 May 2021).
- Francis, R. pophelper: An R package and web app to analyse and visualise population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Guillot, G.; Mortier, F.; Estoup, A. GENELAND: A computer package for landscape genetics. Mol. Ecol. Notes 2005, 5, 712–715. [Google Scholar] [CrossRef]
- ESRI. ArcGis Desktop: Release 10; Environmental Systems Research Institute: Redlands, CA, USA, 2011. [Google Scholar]
- Legendre, P.; Legendre, L. Chapter 13-spatial analysis. In Developments in Environmental Modelling; Legendre, P., Legendre, L., Eds.; Elsevier: Amsterdam, The Netherlands, 2012; Volume 24, pp. 785–858. ISBN 978-0-4445-3868-0. [Google Scholar]
- Reynolds, J.; Weir, B.S.; Cockerham, C.C. Estimation of the coancestry coefficient: Basis for a short-term genetic distance. Genetics 1983, 105, 767–779. [Google Scholar] [CrossRef]
- Smith, A.B. EnmSdm: Tools for Modeling Niches and Distributions of Species, R Package Version 0.3.1.0. 2018. Available online: https://rdrr.io/github/adamlilith/enmSdm/man/enmSdm.html (accessed on 25 January 2022).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5-7. 2020. Available online: https://github.com/vegandevs/vegan (accessed on 2 March 2021).
- Jones, O.R.; Wang, J. COLONY: A program for parentage and sibship inference from multilocus genotype data. Mol. Ecol. Resour. 2010, 10, 551–555. [Google Scholar] [CrossRef]
- Wang, J.; Santure, A.W. Parentage and sibship inference from multilocus genotype data under polygamy. Genetics 2009, 181, 1579–1594. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Bartlett, M.S.; Fowler, R.H. Properties of sufficiency and statistical tests. Proc. R. Soc. Lond. A Math. Phys. Sci. 1937, 160, 268–282. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/ (accessed on 25 February 2022).
- Dunn, O.J. Multiple comparisons using rank sums. Technometrics 1964, 6, 241–252. [Google Scholar] [CrossRef]
- Ogle, D.H.; Doll, J.C.; Wheeler, P.; Dinno, A. FSA: Fisheries Stock Analysis, R Package Version 0.9.3. 2022. Available online: https://github.com/fishR-Core-Team/FSA (accessed on 15 January 2022).
- Mihalik, B.; Stéger, V.; Frank, K.; Szendrei, L.; Kusza, S. Barrier effect of the M3 highway in Hungary on the genetic diversity of wild boar (Sus scrofa) population. Res. J. Biotechnol. 2018, 13, 32–38. [Google Scholar]
- Dellicour, S.; Prunier, J.G.; Piry, S.; Eloy, M.-C.; Bertouille, S.; Licoppe, A.; Frantz, A.C.; Flamand, M.-C. Landscape genetic analyses of Cervus elaphus and Sus scrofa: Comparative study and analytical developments. Heredity 2019, 123, 228–241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Truvé, J.; Lemel, J. Timing and distance of natal dispersal for wild boar Sus scrofa in Sweden. Wildl. Biol. 2003, 9, 51–57. [Google Scholar] [CrossRef]
- Keuling, O.; Lauterbach, K.; Stier, N.; Roth, M. Hunter feedback of individually marked wild boar Sus scrofa L.: Dispersal and efficiency of hunting in northeastern Germany. Eur. J. Wildl. Res. 2010, 56, 159–167. [Google Scholar] [CrossRef]
- Pinto, N.; Keitt, T.H. Beyond the least-cost path: Evaluating corridor redundancy using a graph-theoretic approach. Landsc. Ecol. 2009, 24, 253–266. [Google Scholar] [CrossRef]
