Plant Growth and Development from Biocommunication Perspective
Abstract
1. Introduction
- We cannot understand plant language in communication processes as other plants do.
- We may identify the meaning of plant signaling by observing the habits (growth and development) it produces within real-life habitats.
- To identify the meaning of signaling in plant growth and development, we have to carefully examine the context of signal use within various interaction patterns.
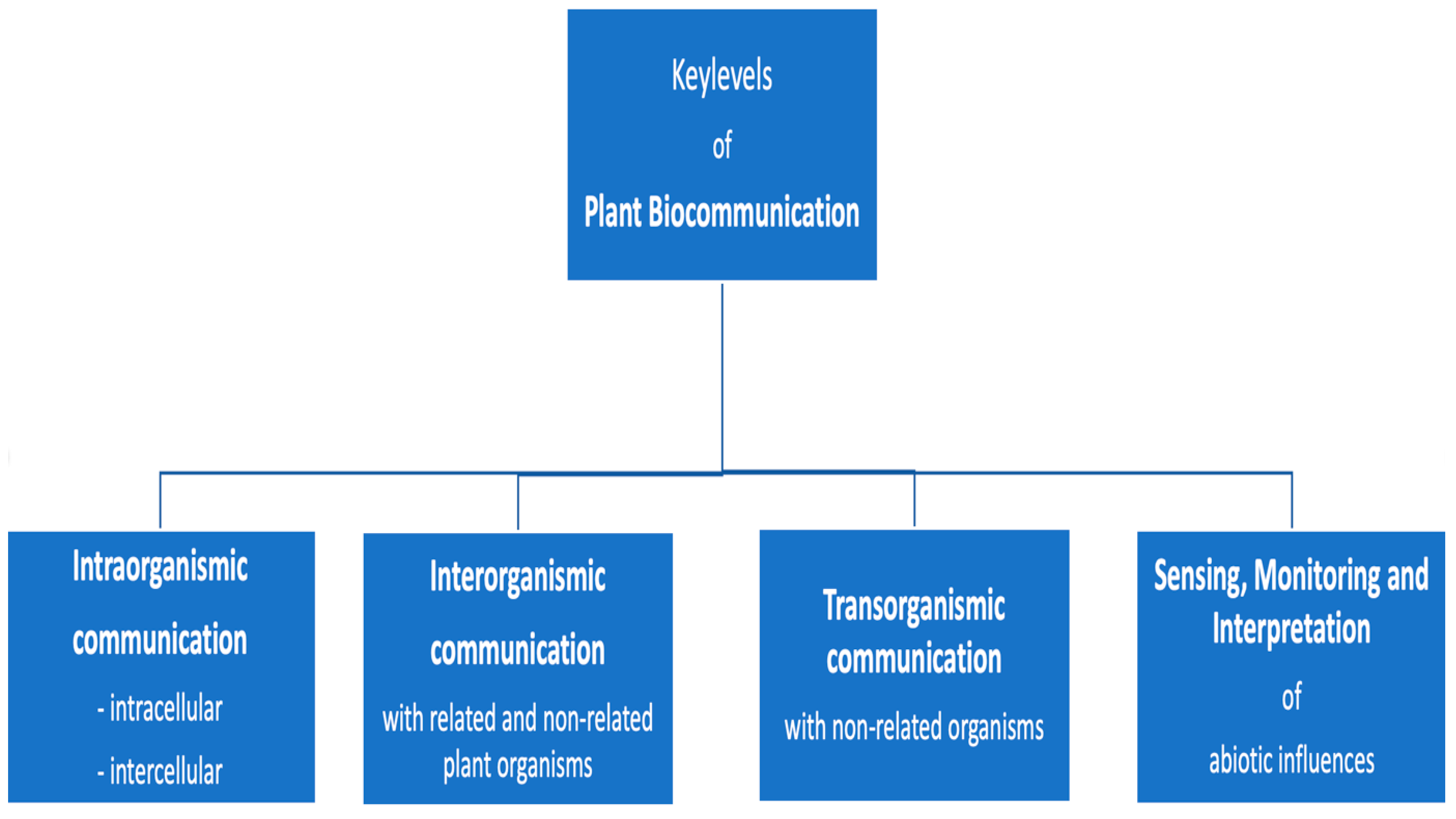
- The context of use may vary according to the four levels of communication of plants: (a) signaling within the plant body, (b) signaling with other plants, (c) signaling with non-plant organisms, and (d) sensing and interpretation of abiotic influences.
