Analysis of ABC Transporter Gene Expression in Atherosclerosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Data Extraction
2.3. Differential Expression Analysis
3. Results
4. Discussion
Limitations of the Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Herrington, W.; Lacey, B.; Sherliker, P.; Armitage, J.; Lewington, S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ. Res. 2016, 118, 535–546. [Google Scholar] [CrossRef] [PubMed]
- Song, P.; Fang, Z.; Wang, H.; Cai, Y.; Rahimi, K.; Zhu, Y.; Fowkes, F.G.R.; Fowkes, F.J.I.; Rudan, I. Global and regional prevalence, burden, and risk factors for carotid atherosclerosis: A systematic review, meta-analysis, and modelling study. Lancet Glob. Health 2020, 8, e721–e729. [Google Scholar] [CrossRef]
- Fitzgerald, M.L.; Mujawar, Z.; Tamehiro, N. ABC transporters, atherosclerosis and inflammation. Atherosclerosis 2010, 211, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Vasiliou, V.; Vasiliou, K.; Nebert, D.W. Human ATP-binding cassette (ABC) transporter family. Hum. Genom. 2008, 3, 281–290. [Google Scholar] [CrossRef]
- Wilkens, S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015, 7, 14. [Google Scholar] [CrossRef]
- Schumacher, T.; Benndorf, R.A. ABC Transport Proteins in Cardiovascular Disease—A Brief Summary. Molecules 2017, 22, 589. [Google Scholar] [CrossRef]
- Tarling, E.J.; Vallim, T.Q.D.A.; Edwards, P.A. Role of ABC transporters in lipid transport and human disease. Trends Endocrinol. Metab. 2013, 24, 342–350. [Google Scholar] [CrossRef]
- Steenman, M.; Espitia, O.; Maurel, B.; Guyomarch, B.; Heymann, M.-F.; Pistorius, M.-A.; Ory, B.; Heymann, D.; Houlgatte, R.; Gouëffic, Y.; et al. Identification of genomic differences among peripheral arterial beds in atherosclerotic and healthy arteries. Sci. Rep. 2018, 8, 3940. [Google Scholar] [CrossRef]
- Zenkova, D.; Kamenev, V.; Sablina, R.; Artyomov, M.; Sergushichev, A. Phantasus: Visual and Interactive Gene Expression Analysis. Available online: https://ctlab.itmo.ru/phantasus/ (accessed on 27 September 2021).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Neumann, J.; Rose-Sperling, D.; Hellmich, U.A. Diverse relations between ABC transporters and lipids: An overview. Biochim. Biophys. Acta Biomembr. 2017, 1859, 605–618. [Google Scholar] [CrossRef]