- Coulon, A.; Guillot, G.; Cosson, J.-F.; Angibault, J.M.A.; Aulagnier, S.; Cargnelutti, B.; Galan, M.; Hewison, A.J.M. Genetic structure is influenced by landscape features: Empirical evidence from a roe deer population. Mol. Ecol. 2006, 15, 1669–1679. [Google Scholar] [CrossRef]

| Marker Name | Marker Type | Forward Primer | Reverse Primer | Ref. | Pooling Group-Label |
|---|---|---|---|---|---|
| SW24 | STR, autosomal chr. | CTTTGGGTGGAGTGTGTGC | ATCCAAATGCTGCAAGCG | [45] | P3-FAM |
| SW122 | STR, autosomal chr. | TTGTCTTTTTATTTTGCTTTTGG | CAAAAAAGGCAAAAGATTGACA | [45] | P2-FAM |
| SW632 | STR, autosomal chr. | TGGGTTGAAAGATTTCCCAA | GGAGTCAGTACTTTGGCTTGA | [45] | P2-HEX |
| SW857 | STR, autosomal chr. | TGAGAGGTCAGTTACAGAAGACC | GATCCTCCTCCAAATCCCAT | [45] | P3-HEX |
| SW911 | STR, autosomal chr. | CTCAGTTCTTTGGGACTGAACC | CATCTGTGGAAAAAAAAAGCC | [45] | P1-ROX |
| SW936 | STR, autosomal chr. | TCTGGAGCTAGCATAAGTGCC | GTGCAAGTACACATGCAGGG | [45] | P1-FAM |
| S0005 | STR, autosomal chr. | TCCTTCCCTCCTGGTAACTA | GCACTTCCTGATTCTGGGTA | [46] | P3-ROX |
| S0097 | STR, autosomal chr. | GACCTATCTAATGTCATTATAGT | TTCCTCCTAGAGTTGACAAACTT | [45] | P2-ROX |
| S0226 | STR, autosomal chr. | GGTTAAACTTTTNCCCCAATACA | GCACTTTTAACTTTCATGATACTCC | [47] | P1-HEX |
| SRYB | sexual gene, sex chr. | TGAACGCTTTCATTGTGTGGTC | GCCAGTAGTCTCTGTGCCTCCT | [48] | - |
| Sample Type | # Jussy | # Les Voirons | # Foron |
|---|---|---|---|
| triplicate | 15 | 0 | 0 |
| tissue | 8 | 14 | 0 |
| feces | 6 | 5 | 0 |
| hair | 1 | 3 | 9 |
| total | 30 | 22 | 9 |
| Locus Name | Ho | Hs | Fis | # Est. Alleles | Length Range |
|---|---|---|---|---|---|
| SW24 | 0.636 | 0.685 | 0.072 | 5.967 | 115–127 |
| SW122 | 0.761 | 0.664 | −0.146 | 4.998 | 97–115 |
| SW632 | 0.842 | 0.807 | −0.044 | 7.902 | 126–160 |
| SW857 | 0.635 | 0.681 | 0.069 | 4.998 | 160–168 |
| SW911 | 0.458 | 0.514 | 0.109 | 3.902 | 160–166 |
| SW936 | 0.537 | 0.486 | −0.104 | 4.902 | 91–107 |
| S0005 | 0.808 | 0.756 | −0.069 | 8.000 | 218–242 |
| S0097 | 0.849 | 0.839 | −0.012 | 15.867 | 219–264 |
| S0226 | 0.743 | 0.687 | −0.081 | 4.000 | 177–187 |
| Locus Name | Allele Length | Freq. Jussy | Freq. Les Voirons | Freq. Foron |
|---|---|---|---|---|
| SW936 | 91 | 0.08 | 0.16 | 0.22 |
| 93 | 0.82 | 0.68 | 0.56 | |
| 101 | 0.06 | |||
| 105 | 0.02 | |||
| 107 | 0.10 | 0.14 | 0.17 | |
| S0226 | 177 | 0.17 | 0.16 | 0.11 |
| 179 | 0.33 | 0.32 | 0.44 | |
| 185 | 0.41 | 0.34 | 0.44 | |
| 187 | 0.09 | 0.18 | ||
| SW911 | 160 | 0.02 | ||
| 162 | 0.65 | 0.52 | 0.44 | |
| 164 | 0.02 | |||
| 166 | 0.35 | 0.43 | 0.56 | |
| SW24 | 115 | 0.03 | 0.02 | |
| 117 | 0.15 | 0.27 | 0.50 | |
| 119 | 0.10 | 0.18 | 0.13 | |
| 123 | 0.58 | 0.41 | 0.13 | |
| 125 | 0.13 | 0.09 | 0.25 | |
| 127 | 0.02 | |||
| SW857 | 160 | 0.02 | 0.02 | |
| 162 | 0.15 | 0.07 | ||
| 164 | 0.10 | 0.27 | 0.28 | |
| 166 | 0.23 | 0.14 | 0.33 | |
| 168 | 0.50 | 0.50 | 0.39 | |
| S0005 | 218 | 0.03 | ||
| 222 | 0.41 | 0.29 | 0.31 | |
| 232 | 0.21 | 0.31 | 0.38 | |
| 234 | 0.02 | |||
| 236 | 0.03 | |||
| 238 | 0.16 | 0.26 | 0.19 | |
| 240 | 0.10 | |||
| 242 | 0.05 | 0.12 | 0.13 | |
| SW122 | 97 | 0.28 | 0.23 | 0.33 |
| 99 | 0.40 | 0.52 | 0.50 | |
| 109 | 0.28 | 0.07 | 0.17 | |
| 111 | 0.03 | 0.14 | ||
| 115 | 0.05 | |||
| SW632 | 126 | 0.18 | 0.05 | 0.06 |
| 148 | 0.10 | 0.09 | 0.11 | |
| 150 | 0.08 | 0.07 | 0.06 | |
| 152 | 0.20 | 0.41 | 0.22 | |