- Actors in plant communication include various types of cells, tissues, and organs, all of which exhibit highly complex interaction patterns coordinated by various signaling processes.
- Biological communication in plants during growth and development is not a mechanistic process; rather, it is highly adaptable and context-dependent.
2. Growth and Development Through Biological Communication of Plants
3. Various Levels of Signaling in Plants
4. Intraorganismic Communication: Intra- and Intercellular Signal-Mediated Interaction
4.1. Intracellular Communication: Non-Coding RNAs Are Key Players
4.2. Epigenetic Programming
4.3. Transgenerational Inheritance of Epigenetic Variations
4.4. Intercellular Communication: Coordination of Tissues and Organs
5. Interorganismic Communication
6. Transorganismic Communication
7. Sensing, Monitoring, and Interpretation of Abiotic Influences
8. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wittgenstein, L. Philosophical Investigations; Basil Blackwell: Oxford, UK, 1953; p. 323. [Google Scholar]
- Peirce, C.S. Collected Papers of Charles Sanders Peirce; Harvard University Press: Harvard, UK, 1974; Volume 5, p. 465. [Google Scholar]
- Friml, J.; Wisniewska, J. Auxin as an intercellular signal. In Intercellular Communication in Plants. Annual Plant Reviews; Fleming, A.J., Ed.; Blackwell Publishing: Oxford, UK, 2005; Volume 16, pp. 1–26. [Google Scholar]
- Witzany, G.; Baluška, F. Life’s code script does not code itself: The machine metaphor for living organisms is outdated. EMBO Rep. 2012, 12, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Sisodia, R.; Bhatla, S.C. Plant Movements. In Plant Physiology, Development and Metabolism; Sisodia, R., Bhatla, S.C., Eds.; Springer: Singapore, 2018; pp. 907–935. [Google Scholar]
- Witzany, G. Plant communication from biosemiotic perspective: Differences in abiotic and biotic signal perception determine content arrangement of response behavior. Context determines meaning of meta-, inter- and intraorganismic plant signaling. Plant Signal Behav. 2006, 1, 169–718. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.; Avesani, S.; Bonato, B.; Dadda, M.; Guerra, S.; Ravazzolo, L.; Simonetti, V.; Castiello, U. Plant behavior: Theoretical and technological advances. Curr. Opin. Psychol. 2025, 64, 102026. [Google Scholar] [CrossRef] [PubMed]
- Lev-Yadun, S. Plant development: Cell movement relative to each other is both common and very important. Plant Signal Behav. 2015, 10, e991566. [Google Scholar] [CrossRef]
- Baluska, F.; Manuso, S.; Volkmann, D. (Eds.) Communication in Plants; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Davis, G.V.; de Souza Moraes, T.; Khanapurkar, S.; Dromiack, H.; Ahmad, Z.; Bayer, E.M.; Bhalerao, R.P.; Walker, S.I.; Bassel, G.W. Toward uncovering an operating system in plant organs. Trends Plant Sci. 2024, 29, 742–753. [Google Scholar] [CrossRef]
- Baluska, F.; Mancuso, S. (Eds.) Signaling in Plants; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar]
- Bonato, B.; Peressotti, F.; Guerra, S.; Wang, Q.; Castiello, U. Cracking the code: A comparative approach to plant communication. Commun. Integr. Biol. 2021, 14, 176–185. [Google Scholar] [CrossRef]
- Howard, M.M.; Bass, E.; Chautá, A.; Mutyambai, D.; Kessler, A. Integrating plant-to-plant communication and rhizosphere microbial dynamics: Ecological and evolutionary implications and a call for experimental rigor. ISME J. 2022, 16, 5–9. [Google Scholar] [CrossRef]
