Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting
Abstract
:1. Introduction
2. Results and Discussion
2.1. 1H-NMR Fingerprinting and Metabolite Identification
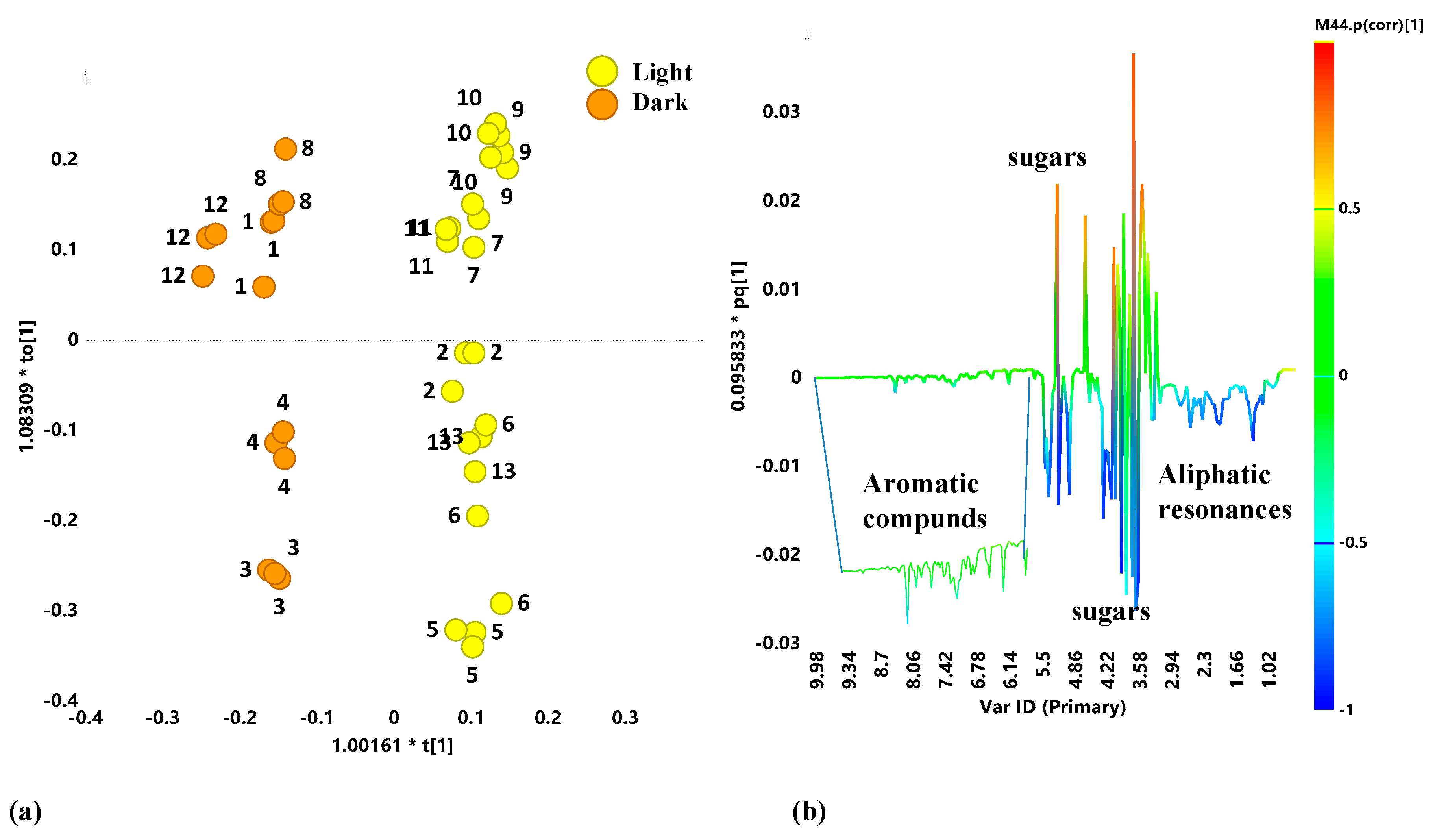
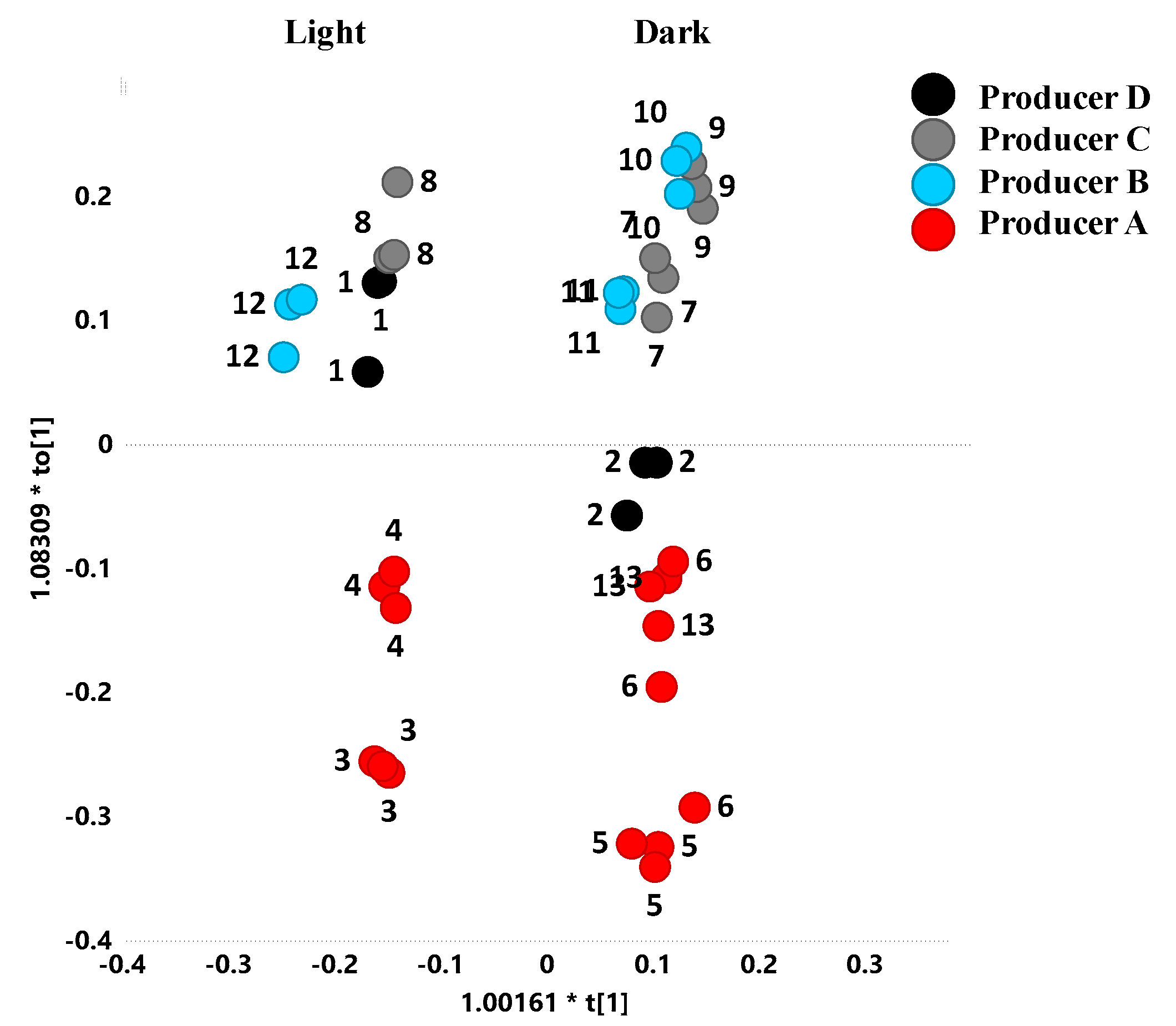
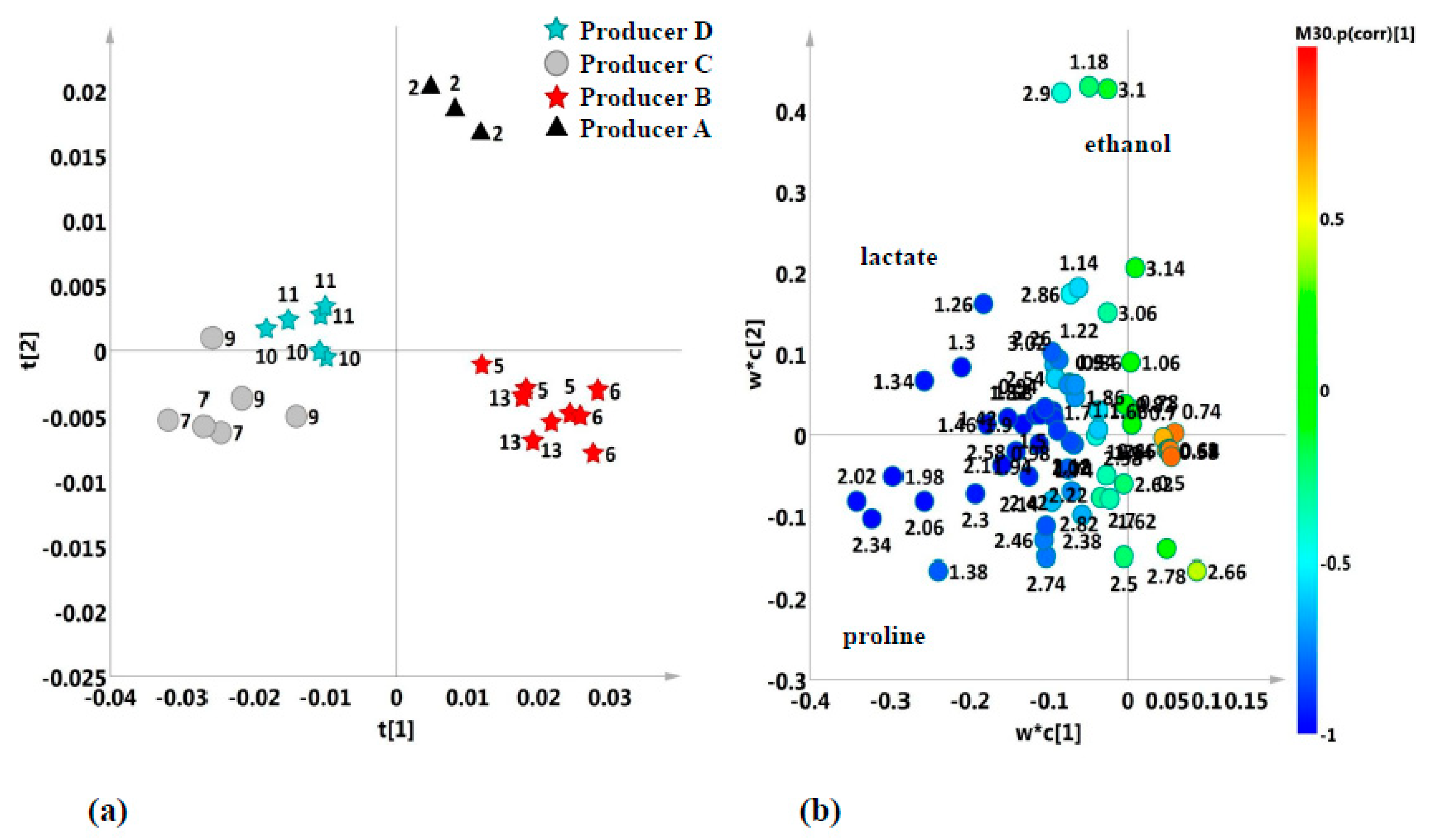
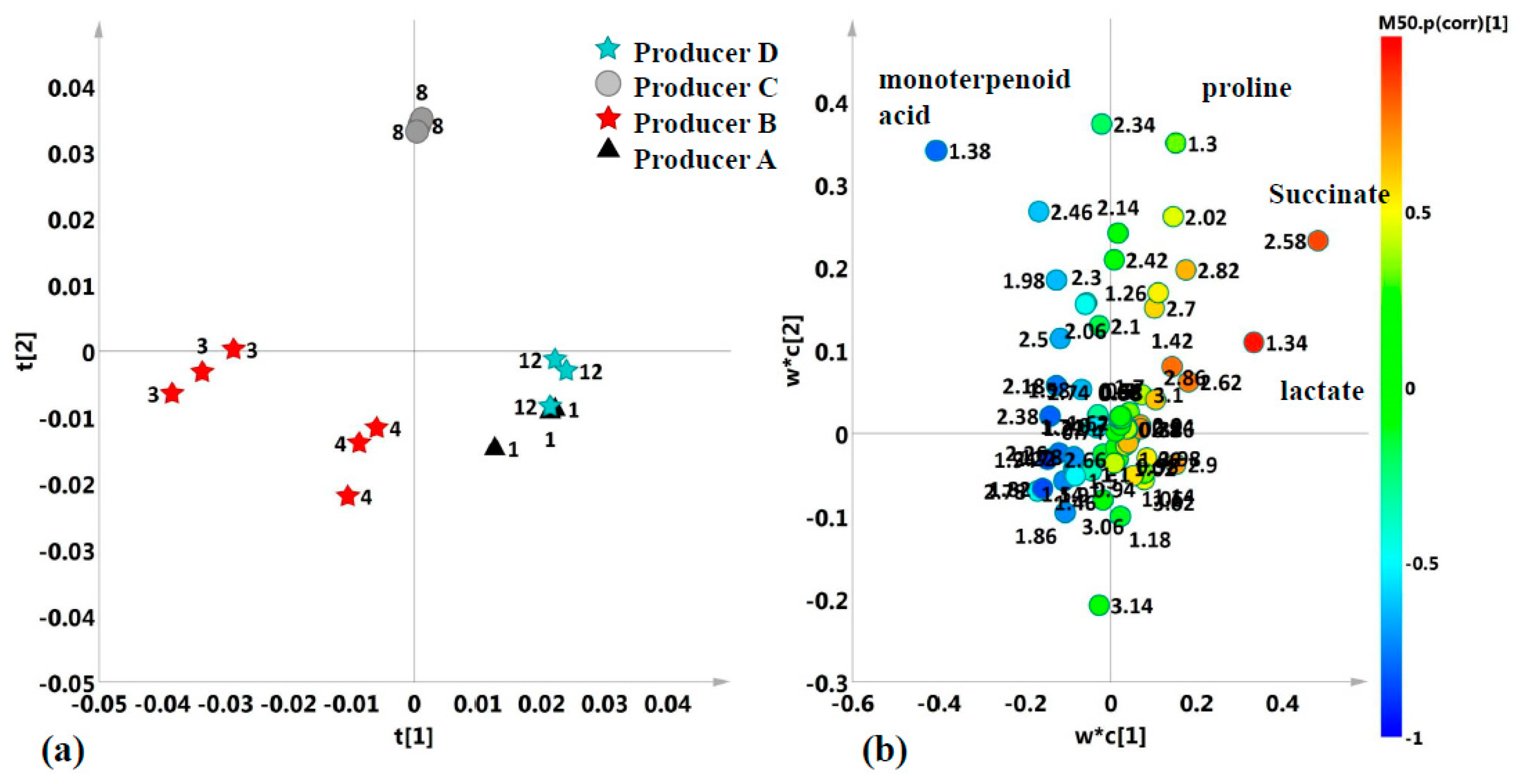
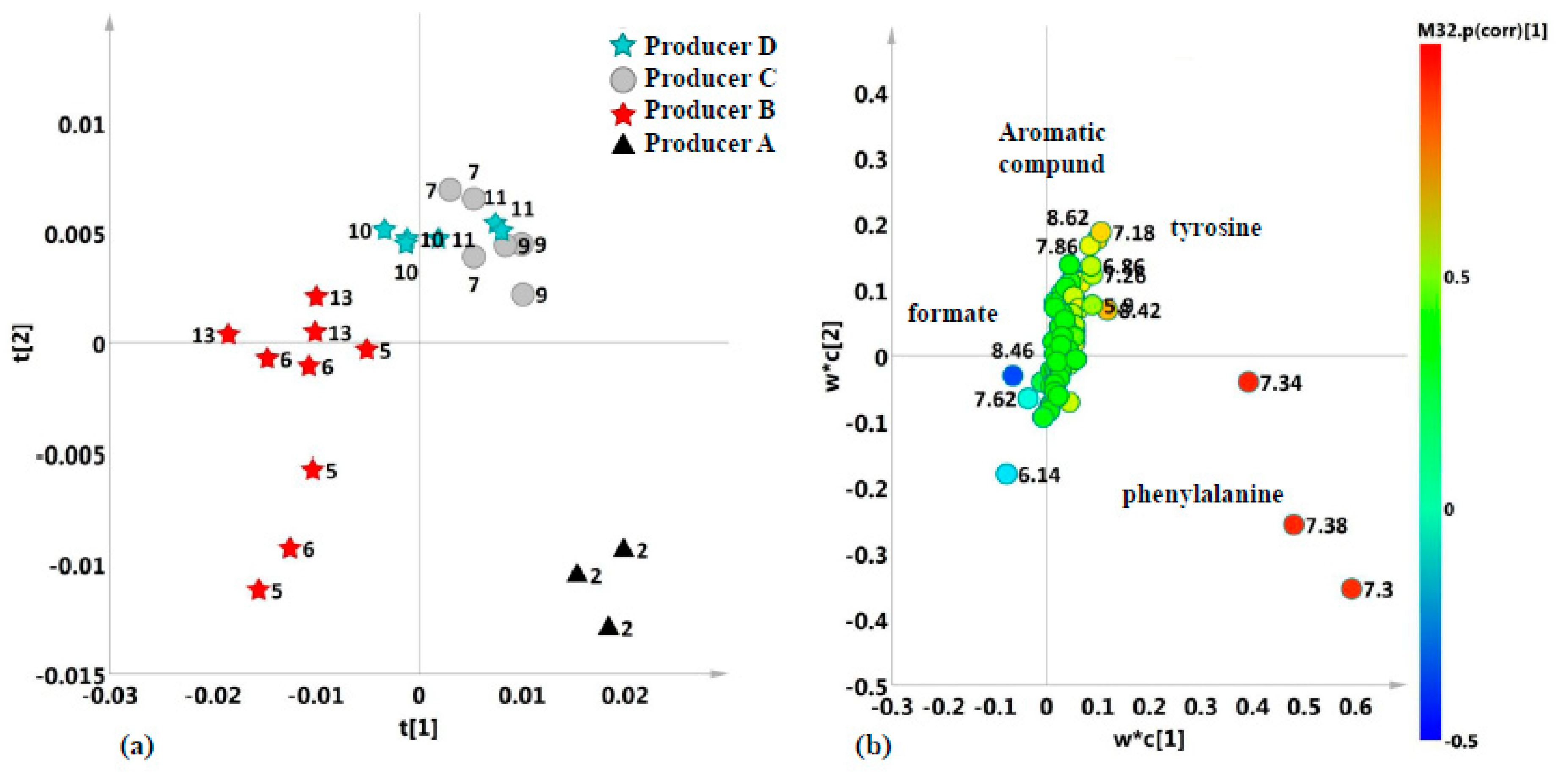
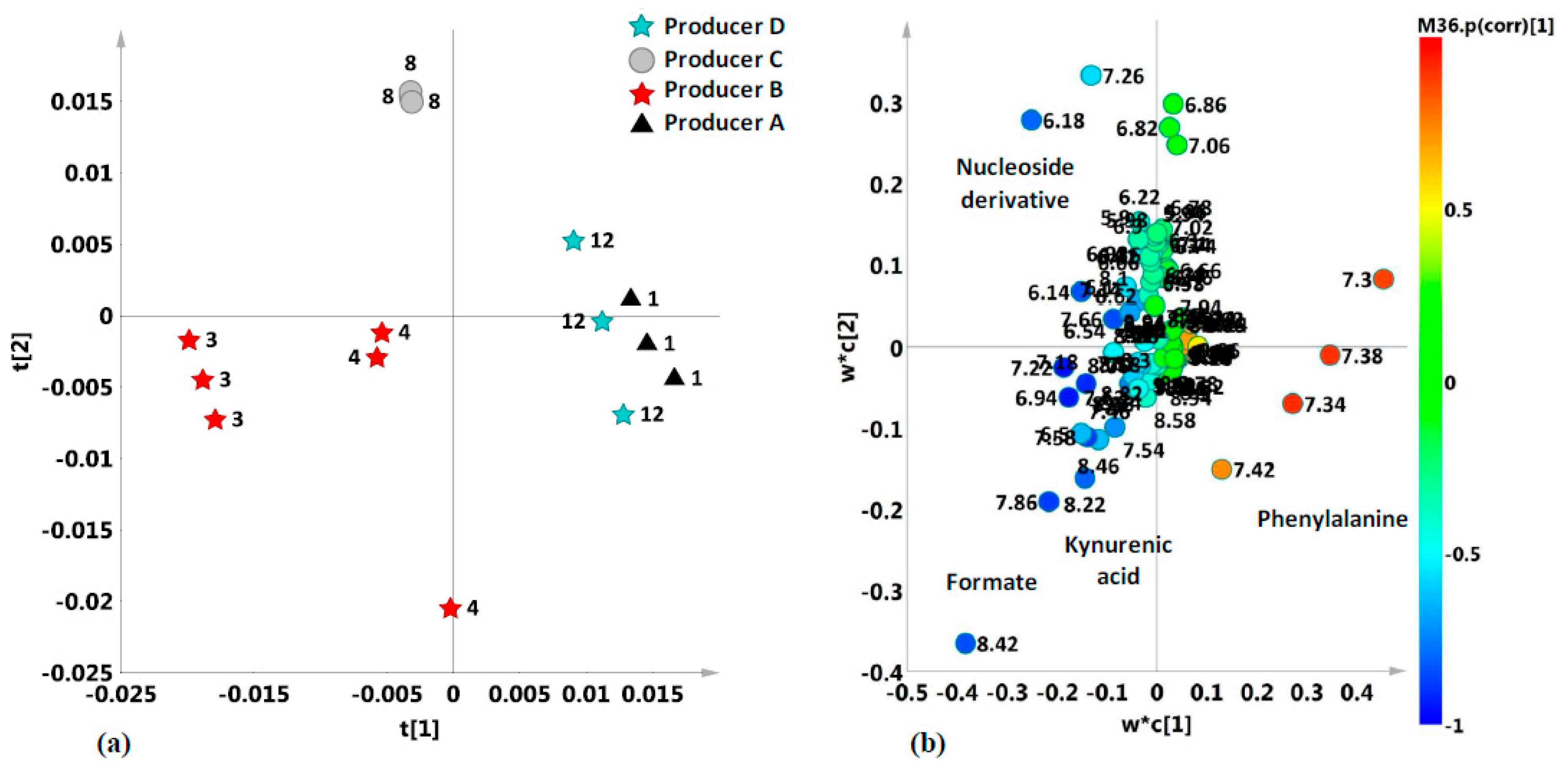
2.2. Unsupervised and Supervised Discriminant Analyses
2.3. Aliphatic Region
2.4. Aromatic Region
2.5. Metabolites Comparison
3. Materials and Methods
3.1. Sampling
3.2. Sample Preparation for NMR Analysis
3.3. 1H-NMR Spectra Acquisition and Processing