- Van Roermund, C.W.T.; Visser, W.F.; Ijlst, L.; van Cruchten, A.; Boek, M.; Kulik, W.; Waterham, H.R.; Wanders, R.J.A. The human peroxisomal ABC half transporter ALDP functions as a homodimer and accepts acyl–CoA esters. FASEB J. 2008, 22, 4201–4208. [Google Scholar] [CrossRef]
- Morita, M.; Imanaka, T. Peroxisomal ABC transporters: Structure, function and role in disease. Biochim. Biophys. Acta Mol. Basis Dis. 2012, 1822, 1387–1396. [Google Scholar] [CrossRef]
- Bisbal, C.; Martinand, C.; Silhol, M.; Lebleu, B.; Salehzada, T. Cloning and Characterization of a RNase L Inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 1995, 270, 13308–13317. [Google Scholar] [CrossRef]
- Hassel, B.; Zhou, A.; Sotomayor, C.; Maran, A.; Silverman, R. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 1993, 12, 3297–3304. [Google Scholar] [CrossRef]
- Le Roy, F.; Bisbal, C.; Silhol, M.; Martinand, C.; Lebleu, B.; Salehzada, T. The 2–5A/RNase L/RNase L Inhibitor (RNI) Pathway Regulates Mitochondrial mRNAs Stability in Interferon α-treated H9 Cells. J. Biol. Chem. 2001, 276, 48473–48482. [Google Scholar] [CrossRef]
- Tian, Y.; Tian, X.; Han, X.; Chen, Y.; Song, C.-Y.; Zhang, Y.-B.; Tian, D.-L. Expression of ATP binding cassette E1 enhances viability and invasiveness of lung adenocarcinoma cells in vitro. Mol. Med. Rep. 2016, 14, 1345–1350. [Google Scholar] [CrossRef]
- Chen, Z.-Q.; Dong, J.; Ishimura, A.; Daar, I.; Hinnebusch, A.G.; Dean, M. The Essential Vertebrate ABCE1 Protein Interacts with Eukaryotic Initiation Factors. J. Biol. Chem. 2006, 281, 7452–7457. [Google Scholar] [CrossRef]
- Pisareva, V.P.; Skabkin, M.A.; Hellen, C.U.T.; Pestova, T.V.; Pisarev, A.V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011, 30, 1804–1817. [Google Scholar] [CrossRef]
- Fabre, O.; Salehzada, T.; Lambert, K.; Seok, Y.B.; Zhou, A.; Mercier, J.; Bisbal, C. RNase L controls terminal adipocyte differentiation, lipids storage and insulin sensitivity via CHOP10 mRNA regulation. Cell Death Differ. 2012, 19, 1470–1481. [Google Scholar] [CrossRef]
- Pennings, M.; Meurs, I.; Ye, D.; Out, R.; Hoekstra, M.; Van Berkel, T.J.; Van Eck, M. Regulation of cholesterol homeostasis in macrophages and consequences for atherosclerotic lesion development. FEBS Lett. 2006, 580, 5588–5596. [Google Scholar] [CrossRef]
- Kotlyarov, S. Participation of ABCA1 Transporter in Pathogenesis of Chronic Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2021, 22, 3334. [Google Scholar] [CrossRef]
