New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata
Abstract
1. Introduction
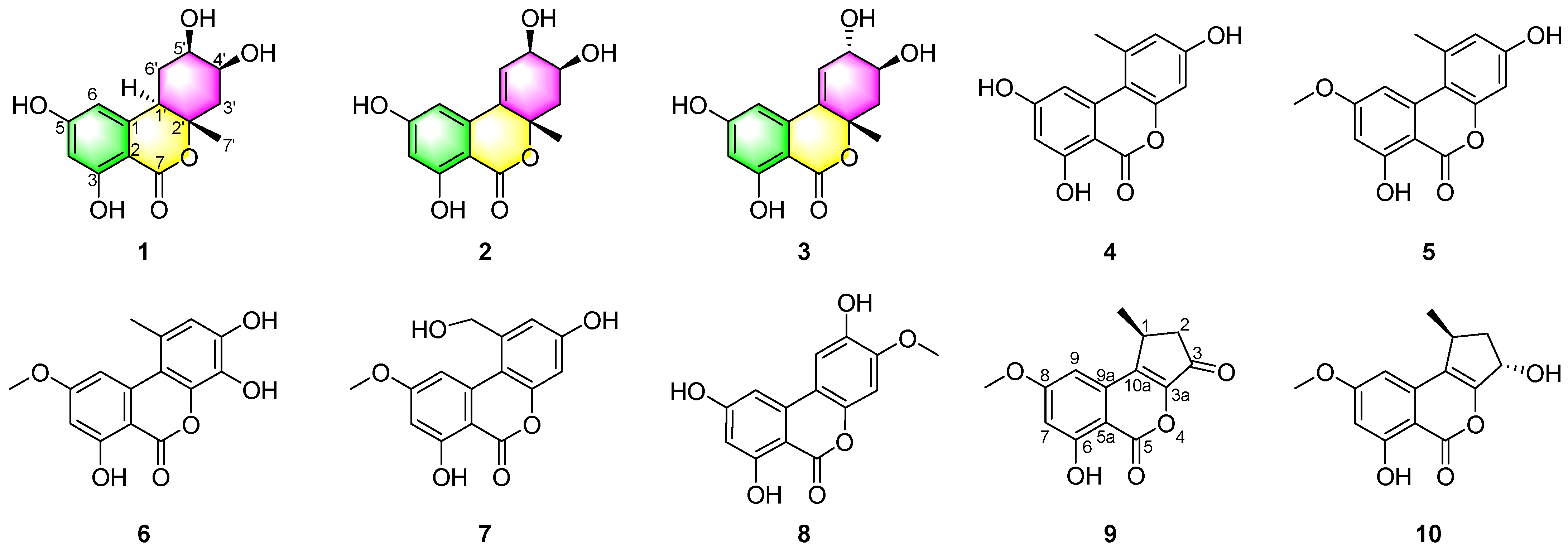
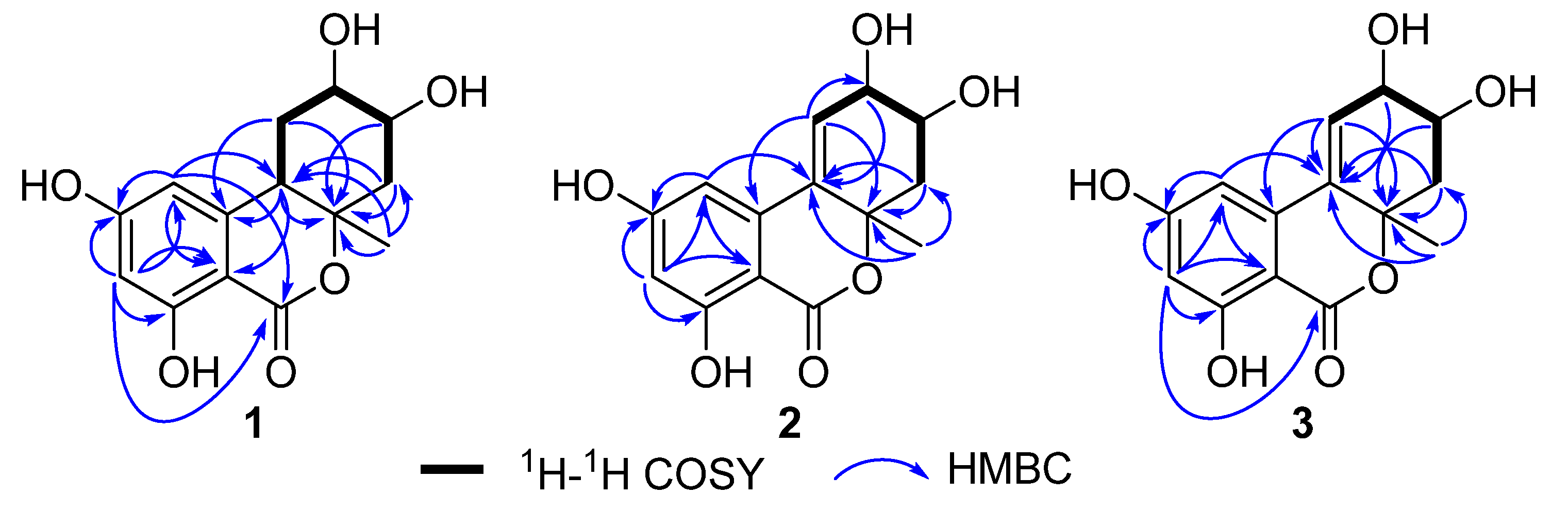
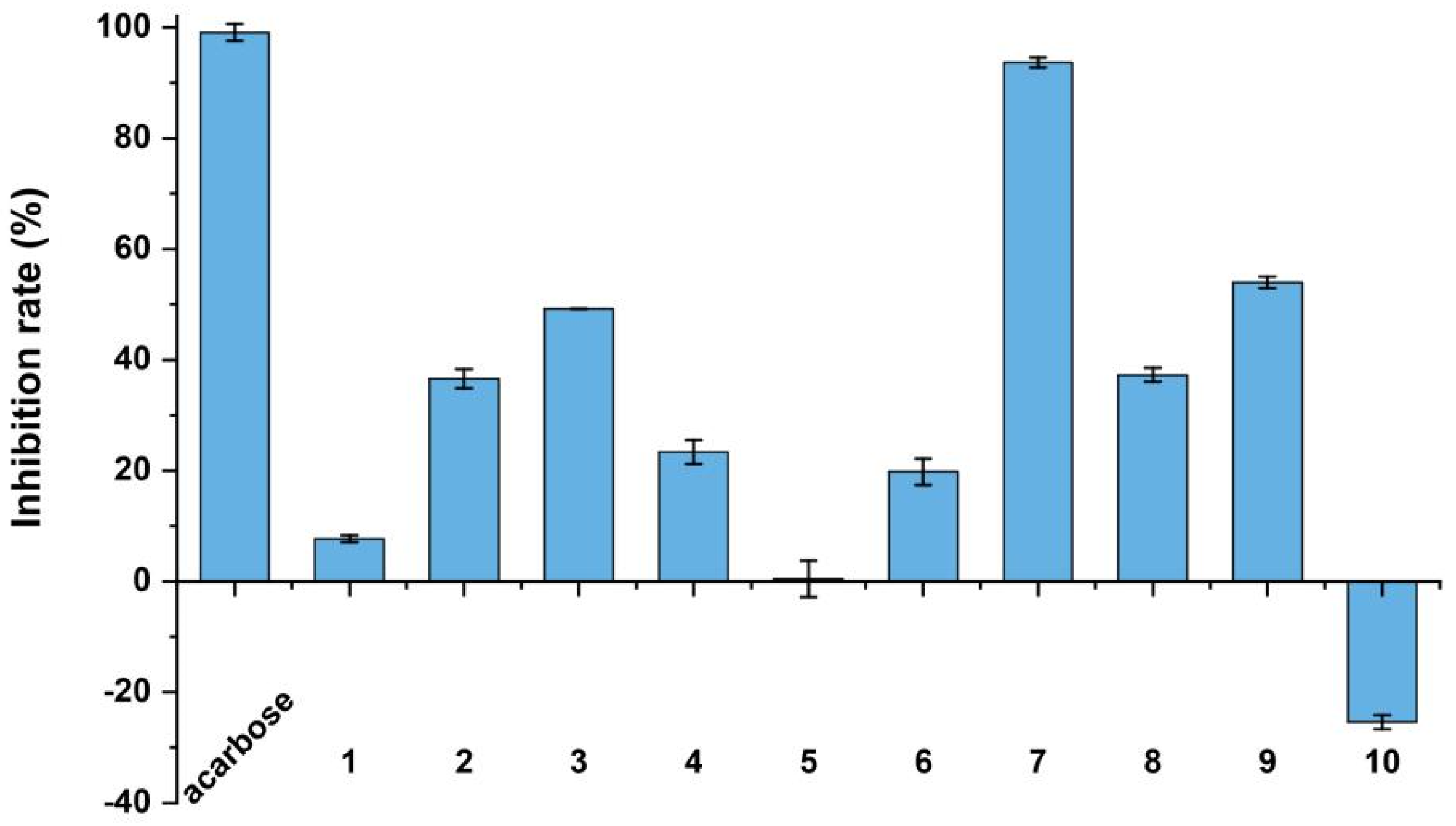
2. Results
3. Experimental Section
3.1. General Experimental Procedure
3.2. Strain and Fermentation
3.3. Extraction and Isolation
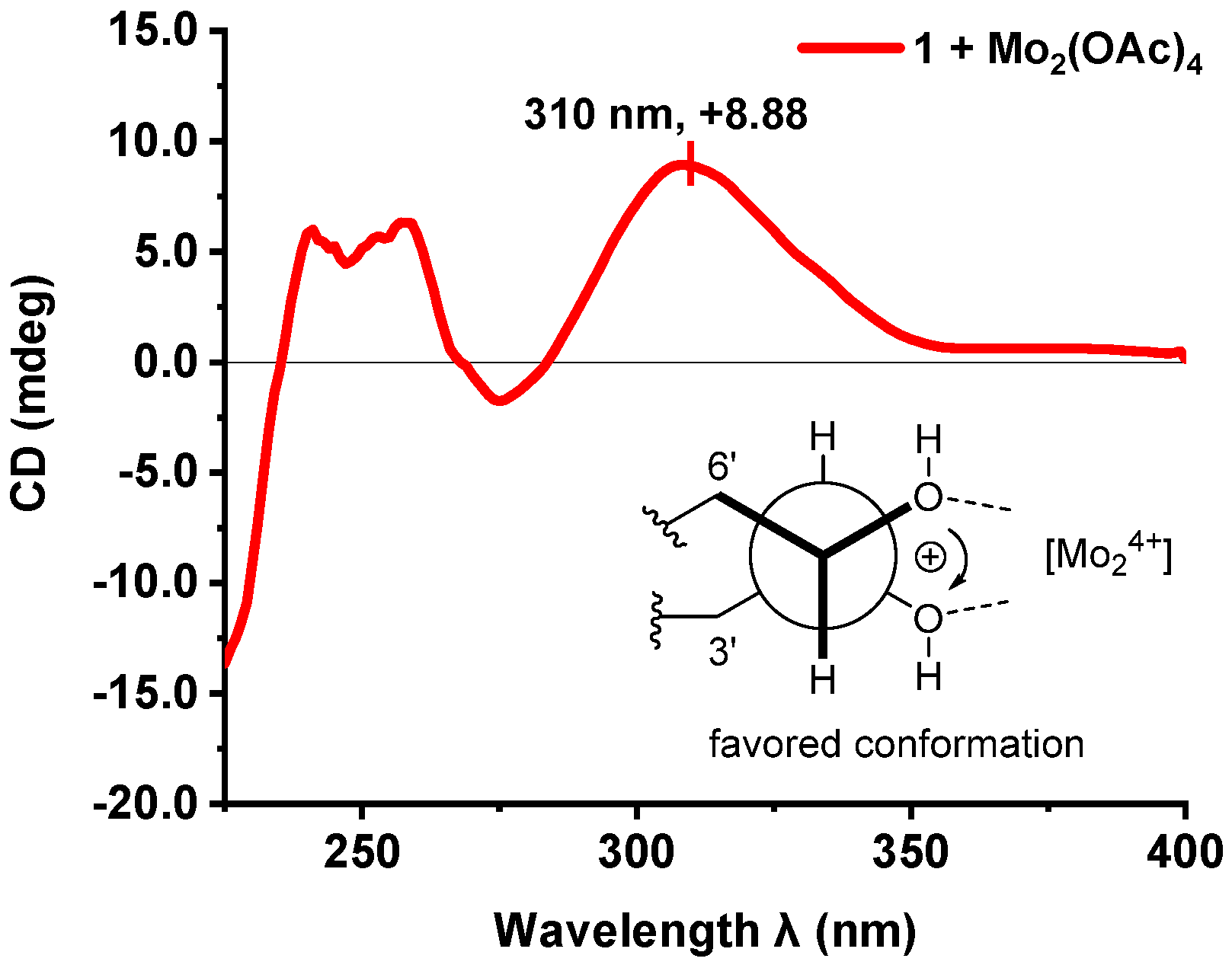
3.4. Absolute Configuration of the 4′,5′-Diol Moiety in 1
3.5. ECD Calculation
3.6. Bioassays for Cytotoxic Activity
3.7. Antioxidant Assay
3.8. Bioassays for α-Glucosidase Inhibition Assay
3.9. Enzyme Kinetics of α-Glucosidase Inhibition Assay
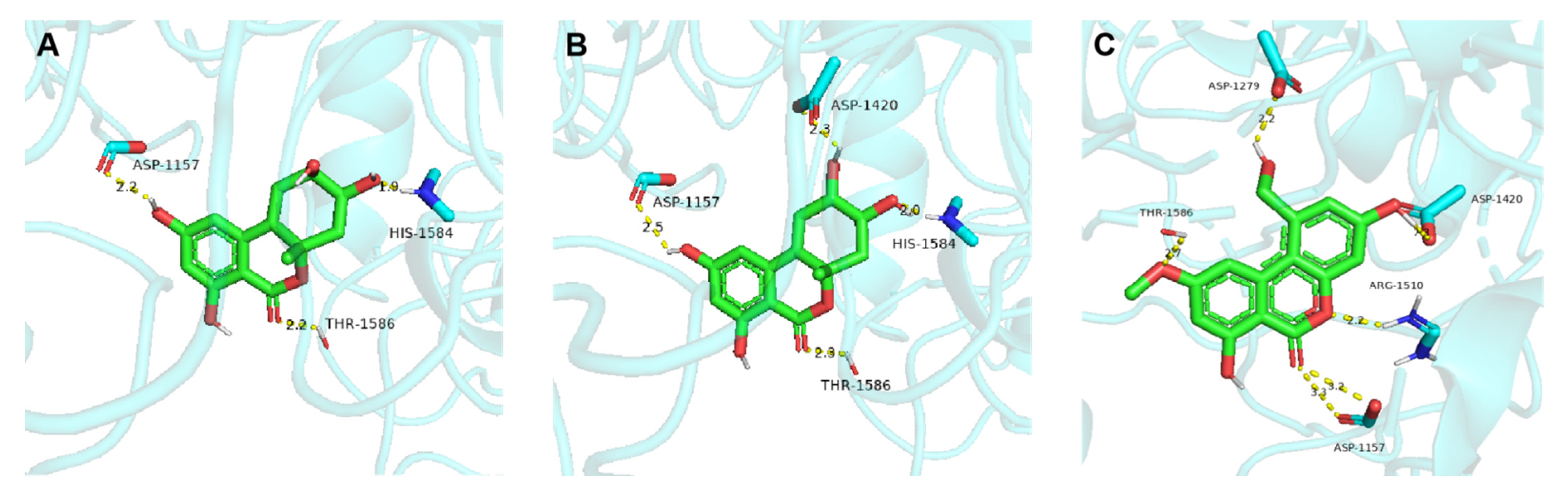
3.10. Molecular Docking Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lai, D.W.; Wang, A.L.; Cao, Y.H.; Zhou, K.Y.; Mao, Z.L.; Dong, X.J.; Tian, J.; Xu, D.; Dai, J.G.; Peng, Y.; et al. Bioactive dibenzo-α-pyrone derivatives from the endophytic fungus Rhizopycnis vagum Nitaf22. J. Nat. Prod. 2016, 79, 2022–2031. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.S.; Tian, K.L.; Li, Y.; Ji, W.X.; Liu, F.; Khan, B.; Yan, W.; Ye, Y.H. Enantiomeric dibenzo-α-pyrone derivatives from Alternaria alternata ZHJG5 and their potential as agrochemicals. J. Agric. Food Chem. 2020, 68, 15115–15122. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Fu, L.Y.; Zhang, Z.H.; Dong, X.J.; Xu, D.; Mao, Z.L.; Liu, Y.; Lai, D.W.; Zhou, L.G. Dibenzo-α-pyrones from the endophytic fungus Alternaria sp. Samif01: Isolation, structure elucidation, and their antibacterial and antioxidant activities. Nat. Prod. Res. 2017, 31, 387–396. [Google Scholar] [CrossRef] [PubMed]
- Shantanu, P.; Stephen, D. Toxicity of the Alternaria spp metabolites, tenuazonic acid, alternariol, altertoxin-I, and alternariol monomethyl ether to brine shrimp (Artemia salina L.) larvae. J. Sci. Food Agric. 1994, 66, 493–496. [Google Scholar]
- Aly, A.H.; Edrada-Ebe, R.; Indriani, I.D.; Wray, V.; Müller, W.E.; Totzke, F.; Zirrgiebel, U.; Schächtele, C.; Kubbutat, M.H.; Lin, W.H.; et al. Cytotoxic metabolites from the fungal endophyte Alternaria sp. and their subsequent detection in its host plant Polygonum senegalense. J. Nat. Prod. 2008, 71, 972–980. [Google Scholar] [CrossRef] [PubMed]
- Schreck, I.; Deigendesch, U.; Burkhardt, B.; Marko, D.; Weiss, C. The Alternaria mycotoxins alternariol and alternariol methyl ether induce cytochrome P450 1A1 and apoptosis in murine hepatoma cells dependent on the aryl hydrocarbon receptor. Arch. Toxicol. 2012, 86, 625–632. [Google Scholar] [CrossRef]
- Aichinger, G. Natural dibenzo-α-pyrones: Friends or foes? Int. J. Mol. Sci. 2021, 22, 13063. [Google Scholar] [CrossRef]