3.4. Multivariate Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- del Campo, G.; Zuriarrain, J.; Zuriarrain, A.; Berregi, I. Quantitative determination of carboxylic acids, amino acids, carbohydrates, ethanol and hydroxymethylfurfural in honey by 1H NMR. Food Chem. 2016, 196, 1031–1039. [Google Scholar] [CrossRef] [PubMed]
- Kazalaki, A.; Misiak, M.; Spyros, A.; Dais, P. Identification and quantitative determination of carbohydrate molecules in Greek honey by employing 13C NMR spectroscopy. Anal. Methods 2015, 7, 5962–5972. [Google Scholar] [CrossRef]
- Consonni, R.; Cagliani, L.R.; Cogliati, C. NMR characterization of saccharides in Italian honeys of different floral sources. J. Agric. Food Chem. 2012, 60, 4526–4534. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Maté, A.; Osés, S.M.; Marcazzan, G.L.; Gardini, S.; Muiño, M.A.F.; Sancho, M.T. Sugar composition and sugar-related parameters of honeys from the northern Iberian Plateau. J. Food Compos. Anal. 2018, 74, 34–43. [Google Scholar] [CrossRef]
- Bogdanov, S.; Jurendic, T.; Sieber, R.; Gallmann, P. Honey for nutrition and health: A review. J. Am. Coll. Nutr. 2008, 27, 677–689. [Google Scholar] [CrossRef]
- Gheldof, N.; Wang, X.-H.; Engeseth, N.J. Identification and quantification of antioxidant components of honeys from various floral sources. J. Agric. Food Chem. 2002, 50, 5870–5877. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Nepi, M.; Pacini, E. Nectaries and Nectar; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Olawode, E.O.; Tandlich, R.; Cambray, G. 1H-NMR Profiling and Chemometric Analysis of Selected Honeys from South Africa, Zambia, and Slovakia. Molecules 2018, 23, 578. [Google Scholar] [CrossRef] [Green Version]
- Commission FAO. Codex Standard for Honey Codex Stan 12-1981 12-1981. 1981, pp. 1–8. Available online: http://www.fao.org/3/w0076e/w0076e30.htm (accessed on 27 November 2019).