- Liu, H.-F.; Cui, K.-F.; Wang, J.-P.; Zhang, M.; Guo, Y.-P.; Li, X.-Y.; Jiang, C. Significance of ABCA1 in human carotid atherosclerotic plaques. Exp. Ther. Med. 2012, 4, 297–302. [Google Scholar] [CrossRef][Green Version]
- Murphy, A.; Bijl, N.; Yvan-Charvet, L.; Welch, C.B.; Bhagwat, N.; Reheman, A.; Wang, Y.; A Shaw, J.; Levine, R.L.; Ni, H.; et al. Cholesterol efflux in megakaryocyte progenitors suppresses platelet production and thrombocytosis. Nat. Med. 2013, 19, 586–594. [Google Scholar] [CrossRef]
- Batetta, B.; Mulas, M.; Petruzzo, P.; Putzolu, M.; Bonatesta, R.; Sanna, F.; Cappai, A.; Brotzu, G.; Dessì, S. Opposite pattern of MDR1 and caveolin-1 gene expression in human atherosclerotic lesions and proliferating human smooth muscle cells. Cell. Mol. Life Sci. 2001, 58, 1113–1120. [Google Scholar] [CrossRef]
- Petruzzo, P.; Cappai, A.; Brotzu, G.; Batetta, B.; Putzolu, M.; Mulas, M.; Bonatesta, R.; Sanna, F.; Dessì, S. Lipid Metabolism and Molecular Changes in Normal andAtherosclerotic Vessels. Eur. J. Vasc. Endovasc. Surg. 2001, 22, 31–36. [Google Scholar] [CrossRef][Green Version]
- Batetta, B.; Dessì, S.; Putzolu, M.; Sanna, F.; Spano, O.; Mulas, M.F.; Petruzzo, P.; Cappai, A.; Brotzu, G. MDR1 Gene Expression in Normal and Atherosclerotic Human Arteries1. J. Vasc. Res. 1999, 36, 261–271. [Google Scholar] [CrossRef]
- Saint-Pol, J.; Candela, P.; Boucau, M.-C.; Fenart, L.; Gosselet, F. Oxysterols decrease apical-to-basolateral transport of Aß peptides via an ABCB1-mediated process in an in vitro Blood-brain barrier model constituted of bovine brain capillary endothelial cells. Brain Res. 2013, 1517, 1–15. [Google Scholar] [CrossRef]
- Wadén, K.; Lengquist, M.; Paulsson-Berne, G.; Hedin, U.; Roy, J.; Matic, L. Abstract 139: Correlation of Clinical Risk Scores for Stroke with Carotid Plaque Gene Expression Profiles in Atherosclerotic Patients. Arterioscler. Thromb. Vasc. Biol. 2019, 39, A139. [Google Scholar] [CrossRef]
- Favre, G.; Laurain, A.; Aranyi, T.; Szeri, F.; Fulop, K.; Le Saux, O.; Duranton, C.; Kauffenstein, G.; Martin, L.; Lefthériotis, G. The ABCC6 Transporter: A New Player in Biomineralization. Int. J. Mol. Sci. 2017, 18, 1941. [Google Scholar] [CrossRef]
- Le Saux, O.; Martin, L.; Aherrahrou, Z.; Leftheriotis, G.; Váradi, A.; Brampton, C.N. The molecular and physiological roles of ABCC6: More than meets the eye. Front. Genet. 2012, 3, 289. [Google Scholar] [CrossRef]