- Mao, Z.L.; Lai, D.W.; Liu, X.D.; Fu, X.X.; Meng, J.J.; Wang, A.L.; Wang, X.H.; Sun, W.B.; Liu, Z.L.; Zhou, L.G.; et al. Dibenzo-α-pyrones: A new class of larvicidal metabolites against Aedes aegypti from the endophytic fungus Hyalodendriella sp. Ponipodef12. Pest Manag. Sci. 2017, 73, 1478–1485. [Google Scholar] [CrossRef]
- Jiao, P.; Gloer, J.B.; Campbell, J.; Shearer, C.A. Altenuene derivatives from an unidentified freshwater fungus in the family Tubeufiaceae. J. Nat. Prod. 2006, 69, 612–615. [Google Scholar] [CrossRef]
- Wang, A.L.; Zhao, S.J.; Gu, G.; Xu, D.; Zhang, X.P.; Lai, D.W.; Zhou, L.G. Rhizovagine A, an unusual dibenzo-α-pyrone alkaloid from the endophytic fungus Rhizopycnis vagum Nitaf22. RSC Adv. 2020, 10, 27894–27898. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.; Miao, J. New antiproliferative dibenzo-α-pyrone from whole plants of Centella asiatica. Nat. Prod. Commun. 2021, 16, 1934578X211003019. [Google Scholar] [CrossRef]
- Koch, K.; Podlech, J.; Pfeiffer, E.; Metzler, M. Total synthesis of alternariol. J. Org. Chem. 2005, 70, 3275–3276. [Google Scholar] [CrossRef] [PubMed]
- Altemöller, M.; Podlech, J. Total synthesis of dehydroaltenuene A. Revision of the structure and total synthesis of dihydroaltenuene B. J. Nat. Prod. 2009, 72, 1288–1290. [Google Scholar] [CrossRef] [PubMed]
- Teske, J.A.; Deiters, A. A cyclotrimerization route to cannabinoids. Org. Lett. 2008, 10, 2195–2198. [Google Scholar] [CrossRef] [PubMed]