- Witzany, G. (Ed.) Biocommunication in Soil Microorganisms; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Witzany, G. (Ed.) Biocommunication of Fungi; Springer: Dordrecht, The Netherlands, 2012. [Google Scholar]
- Witzany, G. (Ed.) Biocommunication of Animals; Springer: Dordrecht, The Netherlands, 2014. [Google Scholar]
- Witzany, G. (Ed.) Biocommunication of Archaea; Springer: Dordrecht, The Netherlands, 2017. [Google Scholar]
- Witzany, G. (Ed.) Biocommunication of Phages; Springer: Cham, Switzerland, 2020. [Google Scholar]
- Witzany, G.; Baluška, F. (Eds.) Biocommunication of Plants; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Witzany, G.; Nowacki, M. (Eds.) Biocommunication of Ciliates; Springer: Dordrecht, The Netherlands, 2016. [Google Scholar]
- Witzany, G. Can mathematics explain the evolution of human language? Commun. Integr. Biol. 2011, 4, 516–520. [Google Scholar] [CrossRef]
- McWatters, H.G.; Roden, L.C.; Staiger, D. Picking out parallels: Plant circadian clocks in context. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2001, 356, 1735–1743. [Google Scholar] [CrossRef]
- Creux, N.; Harmer, S. Circadian Rhythms in Plants. Cold Spring Harb. Perspect. Biol. 2019, 11, a034611. [Google Scholar] [CrossRef]
- Witzany, G. Noncoding RNAs: Persistent viral agents as modular tools for cellular needs. Ann. N. Y. Acad. Sci. 2009, 1178, 244–267. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.; Amaral, P. RNA, the Epicenter of Genetic Information: A New Understanding of Molecular Biology; CRC Press: Abingdon, UK, 2022. [Google Scholar]
- Ohtani, M.; Kurihara, Y.; Seki, M.; Crespi, M. RNA-Mediated Plant Behavior. Plant Cell Physiol. 2019, 60, 1893–1896. [Google Scholar] [CrossRef] [PubMed]
- Bhogireddy, S.; Mangrauthia, S.K.; Kumar, R.; Pandey, A.K.; Singh, S.; Jain, A.; Budak, H.; Varshney, R.K.; Kudapa, H. Regulatory non-coding RNAs: A new frontier in regulation of plant biology. Funct. Integr. Genom. 2021, 21, 313–330. [Google Scholar] [CrossRef] [PubMed]
- Panchal, A.; Maurya, J.; Seni, S.; Singh, R.K.; Prasad, M. An insight into the roles of regulatory ncRNAs in plants: An abiotic stress and developmental perspective. Plant Physiol. Biochem. 2023, 201, 107823. [Google Scholar] [CrossRef]
- Li, S.; Zhao, Y.; Tan, S.; Li, Z. Non-coding RNAs and leaf senescence: Small molecules with important roles. Plant Physiol. Biochem. 2024, 207, 108399. [Google Scholar] [CrossRef]
- Villarreal, L.P. Viruses and the Evolution of Life; ASM Press: Washington, DC, USA, 2005. [Google Scholar]
- Roossinck, M.J. Symbiosis versus competition in plant virus evolution. Nat. Rev. Microbiol. 2005, 3, 917–924. [Google Scholar] [CrossRef]
- Witzany, G. Two genetic codes: Repetitive syntax for active non-coding RNAs; non-repetitive syntax for the DNA archives. Commun. Integr. Biol. 2017, 10, e1297352. [Google Scholar] [CrossRef]
- Goldtzvik, Y.; Sen, N.; Lam, S.D.; Orengo, C. Protein diversification through post-translational modifications, alternative splicing, and gene duplication. Curr. Opin. Struct. Biol. 2023, 81, 102640. [Google Scholar] [CrossRef]
- Jenuwein, T. An RNA-guided pathway for the epigenome. Science 2002, 297, 2215–2218. [Google Scholar] [CrossRef]