- Atzeni, F.; Sarzi-Puttini, P.; Bevilacqua, M. Calcium Deposition and Associated Chronic Diseases (Atherosclerosis, Diffuse Idiopathic Skeletal Hyperostosis, and Others). Rheum. Dis. Clin. N. Am. 2006, 32, 413–426. [Google Scholar] [CrossRef]
- Trip, M.D.; Smulders, Y.M.; Wegman, J.J.; Hu, X.; Boer, J.M.; Brink, J.B.T.; Zwinderman, A.H.; Kastelein, J.J.; Feskens, E.J.; Bergen, A.A. Frequent mutation in the ABCC6 gene (R1141X) is associated with a strong increase in the prevalence of coronary artery disease. Circulation 2002, 106, 773–775. [Google Scholar] [CrossRef] [PubMed]
- De Vilder, E.Y.G.; Hosen, M.J.; Vanakker, O.M. The ABCC6 Transporter as a Paradigm for Networking from an Orphan Disease to Complex Disorders. BioMed Res. Int. 2015, 2015, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Brodsky, J.L.; Conlin, L.K.; Pawel, B.; Glatz, A.C.; Gafni, R.I.; Schurgers, L.; Uitto, J.; Hakonarson, H.; Deardorff, M.A.; et al. Mutations in the ABCC6 Gene as a Cause of Generalized Arterial Calcification of Infancy: Genotypic Overlap with Pseudoxanthoma Elasticum. J. Investig. Dermatol. 2014, 134, 658–665. [Google Scholar] [CrossRef]
- Keppler, D. Multidrug resistance proteins (MRPs, ABCCs): Importance for pathophysiology and drug therapy. Handb. Exp. Pharmacol. 2011, 299–323. [Google Scholar] [CrossRef]
- Sodani, K.; Patel, A.; Kathawala, R.J.; Chen, Z.-S. Multidrug resistance associated proteins in multidrug resistance. Chin. J. Cancer 2012, 31, 58–72. [Google Scholar] [CrossRef]
- Chutkow, W.A.; Pu, J.; Wheeler, M.T.; Wada, T.; Makielski, J.C.; Burant, C.F.; McNally, E.M. Episodic coronary artery vasospasm and hypertension develop in the absence of Sur2 KATP channels. J. Clin. Investig. 2002, 110, 203–208. [Google Scholar] [CrossRef]
- Tian, Q.; Zhang, J.; Chan, E.; Duan, W.; Zhou, S. Multidrug resistance proteins (MRPs) and implication in drug development. Drug Dev. Res. 2005, 64, 1–18. [Google Scholar] [CrossRef]
- Wang, L.; Li, H.; Tang, Y.; Yao, P. Potential Mechanisms and Effects of Efferocytosis in Atherosclerosis. Front. Endocrinol. 2021, 11, 585285. [Google Scholar] [CrossRef]
- Nofer, J.-R.; Herminghaus, G.; Brodde, M.; Morgenstern, E.; Rust, S.; Engel, T.; Seedorf, U.; Assmann, G.; Bluethmann, H.; Kehrel, B.E. Impaired Platelet Activation in Familial High Density Lipoprotein Deficiency (Tangier Disease). J. Biol. Chem. 2004, 279, 34032–34037. [Google Scholar] [CrossRef]