- Council Directive. 2001/110/EC of 20 December 2001 relating to honey. OJEC 2002, 10, 47–52. [Google Scholar]
- Consonni, R.; Cagliani, L.R. Geographical characterization of polyfloral and acacia honeys by nuclear magnetic resonance and chemometrics. J. Agric. Food Chem. 2008, 56, 6873–6880. [Google Scholar] [CrossRef]
- Consonni, R.; Bernareggi, F.; Cagliani, L.R. NMR-based metabolomic approach to differentiate organic and conventional Italian honey. Food Control 2019, 98, 133–140. [Google Scholar] [CrossRef]
- Pontis, J.A.; da Costa, L.A.M.A.; da Silva, S.J.R.; Flach, A. Color, phenolic and flavonoid content, and antioxidant activity of honey from Roraima, Brazil. J. Food Sci. Technol. 2014, 34, 69–73. [Google Scholar] [CrossRef] [Green Version]
- Abrol, D. Beekeeping: A Compressive Guide to Bees and Beekeeping; Scientific Publishers: Jodhpur, India, 2013. [Google Scholar]
- Bogdanov, S.; Ruoff, K.; Oddo, L.P. Physico-chemical methods for the characterisation of unifloral honeys: A review. Apidologie 2004, 35, S4–S17. [Google Scholar] [CrossRef] [Green Version]
- Pita-Calvo, C.; Vázquez, M. Differences between honeydew and blossom honeys: A review. Trends Food Sci. Technol. 2017, 59, 79–87. [Google Scholar] [CrossRef]
- Karabagias, I.K.; Vlasiou, M.; Kontakos, S.; Drouza, C.; Kontominas, M.G.; Keramidas, A.D. Geographical discrimination of pine and fir honeys using multivariate analyses of major and minor honey components identified by 1H NMR and HPLC along with physicochemical data. Eur. Food Res. Technol. 2018, 244, 1249–1259. [Google Scholar] [CrossRef]
- Spiteri, M.; Jamin, E.; Thomas, F.; Rebours, A.; Lees, M.; Rogers, K.M.; Rutledge, D.N. Fast and global authenticity screening of honey using 1H-NMR profiling. Food Chem. 2015, 189, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, N.; Birkenmeier, M.; Kuballa, T.; Ohmenhaeuser, M.; Rohn, S.; Weller, P. Differentiation of the botanical origin of honeys by fast, non-targeted 1H-NMR profiling and chemometric tools as alternative authenticity screening tool. In Proceedings of the XIII International Conference on the Applications of Magnetic Resonance in Food Science, Karlsruhe, Germany, 7–10 June 2016; Guthausen, G., van Duynhoven, J.P.M., Eds.; IM Publications: Chichester, UK, 2018; pp. 33–37. [Google Scholar]
- Schievano, E.; Stocchero, M.; Morelato, E.; Facchin, C.; Mammi, S. An NMR-based metabolomic approach to identify the botanical origin of honey. Metabolomics 2012, 8, 679–690. [Google Scholar] [CrossRef]
- Beretta, G.; Caneva, E.; Regazzoni, L.; Bakhtyari, N.G.; Facino, R.M. A solid-phase extraction procedure coupled to 1H NMR, with chemometric analysis, to seek reliable markers of the botanical origin of honey. Anal. Chim. Acta 2008, 620, 176–182. [Google Scholar] [CrossRef]
- Boffo, E.F.; Tavares, L.A.; Tobias, A.C.; Ferreira, M.M.; Ferreira, A.G. Identification of components of Brazilian honey by 1H NMR and classification of its botanical origin by chemometric methods. LWT–J. Food Sci. Technol. 2012, 49, 55–63. [Google Scholar] [CrossRef] [Green Version]
- Schievano, E.; Peggion, E.; Mammi, S. 1H nuclear magnetic resonance spectra of chloroform extracts of honey for chemometric determination of its botanical origin. J. Agric. Food Chem. 2009, 58, 57–65. [Google Scholar] [CrossRef]
- Girelli, C.R.; Accogli, R.; Del Coco, L.; Angilè, F.; De Bellis, L.; Fanizzi, F.P. 1H-NMR-based metabolomic profiles of different sweet melon (Cucumis melo L.) Salento varieties: Analysis and comparison. Food Res. Int. 2018, 114, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Girelli, C.R.; De Pascali, S.A.; Del Coco, L.; Fanizzi, F.P. Metabolic profile comparison of fruit juice from certified sweet cherry trees (Prunus avium L.) of Ferrovia and Giorgia cultivars: A preliminary study. Food Res. Int. 2016, 90, 281–287. [Google Scholar] [CrossRef]
- Girelli, C.R.; Del Coco, L.; Fanizzi, F.P. 1H NMR spectroscopy and multivariate analysis as possible tool to assess cultivars, from specific geographical areas, in EVOOs. Eur. J. Lipid Sci. Technol. 2016, 118, 1380–1388. [Google Scholar] [CrossRef]
- Schievano, E.; Morelato, E.; Facchin, C.; Mammi, S. Characterization of markers of botanical origin and other compounds extracted from unifloral honeys. J. Agric. Food Chem. 2013, 61, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Barrasso, R.; Bonerba, E.; Savarino, A.; Ceci, E.; Bozzo, G.; Tantillo, G. Simultaneous Quantitative Detection of Six Families of Antibiotics in Honey Using A Biochip Multi-Array Technology. Vet. Sci. 2019, 6, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osservatorio Nazionale Miele. Available online: https://www.informamiele.it/doc-cat/produzione-miele (accessed on 27 November 2019).
- Anagrafe Nazionale Zootecnica–Statistiche. Available online: https://www.vetinfo.it/j6_statistiche/#/report-pbi/45 (accessed on 25 February 2020).
- Di Ciaula, A. Rischi derivanti dall’obbligo d’uso di pesticidi per il controllo della Xylella fastidiosa in Puglia. Available online: https://www.isde.it/rischi-derivanti-dallobbligo-duso-di-pesticidi-per-il-controllo-della-xylella-fastidiosa-in-puglia/ (accessed on 19 November 2019).