- Bhatia, G.; Prall, W.; Sharma, B.; Gregory, B.D. Covalent RNA modifications and their budding crosstalk with plant epigenetic processes. Curr. Opin. Plant Biol. 2022, 69, 102287. [Google Scholar] [CrossRef]
- Assmann, S.M.; Chou, H.L.; Bevilacqua, P.C. Rock, scissors, paper: How RNA structure informs function. Plant Cell 2023, 35, 1671–1707. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gautam, V.; Singh, S.; Sarkar Das, S.; Verma, S.; Mishra, V.; Mukherjee, S.; Sarkar, A.K. Plant small RNAs: Advancement in the understanding of biogenesis and role in plant development. Planta 2018, 248, 545–558. [Google Scholar] [CrossRef]
- Samarfard, S.; Ghorbani, A.; Karbanowicz, T.P.; Lim, Z.X.; Saedi, M.; Fariborzi, N.; McTaggart, A.R.; Izadpanah, K. Regulatory non-coding RNA: The core defense mechanism against plant pathogens. J. Biotechnol. 2022, 359, 82–94. [Google Scholar] [CrossRef]
- Jin, D.; Wang, Y.; Zhao, Y.; Chen, M. MicroRNAs and their cross-talks in plant development. J. Genet. Genom. 2013, 40, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Curaba, J.; Singh, M.B.; Bhalla, P.L. miRNAs in the crosstalk between phytohormone signalling pathways. J. Exp. Bot. 2014, 65, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, B. MicroRNAs in Control of Plant Development. J. Cell Physiol. 2016, 231, 303–313. [Google Scholar] [CrossRef]
- Teotia, S.; Tang, G. To bloom or not to bloom: Role of microRNAs in plant flowering. Mol. Plant 2015, 8, 359–377. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Dong, Q.; Hu, B.; Zhang, C. microRNAs and Their Roles in Plant Development. Front. Plant Sci. 2022, 13, 824240. [Google Scholar] [CrossRef]
- Bayer, E.M.; Benitez-Alfonso, Y. Plasmodesmata: Channels Under Pressure. Annu. Rev. Plant Biol. 2024, 75, 291–317. [Google Scholar] [CrossRef]
- Ambrosone, A.; Barbulova, A.; Cappetta, E.; Cillo, F.; De Palma, M.; Ruocco, M.; Pocsfalvi, G. Plant Extracellular Vesicles: Current Landscape and Future Directions. Plants 2023, 12, 4141. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Xu, L.G.; Duan, C.G. The trans-kingdom communication of noncoding RNAs in plant-environment interactions. Plant Genome 2023, 16, e20289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Witzany, G. Do we eat gene regulators? Commun. Integr. Biol. 2012, 5, 230–232. [Google Scholar] [CrossRef]
- Sanchita; Trivedi, R.; Asif, M.H.; Trivedi, P.K. Dietary plant miRNAs as an augmented therapy: Cross-kingdom gene regulation. RNA Biol. 2018, 15, 1433–1439. [Google Scholar] [CrossRef]
- Huang, C.Y.; Jin, H. Coordinated Epigenetic Regulation in Plants: A Potent Managerial Tool to Conquer Biotic Stress. Front. Plant Sci. 2022, 12, 795274. [Google Scholar] [CrossRef]
- Novoplansky, A. What plant roots know? Semin. Cell Dev. Biol. 2019, 92, 126–133. [Google Scholar] [CrossRef]
- Chivasa, S.; Goodman, H.L. Stress-adaptive gene discovery by exploiting collective decision-making of decentralized plant response systems. New Phytol. 2020, 225, 2307–2313. [Google Scholar] [CrossRef]
- Baluska, F.; Gagliano, M.; Witzany, G. (Eds.) Memory and Learning in Plants; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Lolle, S.J.; Victor, J.L.; Young, J.M.; Pruitt, R.E. Genome-wide non-mendelian inheritance of extra-genomic information in Arabidopsis. Nature 2005, 434, 505–509. [Google Scholar] [CrossRef]