- Kotlyarov, S.; Kotlyarova, A. The Role of ABC Transporters in Lipid Metabolism and the Comorbid Course of Chronic Obstructive Pulmonary Disease and Atherosclerosis. Int. J. Mol. Sci. 2021, 22, 6711. [Google Scholar] [CrossRef]
- Low, F.G.; Shabir, K.; Brown, J.E.; Bill, R.M.; Rothnie, A.J. Roles of ABCC1 and ABCC4 in Proliferation and Migration of Breast Cancer Cell Lines. Int. J. Mol. Sci. 2020, 21, 7664. [Google Scholar] [CrossRef]
- Weigert, A.; Weis, N.; Brüne, B. Regulation of macrophage function by sphingosine-1-phosphate. Immunobiology 2009, 214, 748–760. [Google Scholar] [CrossRef]
- Takabe, K.; Kim, R.H.; Allegood, J.C.; Mitra, P.; Ramachandran, S.; Nagahashi, M.; Harikumar, K.; Hait, N.C.; Milstien, S.; Spiegel, S. Estradiol Induces Export of Sphingosine 1-Phosphate from Breast Cancer Cells via ABCC1 and ABCG2. J. Biol. Chem. 2010, 285, 10477–10486. [Google Scholar] [CrossRef]
- Yamada, A.; Nagahashi, M.; Aoyagi, T.; Huang, W.-C.; Lima, S.; Hait, N.C.; Maiti, A.; Kida, K.; Terracina, K.P.; Miyazaki, H.; et al. ABCC1-Exported Sphingosine-1-phosphate, Produced by Sphingosine Kinase 1, Shortens Survival of Mice and Patients with Breast Cancer. Mol. Cancer Res. 2018, 16, 1059–1070. [Google Scholar] [CrossRef]
- Nagahashi, M.; Takabe, K.; Terracina, K.P.; Soma, D.; Hirose, Y.; Kobayashi, T.; Matsuda, Y.; Wakai, T. Sphingosine-1-Phosphate Transporters as Targets for Cancer Therapy. BioMed Res. Int. 2014, 2014, 1–7. [Google Scholar] [CrossRef]
- Olivera, A.; Spiegel, S. Sphingosine kinase: A mediator of vital cellular functions. Prostaglandins Other Lipid Mediat. 2001, 64, 123–134. [Google Scholar] [CrossRef]
- Pyne, S.; Pyne, N.J. Sphingosine 1-phosphate signalling and termination at lipid phosphate receptors. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2002, 1582, 121–131. [Google Scholar] [CrossRef]
- Spiegel, S.; Milstien, S. Exogenous and intracellularly generated sphingosine 1-phosphate can regulate cellular processes by divergent pathways. Biochem. Soc. Trans. 2003, 31, 1216–1219. [Google Scholar] [CrossRef]
- Spiegel, S.; Milstien, S. Sphingosine-1-phosphate: An enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003, 4, 397–407. [Google Scholar] [CrossRef]