- Di Serio, F.; Bodino, N.; Cavalieri, V.; Demichelis, S.; Di Carolo, M.; Dongiovanni, C.; Fumarola, G.; Gilioli, G.; Guerrieri, E.; Picciotti, U. Collection of Data and Information on Biology and Control of Vectors of Xylella fastidiosa. EFSA Support Publ. 2019, 16, e0232363. [Google Scholar] [CrossRef]
- Goulson, D. Call to restrict neonicotinoids. Science 2018, 360, 973. [Google Scholar]
- Cimino, A.M.; Boyles, A.L.; Thayer, K.A.; Perry, M.J. Effects of neonicotinoid pesticide exposure on human health: a systematic review. Environ. Health Perspect. 2017, 125, 155–162. [Google Scholar] [CrossRef] [Green Version]
- Kurwadkar, S.; Evans, A. Neonicotinoids: Systemic Insecticides and Systematic Failure. Bull. Environ. Contam. Toxicol. 2016, 97, 745–748. [Google Scholar] [CrossRef] [Green Version]
- Authority, E.F.S. Conclusion on the peer review of the pesticide risk assessment for bees for the active substance imidacloprid considering all uses other than seed treatments and granules. EFSA J. 2015, 13, 4211. [Google Scholar] [CrossRef]
- European Commission. Commission Implementing Regulation (EU) 2018/783 of 29 May 2018 Amending Implementing Regulation (EU) No 540/2011 as Regards the Conditions of Approval of the Active Substance Imidacloprid. OJEU 2018, 61, 31–34. [Google Scholar]
- Dongiovanni, C.; Altamura, G.; Di Carolo, M.; Fumarola, G.; Saponari, M.; Cavalieri, V. Evaluation of efficacy of different insecticides against Philaenus spumarius L., vector of Xylella fastidiosa in olive orchards in Southern Italy, 2015–2017. Arthropod. Manag. Tests 2018, 43, tsy034. [Google Scholar] [CrossRef] [Green Version]
- MIPAAF. Decreto n. 4999 del 13/02/2018 Misure di Emergenza per la Prevenzione, il Controllo e L’eradicazione di Xylella Fastidiosa (Well et al.) nel Territorio della Repubblica Italiana; GU Serie Generale n.80 del 06-04-2018; Gazzetta Ufficiale della Repubblica Italiana: Roma, Italy, 2018. [Google Scholar]
- Karabagias, I.K.; Maia, M.; Karabagias, V.K.; Gatzias, I.; Badeka, A.V. Characterization of eucalyptus, chestnut and heather honeys from Portugal using multi-parameter analysis and chemo-calculus. Foods 2018, 7, 194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shamsudin, S.; Selamat, J.; Sanny, M.; AR, S.B.; Jambari, N.N.; Khatib, A. A Comparative Characterization of Physicochemical and Antioxidants Properties of Processed Heterotrigona itama Honey from Different Origins and Classification by Chemometrics Analysis. Molecules 2019, 24, 3898. [Google Scholar] [CrossRef] [Green Version]
- Kortesniemi, M.; Slupsky, C.M.; Ollikka, T.; Kauko, L.; Spevacek, A.R.; Sjövall, O.; Yang, B.; Kallio, H. NMR profiling clarifies the characterization of Finnish honeys of different botanical origins. Food Res. Int. 2016, 86, 83–92. [Google Scholar] [CrossRef] [Green Version]
- Berregi, I.; del Campo, G.; Caracena, R.; Miranda, J.I. Quantitative determination of formic acid in apple juices by 1H NMR spectrometry. Talanta 2007, 72, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- White, J.W. Composition of honey. Honey: A Comprehensive Survey, 1st ed.; Heinemann [for] the Bee Research Association: London, UK, 1975; pp. 157–206. [Google Scholar]
- Satta, A.; Floris, I.; Eguaras, M.; Cabras, P.; Garau, V.L.; Melis, M. Formic acid-based treatments for control of Varroa destructor in a Mediterranean area. J. Econ. Entomol. 2005, 98, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Donarski, J.A.; Jones, S.A.; Charlton, A.J. Application of cryoprobe 1H nuclear magnetic resonance spectroscopy and multivariate analysis for the verification of Corsican honey. J. Agric. Food Chem. 2008, 56, 5451–5456. [Google Scholar] [CrossRef]
- Ribeiro, R.d.O.R.; Carneiro, C.d.S.; Mársico, E.T.; Cunha, F.L.; Conte Junior, C.A.; Mano, S.B. Influence of the time/temperature binomial on the hydroxymethylfurfural content of floral honeys subjected to heat treatment. Ciên. Agrotec. 2012, 36, 204–209. [Google Scholar] [CrossRef] [Green Version]