- Weigel, D.; Jürgens, G. Hotheaded healer. Nature 2005, 434, 443. [Google Scholar] [CrossRef]
- Cao, S.; Chen, Z.J. Transgenerational epigenetic inheritance during plant evolution and breeding. Trends Plant Sci. 2024, 29, 1203–1223. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C.; Dean, C. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 2014, 6, a019471. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Lao, S.; Fan, L. De-Domestication: An Extension of Crop Evolution. Trends Plant Sci. 2021, 26, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef]
- Mano, H.; Hasebe, M. Rapid movements in plants. J. Plant Res. 2021, 134, 3–17. [Google Scholar] [CrossRef]
- Freytes, S.N.; Canelo, M.; Cerdán, P.D. Regulation of Flowering Time: When and Where? Curr. Opin. Plant Biol. 2021, 63, 102049. [Google Scholar] [CrossRef]
- Johns, S.; Hagihara, T.; Toyota, M.; Gilroy, S. The fast and the furious: Rapid long-range signaling in plants. Plant Physiol. 2021, 185, 694–706. [Google Scholar] [CrossRef]
- Fleming, A. Intercellular Communication in Plants. Annual Plant Reviews; Wiley Blackwell: Hoboken, NJ, USA, 2005; Volume 16. [Google Scholar]
- Davière, J.M.; Achard, P. Organ communication: Cytokinins on the move. Nat. Plants 2017, 3, 17116. [Google Scholar] [CrossRef]
- Takahashi, F.; Shinozaki, K. Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 2019, 47, 106–111. [Google Scholar] [CrossRef]
- Sun, T.; Zhang, Y. Short- and long-distance signaling in plant defense. Plant J. 2021, 105, 505–517. [Google Scholar] [CrossRef]
- Beveridge, C.A. Axillary bud outgrowth: Sending a message. Curr. Opin. Plant Biol. 2006, 9, 35–40. [Google Scholar] [CrossRef] [PubMed]
- López-Salmerón, V.; Cho, H.; Tonn, N.; Greb, T. The Phloem as a Mediator of Plant Growth Plasticity. Curr. Biol. 2019, 29, R173–R181. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Fujita, M.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Inter-tissue and inter-organ signaling in drought stress response and phenotyping of drought tolerance. Plant J. 2022, 109, 342–358. [Google Scholar] [CrossRef] [PubMed]
- Evert, R. Esau’s Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development, 3rd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Baluska, F.; Salaj, J.; Mathur, J.; Braun, M.; Jasper, F.; Samaj, J.; Chua, N.H.; Barlow, P.W.; Volkmann, D. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev. Biol. 2000, 227, 618–632. [Google Scholar] [CrossRef]
- Ketelaar, T. The actin cytoskeleton in root hairs: All is fine at the tip. Curr. Opin. Plant Biol. 2013, 16, 749–756. [Google Scholar] [CrossRef]
- Golz, J.F. Signalling between the shoot apical meristem and developing lateral organs. Plant Mol. Biol. 2006, 60, 889–903. [Google Scholar] [CrossRef]
- Munné-Bosch, S. Limits to Tree Growth and Longevity. Trends Plant Sci. 2018, 23, 985–993. [Google Scholar] [CrossRef]
- Zweifel, R.; Sterck, F.; Braun, S.; Buchmann, N.; Eugster, W.; Gessler, A.; Häni, M.; Peters, R.L.; Walthert, L.; Wilhelm, M.; et al. Why trees grow at night. New Phytol. 2021, 231, 2174–2185. [Google Scholar] [CrossRef]
- Novoplansky, A. Picking battles wisely: Plant behaviour under competition. Plant Cell Environ. 2009, 32, 726–741. [Google Scholar] [CrossRef]
- Novoplansky, A.; Souza, G.M.; Brenner, E.D.; Bhatla, S.C.; Van Volkenburgh, E. Exploring the complex information processes underlying plant behavior. Plant Signal Behav. 2024, 19, 2411913. [Google Scholar] [CrossRef]