- Usatyuk, P.V.; He, D.; Bindokas, V.; Gorshkova, I.A.; Berdyshev, E.V.; Garcia, J.G.N.; Natarajan, V. Photolysis of caged sphingosine-1-phosphate induces barrier enhancement and intracellular activation of lung endothelial cell signaling pathways. Am. J. Physiol. Cell. Mol. Physiol. 2011, 300, L840–L850. [Google Scholar] [CrossRef] [PubMed]
- Osgood, D.; Miller, M.C.; Messier, A.A.; Gonzalez, L.; Silverberg, G.D. Aging alters mRNA expression of amyloid transporter genes at the blood-brain barrier. Neurobiol. Aging 2017, 57, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Toornvliet, R.; Vanberckel, B.; Luurtsema, G.; Lubberink, M.; Geldof, A.A.; Bosch, T.M.; Oerlemans, R.; Lammertsma, A.A.; Franssen, E.J.; Berckel, B.N. Effect of age on functional P-glycoprotein in the blood-brain barrier measured by use of (R)-[11C]verapamil and positron emission tomography. Clin. Pharmacol. Ther. 2006, 79, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Bartels, A.L.; Kortekaas, R.; Bart, J.; Willemsen, A.T.; de Klerk, O.L.; de Vries, J.J.; van Oostrom, J.C.; Leenders, K.L. Blood–brain barrier P-glycoprotein function decreases in specific brain regions with aging: A possible role in progressive neurodegeneration. Neurobiol. Aging 2009, 30, 1818–1824. [Google Scholar] [CrossRef]
- Billington, S.; Salphati, L.; Hop, C.E.C.A.; Chu, X.; Evers, R.; Burdette, D.; Rowbottom, C.; Lai, Y.; Xiao, G.; Humphreys, W.G.; et al. Interindividual and Regional Variability in Drug Transporter Abundance at the Human Blood–Brain Barrier Measured by Quantitative Targeted Proteomics. Clin. Pharmacol. Ther. 2019, 106, 228–237. [Google Scholar] [CrossRef]
- Erdő, F.; Krajcsi, P. Age-Related Functional and Expressional Changes in Efflux Pathways at the Blood-Brain Barrier. Front. Aging Neurosci. 2019, 11, 196. [Google Scholar] [CrossRef]
- Guay, S.-P.; Légaré, C.; Houde, A.-A.; Mathieu, P.; Bossé, Y.; Bouchard, L. Acetylsalicylic acid, aging and coronary artery disease are associated with ABCA1 DNA methylation in men. Clin. Epigenet. 2014, 6, 14. [Google Scholar] [CrossRef]
- Houde, A.-A.; Guay, S.-P.; Desgagné, V.; Hivert, M.-F.; Baillargeon, J.-P.; St-Pierre, J.; Perron, P.; Gaudet, D.; Brisson, D.; Bouchard, L. Adaptations of placental and cord bloodABCA1 DNA methylation profile to maternal metabolic status. Epigenetics 2013, 8, 1289–1302. [Google Scholar] [CrossRef]
- Liang, Y.; Yang, X.; Ma, L.; Cai, X.; Wang, L.; Yang, C.; Li, G.; Zhang, M.; Sun, W.; Jiang, Y. Homocysteine-mediated cholesterol efflux via ABCA1 and ACAT1 DNA methylation in THP-1 monocyte-derived foam cells. Acta Biochim. Biophys. Sin. 2013, 45, 220–228. [Google Scholar] [CrossRef]