- Nandaluru, P.R.; Bodwell, G.J. Multicomponent synthesis of 6H-dibenzo[b,d]pyran-6-ones and a total synthesis of cannabinol. Org. Lett. 2012, 14, 310–313. [Google Scholar] [CrossRef]
- Edwards, J.P.; West, S.J.; Marschke, K.B.; Mais, D.E.; Gottardis, M.M.; Jones, T.K. 5-Aryl-1,2-dihydro-5H-chromeno[3,4-f]quinolines as potent, orally active, nonsteroidal progesterone receptor agonists: The effect of D-ring substituents. J. Med. Chem. 1998, 41, 303–310. [Google Scholar] [CrossRef]
- Coghlan, M.J.; Kym, P.R.; Elmore, S.W.; Wang, A.X.; Luly, J.R.; Wilcox, D.; Stashko, M.; Lin, C.W.; Miner, J.; Tyree, C.; et al. Synthesis and characterization of non-steroidal ligands for the glucocorticoid receptor: Selective quinoline derivatives with prednisolone-equivalent functional activity. J. Med. Chem. 2001, 44, 2879–2885. [Google Scholar] [CrossRef]
- Mao, Z.L.; Sun, W.B.; Fu, L.Y.; Luo, H.Y.; Lai, D.W.; Zhou, L.G. Natural dibenzo-α-pyrones and their bioactivities. Molecules 2014, 19, 5088–5108. [Google Scholar] [CrossRef]
- Baron, A.D. Postprandial hyperglycaemia and alpha-glucosidase inhibitors. Diabetes Res. Clin. Pract. 1998, 40, 51–55. [Google Scholar] [CrossRef]
- Heacock, P.M.; Hertzler, S.R.; Williams, J.A.; Wolf, B.W. Effects of a medical food containing an herbal alpha-glucosidase inhibitor on postprandial glycemia and insulinemia in healthy adults. J. Am. Diet. Assoc. 2005, 105, 65–71. [Google Scholar] [CrossRef]