- Falik, O.; Hoffmann, I.; Novoplansky, A. Say it with flowers: Flowering acceleration by root communication. Plant Signal Behav. 2014, 9, e28258. [Google Scholar] [CrossRef] [PubMed]
- Pare, P.W.; Tumlinson, J.H. Plant volatiles as a defense against insect herbivores. Plant Physiol. 1999, 121, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Van der Putten, W.H.; Vet, L.E.M.; Harvey, J.A.; Wackers, F.L. Linking above and belowground multitrophic interactions of plants, herbivores, pathogens, and their antagonists. Trends Ecol. Evol. 2001, 16, 547–554. [Google Scholar] [CrossRef]
- Pearce, G.; Ryan, C.A. Systemic signaling in tomato plants for defense against herbivores. Isolation and characterization of three novel defense-signaling glycopeptide hormones coded in a single precursor gene. J. Biol. Chem. 2003, 278, 30044–30050. [Google Scholar] [CrossRef]
- Vivaldo, G.; Masi, E.; Taiti, C.; Caldarelli, G.; Mancuso, S. The network of plants volatile organic compounds. Sci. Rep. 2017, 7, 11050. [Google Scholar] [CrossRef]
- Gianoli, E.; Carrasco-Urra, F. Leaf mimicry in a climbing plant protects against herbivory. Curr. Biol. 2014, 24, 984–987. [Google Scholar] [CrossRef]
- Pannell, J.R.; Farmer, E.E. Mimicry in plants. Curr. Biol. 2016, 26, R784–R785. [Google Scholar] [CrossRef]
- Sharifi, R.; Jeon, J.S.; Ryu, C.M. Belowground plant-microbe communications via volatile compounds. J. Exp. Bot. 2022, 73, 463–486. [Google Scholar] [CrossRef]
- Kistner, C.; Parniske, M. Evolution of signal transduction in intracellular symbiosis. Trends Plant Sci. 2002, 7, 511–518. [Google Scholar] [CrossRef]
- Hirsch, A.M.; Bauer, W.D.; Bird, D.M.; Cullimore, J.; Tyler, B.; Yoder, J.I. Molecular signals and receptors: Controlling rhizosphere interacting between plants and other organisms. Ecology 2003, 84, 858–868. [Google Scholar] [CrossRef]
- Sharma, A.; Ahgal, M.; Johri, B.N. Microbial communication in the rhizosphere: Operation of quorum sensing. Curr. Sci. 2003, 85, 1164–1172. [Google Scholar]
- Schultz, J.C.; Appel, H.M. Cross kingdom cross talk: Hormones shared by plants and their insect herbivores. Ecology 2004, 85, 70–77. [Google Scholar] [CrossRef]
- Ho-Plágaro, T.; García-Garrido, J.M. Molecular Regulation of Arbuscular Mycorrhizal Symbiosis. Int. J. Mol. Sci. 2022, 23, 5960. [Google Scholar] [CrossRef] [PubMed]
- Oldroyd, G.E.; Murray, J.D.; Poole, P.S.; Downie, J.A. The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 2011, 45, 119–144. [Google Scholar] [CrossRef]
- Rosier, A.; Bishnoi, U.; Lakshmanan, V.; Sherrier, D.J.; Bais, H.P. A perspective on inter-kingdom signaling in plant-beneficial microbe interactions. Plant Mol. Biol. 2016, 90, 537–548. [Google Scholar] [CrossRef]
- Stephens, A.E.A.; Westoby, M. Effects of insect attack to stems on plant survival, growth, reproduction and photosynthesis. Oikos 2015, 124, 266–273. [Google Scholar] [CrossRef]
- Shen, G.; Zhang, J.; Lei, Y.; Xu, Y.; Wu, J. Between-Plant Signaling. Annu. Rev. Plant Biol. 2023, 74, 367–386. [Google Scholar] [CrossRef]
- Munns, R.; Millar, A.H. Seven plant capacities to adapt to abiotic stress. J. Exp. Bot. 2023, 74, 4308–4323. [Google Scholar] [CrossRef]