- Borenstein, A.R.; Mortimer, J.A. Chapter 10—Family History, Genetics, and Down Syndrome. In Alzheimer’s Disease; Borenstein, A.R., Mortimer, J.A., Eds.; Academic Press: San Diego, CA, USA, 2016; pp. 107–120. [Google Scholar]
- Takechi, T.; Hirota, T.; Sakai, T.; Maeda, N.; Kobayashi, D.; Ieiri, I. Interindividual Differences in the Expression of ATP-Binding Cassette and Solute Carrier Family Transporters in Human Skin: DNA Methylation Regulates Transcriptional Activity of the Human ABCC3 Gene. Drug Metab. Dispos. 2018, 46, 628–635. [Google Scholar] [CrossRef]
- Riches, Z.; Abanda, N.; Collier, A.C. BCRP protein levels do not differ regionally in adult human livers, but decline in the elderly. Chem. Interact. 2015, 242, 203–210. [Google Scholar] [CrossRef]
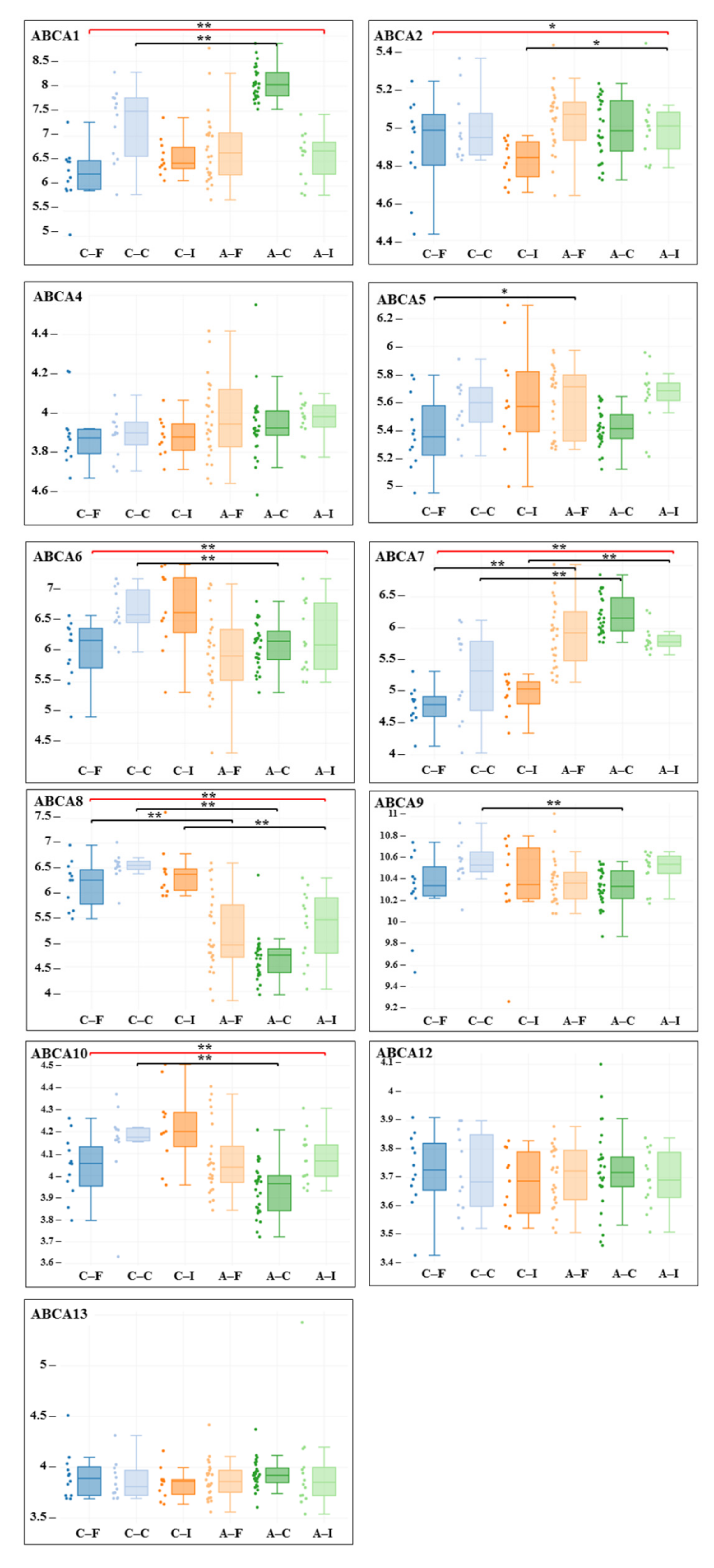
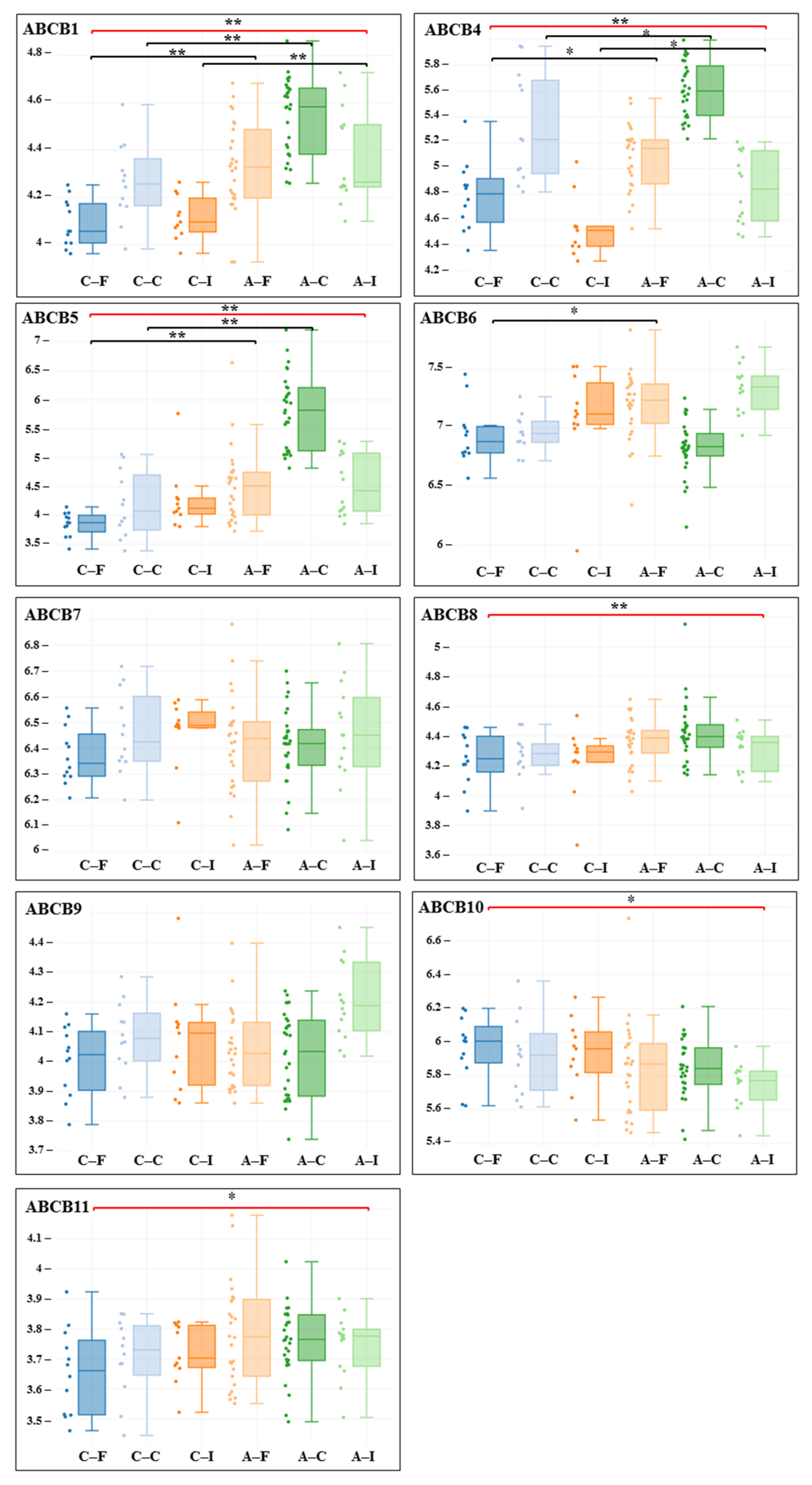
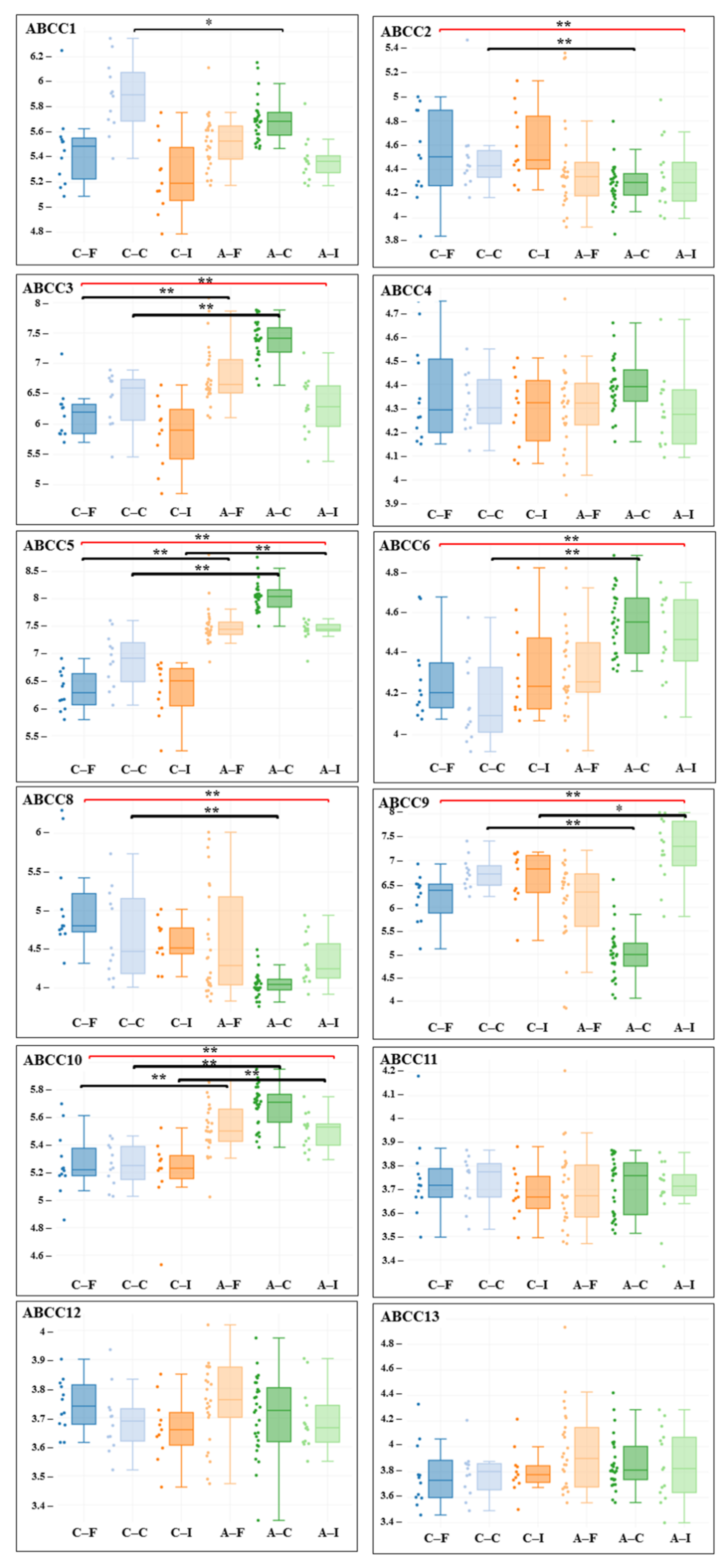
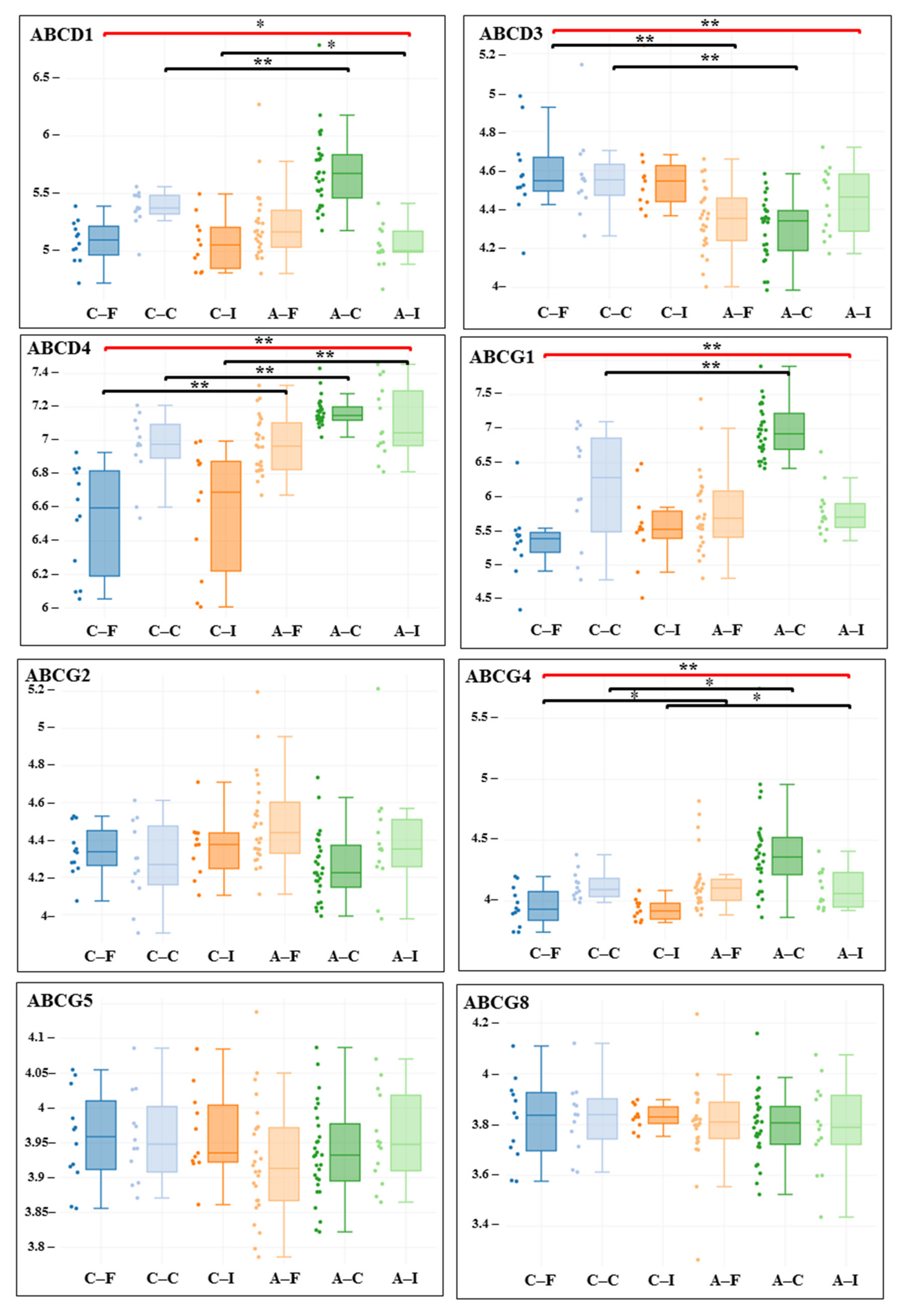
| Gene adj. P. Value LogFC | Carotid Artery | Femoral Artery | Infrapopliteal Artery | All Control/ All Atherosclerosis |
|---|---|---|---|---|
| gene | ABCC5 | ABCC5 | ABCA7 | ABCC5 |
| adj. P. Value | 1.59 × 10−11 | 1.98 × 10−9 | 1.44 × 10−6 | 2.48 × 10−21 |
| logFC | 1.1592 | 1.1792 | 0.8770 | 1.1935 |
| gene | ABCC3 | ABCA7 | ABCC5 | ABCA7 |
| adj. P.Value | 3.46 × 10−9 | 4.71 × 10−7 | 8.04 × 10−6 | 1.26 × 10−17 |
| logFC | 0.9778 | 1.135 | 1.0856 | 1.0271 |
| gene | ABCC10 | ABCF3 | ABCC10 | ABCF3 |
| adj. P.Value | 3.86 × 10−9 | 8.87 × 10−6 | 6.91 × 10−4 | 5.97 × 10−14 |
| logFC | 0.42413 | 0.4166 | 0.3789 | 0.4086 |
| gene | ABCB5 | ABCD4 | ABCD4 | ABCC10 |
| adj. P.Value | 9.57 × 10−9 | 3.83 × 10−5 | 0.00215 | 1.44 × 10−12 |
| logFC | 1.6144 | 0.4432 | 0.5238 | 0.3407 |
| gene | ABCF3 | ABCB1 | ABCF3 | ABCD4 |
| adj. P.Value | 2.08 × 10−7 | 3.33 × 10−4 | 0.00341 | 1.93 × 10−10 |
| logFC | 0.3565 | 0.4041 | 0.4214 | 0.3901 |
| gene | ABCC6 | ABCC3 | ABCB1 | ABCB1 |
| adj. P.Value | 2.40 × 10−7 | 0.00188 | 0.00596 | 2.74 × 10−9 |
| logFC | 0.3966 | 0.6402 | 0.2614 | 0.2710 |
| gene | ABCA7 | ABCC10 | ABCC9 | ABCC3 |
| adj. P.Value | 8.71 × 10−7 | 0.00455 | 0.01277 | 3.70 × 10−9 |
| logFC | 0.9644 | 0.2812 | 0.5452 | 0.8043 |
| gene | ABCA1 | ABCB5 | ABCG4 | ABCB5 |
| adj. P.Value | 3.28 × 10−5 | 0.00596 | 0.02498 | 1.50 × 10−7 |
| logFC | 0.8161 | 0.6618 | 0.1732 | 0.9613 |