- Sodré, G.d.S.; Marchini, L.C.; Moreti, A.C.d.C.C.; Otsuk, I.P.; da Carvalho, C.A.L. Physico-chemical characteristics of honey produced by Apis mellifera in the Picos region, state of Piauí, Brazil. R. Bras. Zootec. 2011, 40, 1837–1843. [Google Scholar] [CrossRef] [Green Version]
- Turski, M.P.; Turska, M.; Paluszkiewicz, P.; Parada-Turska, J.; Oxenkrug, G.F. Kynurenic Acid in the Digestive System–-New Facts, New Challenges. IJTR 2013, 6, S12536. [Google Scholar] [CrossRef] [Green Version]
- Bohár, Z.; Toldi, J.; Fülöp, F.; Vécsei, L. Changing the face of kynurenines and neurotoxicity: Therapeutic considerations. Int. J. Mol. Sci. 2015, 16, 9772–9793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.; Hu, D.; Lv, M.; Jin, L.; Wu, J.; Zeng, S.; Yang, S.; Song, B. Synthesis, insecticidal, and antibacterial activities of novel neonicotinoid analogs with dihydropyridine. Chem. Cent. J. 2013, 7, 76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Issaq, H.J.; Veenstra, T.D. Proteomic and Metabolomic Approaches to Biomarker Discovery, 1st ed.; Academic Press: San Diego, CA, USA, 2019. [Google Scholar]
- Dominguez, R.Q.; Reyes-Carrillo, J.L.; de la Cruz-Larios, L.; González-Eguiarte, D.R. Bee honey color variation throughout the year in Hejotitán, Jalisco, México. SAFER 2018, 6. [Google Scholar]
- Czipa, N.; Borbély, M.; Győri, Z. Proline content of different honey types. Acta Aliment. 2012, 41, 26–32. [Google Scholar] [CrossRef]
- Manzanares, A.B.; García, Z.H.; Galdón, B.R.; Rodríguez, E.R.; Romero, C.D. Differentiation of blossom and honeydew honeys using multivariate analysis on the physicochemical parameters and sugar composition. Food Chem. 2011, 126, 664–672. [Google Scholar] [CrossRef]
- Bogdanov, S. Honey as nutrient and functional food. J. Am. Coll. Nutr. 2010, 27, 677–689. [Google Scholar] [CrossRef]
- Ruoff, K.; Luginbühl, W.; Kilchenmann, V.; Bosset, J.O.; von der Ohe, K.; Von Der Ohe, W.; Amadò, R. Authentication of the botanical origin of honey using profiles of classical measurands and discriminant analysis. Apidologie 2007, 38, 438–452. [Google Scholar] [CrossRef]
- Mannina, L.; Sobolev, A.P.; Viel, S. Liquid state 1H high field NMR in food analysis. Prog. Nucl. Magn. Reason. Spectrosc. 2012, 66, 1. [Google Scholar] [CrossRef]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Jackson, J.E. A User’s Guide to Principal Components; John Wiley & Sons: Hoboken, NJ, USA, 2005; Volume 587. [Google Scholar]
- Kettaneh, N.; Berglund, A.; Wold, S. PCA and PLS with very large data sets. CSDA 2005, 48, 69–85. [Google Scholar] [CrossRef]
- Wold, S.; Eriksson, L.; Trygg, J.; Kettaneh, N. The PLS method–Partial Least Squares Projections to Latent Structures–and Its Applications in Industrial RDP (Research, Development, and Production); Unea University: Umeå, Sweden, 2004. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Eriksson, L.; Byrne, T.; Johansson, E.; Trygg, J.; Vikström, C. Multi-and Megavariate Data Analysis Basic Principles and Applications, 3rd ed.; Umetrics Academy: Malmö, Sweden, 2013; Volume 1. [Google Scholar]
- Wheelock, Å.M.; Wheelock, C.E. Trials and tribulations of ‘omics data analysis: assessing quality of SIMCA-based multivariate models using examples from pulmonary medicine. Mol. Biosyst. 2013, 9, 2589–2596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic. Acids. Res. 2018, 46, W486–CW494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
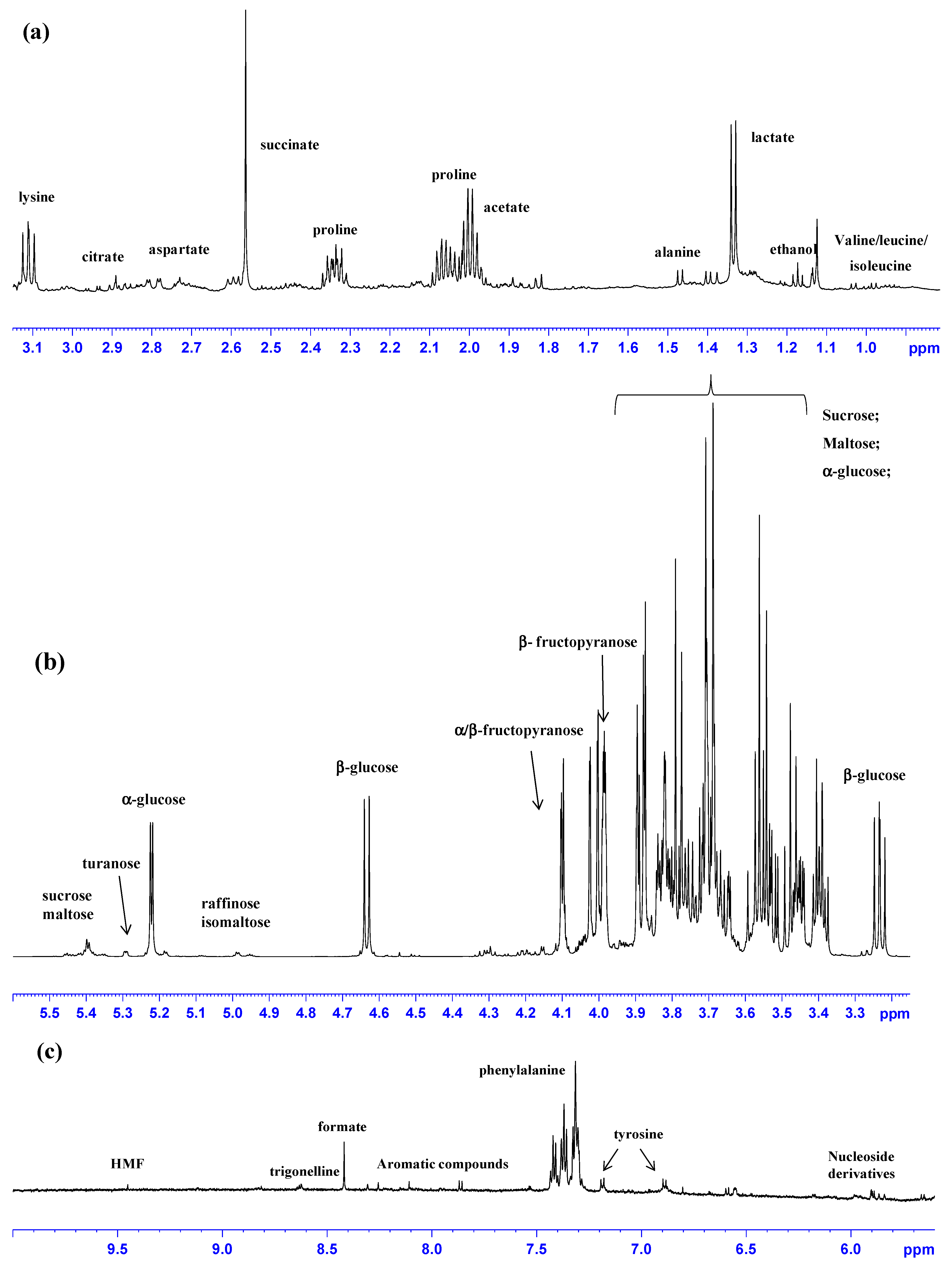
| δ (ppm) | Metabolite | Assignment | Multiplicity |
|---|---|---|---|
| 9.45 | HMF | H1 | s |
| 9.13; 8.84 | trigonelline | N−CH; CH−3.5 | s; t |
| 8.42−8.46 * | formate | HCOOH | s |
| 7.90; 5.90 | uridine | ||
| 8.22, 7.86, 7.55, 6.94 | kynurenic acid (only in dark honey samples) | d, d, d, s | |
| 7.42; 7.38; 7.32 | phenylalanine | CH–3,5 ring; CH4 ring; CH–2,6 ring; | m; m; m; |