- Van Volkenburgh, E.; Mirzaei, K.; Ybarra, Y. Understanding Plant Behavior: A Student Perspective. Trends Plant Sci. 2021, 26, 423–425. [Google Scholar] [CrossRef]
- Lohmann, J.U.; Nemhauser, J.L. Cell signalling and gene regulation. Curr. Opin. Plant Biol. 2009, 12, 517–519. [Google Scholar] [CrossRef]
- Singh, A.P.; Savaldi-Goldstein, S. Growth control: Brassinosteroid activity gets context. J. Exp. Bot. 2015, 66, 1123–1132. [Google Scholar] [CrossRef] [PubMed]
- Brackmann, K.; Greb, T. Long- and short-distance signaling in the regulation of lateral plant growth. Physiol. Plant 2014, 151, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Welchen, E.; Gonzalez, D.H. Breaking boundaries: Exploring short- and long-distance mitochondrial signalling in plants. New Phytol. 2021, 232, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Arsova, B.; Foster, K.J.; Shelden, M.C.; Bramley, H.; Watt, M. Dynamics in plant roots and shoots minimize stress, save energy and maintain water and nutrient uptake. New Phytol. 2020, 225, 1111–1119. [Google Scholar] [CrossRef]
- Colmer, T.D.; Voesenek, L.A.C.J. Flooding tolerance: Suites of plant traits in variable environments. Funct. Plant Biol. 2009, 36, 665–681. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Colmer, T.D. Plant tolerance of flooding stress--recent advances. Plant Cell Environ. 2014, 37, 211–215. [Google Scholar] [CrossRef]
- Atkin, O.K.; Tjoelker, M.G. Thermal acclimation and the dynamic response of plant respiration to temperature. Trends Plant Sci. 2003, 8, 343–351. [Google Scholar] [CrossRef]
- Niu, X.; Lu, H.; Fan, Y.; Wang, W.; Yuan, Y.; Hawkins, M.; Zhang, J.; Ye, Z.; Miao, M.; Liu, Y.; et al. Manipulation of the transcription factor SlNAC1 for improved tolerance to abiotic stress in tomato. Plant Cell Environ. 2022, 45, 3537–3550. [Google Scholar] [CrossRef]
- Negrão, S.; Schmöckel, S.M.; Tester, M. Evaluating physiological responses of plants to salinity stress. Ann. Bot. 2017, 119, 1–11. [Google Scholar] [CrossRef]
- Zou, Y.; Zhang, Y.; Testerink, C. Root dynamic growth strategies in response to salinity. Plant Cell Environ. 2022, 45, 695–704. [Google Scholar] [CrossRef]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Atkin, O.K.; Macherel, D. The crucial role of plant mitochondria in orchestrating drought tolerance. Ann. Bot. 2009, 103, 581–597. [Google Scholar] [CrossRef] [PubMed]
- Phua, S.Y.; De Smet, B.; Remacle, C.; Chan, K.X.; Van Breusegem, F. Reactive oxygen species and organellar signaling. J. Exp. Bot. 2021, 72, 5807–5824. [Google Scholar] [CrossRef] [PubMed]
- Karban, R. Plant Sensing & Communication; The University of Chicago Press: Chicago, IL, USA, 2015. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Witzany, G. Plant Growth and Development from Biocommunication Perspective. Int. J. Plant Biol. 2025, 16, 63. https://doi.org/10.3390/ijpb16020063
Witzany G. Plant Growth and Development from Biocommunication Perspective. International Journal of Plant Biology. 2025; 16(2):63. https://doi.org/10.3390/ijpb16020063
Chicago/Turabian StyleWitzany, Guenther. 2025. "Plant Growth and Development from Biocommunication Perspective" International Journal of Plant Biology 16, no. 2: 63. https://doi.org/10.3390/ijpb16020063
APA StyleWitzany, G. (2025). Plant Growth and Development from Biocommunication Perspective. International Journal of Plant Biology, 16(2), 63. https://doi.org/10.3390/ijpb16020063