| gene | ABCB1 | ABCB4 | ABCB4 | ABCC6 |
| adj. P.Value | 6.07 × 10−5 | 0.01090 | 0.03594 | 5.94 × 10−5 |
| logFC | 0.2798 | 0.2947 | 0.2996 | 0.2018 |
| gene | ABCD4 | ABCA5 | ABCA2 | ABCG4 |
| adj. P.Value | 1.30 × 10−4 | 0.03991 | 0.04503 | 1.19 × 10−4 |
| logFC | 0.2161 | 0.2292 | 0.1712 | 0.2461 |
| Gene adj. P.Value LogFC | Carotid Artery | Femoral Artery | Infrapopliteal Artery | All Control/ All Atherosclerosis |
|---|---|---|---|---|
| gene | ABCA8 | ABCD3 | ABCA8 | ABCA8 |
| adj. P.Value | 1.67 × 10−14 | 0.00106 | 0.00220 | 6.86 × 10−17 |
| LogFC | −1.8533 | −0.3349 | −1.067 | −1.3643 |
| gene | ABCC9 | ABCA8 | ABCD1 | ABCD3 |
| adj. P.Value | 2.20 × 10−12 | 0.00124 | 0.02662 | 2.10 × 10−7 |
| LogFC | −0.7550 | −0.9879 | −0.1222 | −0.2309 |
| gene | ABCC8 | ABCF2 | - | ABCF2 |
| adj. P.Value | 6.57 × 10−6 | 0.00212 | 2.00 × 10−5 | |
| LogFC | −0.6352 | −0.2322 | −0.2194 | |
| gene | ABCA10 | ABCE1 | - | ABCC8 |
| adj. P.Value | 2.20 × 10−4 | 0.01666 | 1.33 × 10−4 | |
| LogFC | −0.2184 | −0.2573 | −0.4701 | |
| gene | ABCD3 | - | - | ABCA10 |
| adj. P.Value | 2.39 × 10−4 | 0.00125 | ||
| LogFC | −0.2705 | −0.1215 | ||
| gene | ABCA9 | - | - | ABCA6 |
| adj. P.Value | 0.00239 | 0.00302 | ||
| LogFC | −0.2275 | −0.3863 | ||
| gene | ABCF2 | - | - | ABCC9 |
| adj. P.Value | 0.00842 | 0.00321 | ||
| LogFC | −0.2754 | −0.6535 | ||
| gene | ABCC2 | - | - | ABCC2 |
| adj. P.Value | 0.017797 | 0.0094999 | ||
| LogFC | −0.222 | −0.18827 | ||
| gene | ABCC1 | - | - | ABCE1 |
| adj. P.Value | 0.034844 | 0.013257 | ||
| LogFC | −0.18642 | −0.11818 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kotlyarov, S.; Kotlyarova, A. Analysis of ABC Transporter Gene Expression in Atherosclerosis. Cardiogenetics 2021, 11, 204-218. https://doi.org/10.3390/cardiogenetics11040021
Kotlyarov S, Kotlyarova A. Analysis of ABC Transporter Gene Expression in Atherosclerosis. Cardiogenetics. 2021; 11(4):204-218. https://doi.org/10.3390/cardiogenetics11040021
Chicago/Turabian StyleKotlyarov, Stanislav, and Anna Kotlyarova. 2021. "Analysis of ABC Transporter Gene Expression in Atherosclerosis" Cardiogenetics 11, no. 4: 204-218. https://doi.org/10.3390/cardiogenetics11040021
APA StyleKotlyarov, S., & Kotlyarova, A. (2021). Analysis of ABC Transporter Gene Expression in Atherosclerosis. Cardiogenetics, 11(4), 204-218. https://doi.org/10.3390/cardiogenetics11040021