| 154 | 0.18 | 0.23 | 0.28 | |
| 156 | 0.02 | |||
| 158 | 0.22 | 0.16 | 0.28 | |
| 160 | 0.02 | |||
| S0097 | 219 | 0.12 | 0.07 | |
| 224 | 0.10 | 0.25 | 0.38 | |
| 230 | 0.05 | 0.02 | ||
| 232 | 0.13 | 0.02 | ||
| 234 | 0.17 | 0.11 | 0.25 | |
| 236 | 0.23 | 0.23 | 0.25 | |
| 240 | 0.02 | |||
| 242 | 0.05 | 0.05 | ||
| 244 | 0.02 | 0.05 | ||
| 250 | 0.02 | 0.05 | ||
| 252 | 0.08 | |||
| 254 | 0.02 | 0.13 | ||
| 256 | 0.02 | |||
| 260 | 0.02 | |||
| 262 | 0.02 | |||
| 264 | 0.09 |
| Areas | Ho | Hs | Fst | Fis * | # Priv. All. | # Allel. Richness. |
|---|---|---|---|---|---|---|
| Jussy | 0.672 | 0.660 | 0.063 | −0.018 | 8 | 39.457 |
| Foron | 0.773 | 0.682 | −0.017 | −0.114 | 1 | 32.654 |
| Les Voirons | 0.645 | 0.695 | 0.007 | 0.075 | 9 | 40.460 |
| overall | 0.697 | 0.680 | 0.017 | −0.019 | - | - |
| Locus Name | SW936 | S0226 | SW911 | SW24 | SW857 | S0005 | SW122 | SW632 | S0097 |
|---|---|---|---|---|---|---|---|---|---|
| SW936 | - | - | - | - | - | - | - | - | |
| S0226 | - | + | + | - | - | - | - | + | |
| SW911 | - | - | - | - | - | - | - | + | |
| SW24 | - | - | - | - | - | - | - | + | |
| SW857 | - | - | + | - | + | - | - | + | |
| S0005 | - | - | - | - | - | - | + | + | |
| SW122 | - | - | - | + | - | - | + | + | |
| SW632 | - | + | + | - | + | + | - | - | |
| S0097 | - | - | - | + | + | - | + | - |
| Model | Dataset | N | Freqscoor | Best K (ΔK) | Prob. K |
|---|---|---|---|---|---|
| admixture | with kin | 61 | on | 4 | 1 |
| off | 4 | 1 | |||
| without kin | 50 | on | 2 | 1 | |
| off | 2 | 1 | |||
| prior information (region) | with kin | 61 | on | 5 | 1 |
| off | 2 | 1 | |||
| without kin | 50 | on | 2 | 1 | |
| off | 2 | 1 |
| f. s. Family ID | Prob (Inc.) | Prob (Exc.) | # Memb. Jussy | # Memb. Voirons | # Memb. Foron |
|---|---|---|---|---|---|
| 1 | 0.960 | 0.570 | 2 | 0 | 0 |
| 2 | 0.972 | 0.610 | 0 | 2 | 0 |
| 3 | 0.819 | 0.431 | 0 | 0 | 3 |
| 4 | 0.913 | 0.371 | 0 | 2 | 0 |
| 5 | 0.940 | 0.940 | 2 | 0 | 2 |
| 6 | 0.992 | 0.183 | 0 | 2 | 0 |
| 7 | 0.868 | 0.222 | 2 | 0 | 0 |
| 8 | 0.998 | 0.641 | 0 | 2 | 0 |
| Parentage Cluster | Prob | # Memb. Jussy | # Memb. Voirons | # Memb. Foron |
|---|---|---|---|---|
| 1 | 0.427 | 7 | 5 | 3 |
| 2 | 0.211 | 13 | 10 | 6 |
| 3 | 0.519 | 9 | 4 | 0 |
| 4 | 0.279 | 1 | 2 | 0 |
| 5 | 0.728 | 0 | 1 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kupferschmid, F.A.L.; Crovadore, J.; Fischer, C.; Lefort, F. Shall the Wild Boar Pass? A Genetically Assessed Ecological Corridor in the Geneva Region. Sustainability 2022, 14, 7463. https://doi.org/10.3390/su14127463
Kupferschmid FAL, Crovadore J, Fischer C, Lefort F. Shall the Wild Boar Pass? A Genetically Assessed Ecological Corridor in the Geneva Region. Sustainability. 2022; 14(12):7463. https://doi.org/10.3390/su14127463
Chicago/Turabian StyleKupferschmid, Fanny Alexandra Laura, Julien Crovadore, Claude Fischer, and François Lefort. 2022. "Shall the Wild Boar Pass? A Genetically Assessed Ecological Corridor in the Geneva Region" Sustainability 14, no. 12: 7463. https://doi.org/10.3390/su14127463
APA StyleKupferschmid, F. A. L., Crovadore, J., Fischer, C., & Lefort, F. (2022). Shall the Wild Boar Pass? A Genetically Assessed Ecological Corridor in the Geneva Region. Sustainability, 14(12), 7463. https://doi.org/10.3390/su14127463