- Asano, N. Glycosidase inhibitors: Update and perspectives on practical use. Glycobiology 2003, 13, 93R–104R. [Google Scholar] [CrossRef] [PubMed]
- Usman, B.; Sharma, N.; Satija, S.; Mehta, M.; Vyas, M.; Khatik, G.L.; Khurana, N.; Hansbro, P.M.; Williams, K.; Dua, K. Recent developments in alpha-glucosidase inhibitors for management of type-2 diabetes: An update. Curr. Pharm. Des. 2019, 25, 2510–2525. [Google Scholar] [CrossRef] [PubMed]
- Shah, B.; Sartaj, L.; Ali, F.; Shah, A.; Khan, T. Plant extracts are the potential inhibitors of α-amylase: A review. MOJ Bioequiv. Bioavailab. 2018, 5, 270–273. [Google Scholar]
- Wang, X.J.; Li, J.Y.; Shang, J.Q.; Bai, J.; Wu, K.; Liu, J.; Yang, Z.J.; Ou, H.; Shao, L. Metabolites extracted from microorganisms as potential inhibitors of glycosidases (α-glucosidase and α-amylase): A review. Front. Microbiol. 2022, 13, 1050869. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, Y.; Miki, K.; Suzuki, T.; Nishio, K.; Sugita, T.; Kinoshita, K.; Takahashi, K.; Koyama, K. Antiangiogenic metabolites from a marine-derived fungus, Hypocrea vinosa. J. Nat. Prod. 2010, 73, 579–582. [Google Scholar] [CrossRef]
- Ren, J.W.; Huo, R.Y.; Liu, G.R.; Liu, L. New andrastin-type meroterpenoids from the marine-derived fungus Penicillium sp. Mar. Drugs 2021, 19, 189. [Google Scholar] [CrossRef]