| 7.28, 6.18 | nucleoside derivatives | ||
| 7.19; 6.90 | tyrosine | CH–2,6 ring; CH–3,5 ring; | m; m; |
| 5.41; 3.56 | sucrose | CH1; CH2 | d; dd; |
| 5.40 | maltose | ||
| 5.30; 3.58 | turanose | CH1 | |
| 5.23; 3.52 | α-glucose | CH1; CH2 | d; dd; |
| 4.99 | raffinose | ||
| 4.94 | isomaltose | ||
| 4.64; 3.24 | β-glucose | CH1; CH2 | d; dd; |
| 4.10 | α/β- fructofuranose | CH3 | d; |
| 4.01 | β- fructopyranose | CH5 | d; |
| 3.12 | lysine | γ−CH2 | t |
| 2.70; 2.90 | citrate | Half−CH2; half−CH2 | d; d |
| 2.55; 2.80 | aspartate | half β−CH2; half β−CH2; γ − | dd; dd |
| 2.56 | succinate | α−β - CH2 | s |
| 2.34; 2.07; 2.01 | proline | half β −CH2; half β-CH2; γ−CH2 | m; m; m; |
| 1.95 | acetate | α−CH3 | s |
| 1.47 | alanine | β−CH3 | d |
| 1.38 | monoterpenoid acid (only in polyfloral honeys) | -C(CH3)2OH; | s |
| 1.32 | lactate | β−CH3 | d |
| 1.17 | ethanol | CH3 | t |
| 1.03; 2.28 | valine | γ−CH3; β−CH | d; m; |
| 1.00 | isoleucine | γ-CH3; | d |
| 0.97; 1.7 | leucine | δ-CH3; γ-CH | d; m; |
| Metabolite | Chemical Shift (ppm) | F Value 1 | P-Value 2 | FDR 3 | Tukey’s HSD 4 |
|---|---|---|---|---|---|
| Light honeys | |||||
| Proline | 2.34 | 100.38 | 3.1531 × e−12 | 1.4709 × e−11 | C–A; D–A;C–B; D–B; D–C; |
| monoterpenoid acid | 1.37 | 97.344 | 4.2025 × e−12 | 1.4709 × e−11 | C–A; C–B; D–C; |
| Lactate | 1.34 | 37.096 | 2.3111 × e−8 | 5.3937 × e−8 | C–A;D–A; C–B;D–B; |
| Ethanol | 1.17 | 16.166 | 1.4282 × e−5 | 2.4993 × e−4 | B–A; C–A; D–B; D–C; |
| Phenylalanine | 7.42 | 6.8538 | 2.3633 × e−3 | 3.3087 × e−3 | C–B; D–B; |
| Lysine | 2.90 | 3.5899 | 3.1792 × e−2 | 3.7091 × e−2 | B–A |
| Dark honeys | |||||
| Formate | 8.45 | 3239.5 | 1.2559 × e−12 | 7.937 × e−12 | B-A; C–A; D–A; B–C; B–D; |
| monoterpenoid acid | 1.37 | 2811.2 | 1.9843 × e−12 | 7.937 × e−12 | B–A; C–A; C–B; B–D; C–D; |
| Proline | 2.34 | 172.71 | 1.3075 × e−7 | 3.4866 × e−7 | C–A; C–B; C–D; |
| Lactate | 1.34 | 96.224 | 1.2871 × e−6 | 2.5742 × e−6 | A–B; A–C; D–A; C–B; D–B; D–C; |
| Phenylalanine | 7.42 | 64.339 | 6.0743 × e−6 | 9.7188 × e−6 | A–C; A–D; B–C; B–D; D–C; |
| Nucleoside Derivatives | 6.18 | 57.138 | 95529 2 × e−6 | 1.2737 × e−5 | B–A; C–A; C–B; C–D; |
| Succinate | 2.54 | 4.3187 | 4.3517 × e−2 | 4.4375 × e−2 | D–B; D–C; |
| KYNA | 6.94 | 4.2827 | 4.4375 × e−2 | 4.4375 × e−2 | B–A; C–A; B–D; |
| Sample | Producers | Description | Colour | Area Production |
|---|---|---|---|---|
| 1 | A | summer polyfloral | dark | Ugento (Lecce Province) |
| 2 | A | spring polyfloral | light | Ugento (Lecce Province) |
| 3 1 | B | polyfloral | dark | Taranto Province |
| 4 1 | B | polyfloral | dark | Taranto Province |
| 5 | B | polyfloral | light | Trepuzzi (Lecce Province) |
| 6 | B | monofloral (orange) | light | Taranto Province |
| 7 | C | spring polyfloral | light | Copertino (Lecce Province) |
| 8 | C | summer honeydew | dark | Copertino (Lecce Province) |
| 9 | C | spring polyfloral | light | Nardò Copertino (Lecce Province) |
| 10 | D | monofloral (acacia) | light | Surbo (Lecce Province) |
| 11 | D | polyfloral | light | Surbo (Lecce Province) |
| 12 | D | honeydew | dark | Surbo (Lecce Province) |
| 13 | B | monofloral (citrus) | light | Trepuzzi (Lecce Province) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Girelli, C.R.; Schiavone, R.; Vilella, S.; Fanizzi, F.P. Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting. Sustainability 2020, 12, 5009. https://doi.org/10.3390/su12125009
Girelli CR, Schiavone R, Vilella S, Fanizzi FP. Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting. Sustainability. 2020; 12(12):5009. https://doi.org/10.3390/su12125009
Chicago/Turabian StyleGirelli, Chiara Roberta, Roberta Schiavone, Sebastiano Vilella, and Francesco Paolo Fanizzi. 2020. "Salento Honey (Apulia, South-East Italy): A Preliminary Characterization by 1H-NMR Metabolomic Fingerprinting" Sustainability 12, no. 12: 5009. https://doi.org/10.3390/su12125009