- Chen, X.H.; Zhou, G.L.; Sun, C.X.; Zhang, X.M.; Zhang, G.J.; Zhu, T.J.; Li, J.; Che, Q.; Li, D.H. Penicacids E-G, three new mycophenolic acid derivatives from the marine-derived fungus Penicillium parvum HDN17-478. Chin. J. Nat. Med. 2020, 18, 850–854. [Google Scholar] [CrossRef]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine nature products. Nat. Prod. Rep. 2020, 37, 175–223. [Google Scholar] [CrossRef]
- Thomma, B.P. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef]
- Lou, J.F.; Fu, L.Y.; Peng, Y.L.; Zhou, L.G. Metabolites from Alternaria fungi and their bioactivities. Molecules 2013, 18, 5891–5935. [Google Scholar] [CrossRef]
- Shi, Z.Z.; Miao, F.P.; Fang, S.T.; Liu, X.H.; Yin, X.L.; Ji, N.Y. Sesteralterin and tricycloalterfurenes A-D: Terpenes with rarely occurring frameworks from the marine-alga-epiphytic fungus Alternaria alternata k21-1. J. Nat. Prod. 2017, 80, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Z.; Fang, S.T.; Miao, F.P.; Ji, N.Y. Two new tricycloalternarene esters from an alga-epiphytic isolate of Alternaria alternata. Nat. Prod. Res. 2018, 32, 2523–2528. [Google Scholar] [CrossRef] [PubMed]
- Li, F.L.; Ye, Z.; Huang, Z.Y.; Chen, X.; Sun, W.G.; Gao, W.X.; Zhang, S.T.; Cao, F.; Wang, J.P.; Hu, Z.X.; et al. New α-pyrone derivatives with herbicidal activity from the endophytic fungus Alternaria brassicicola. Bioorg. Chem. 2021, 117, 105452. [Google Scholar] [CrossRef] [PubMed]
- Kustrzeba-Wójcicka, I.; Siwak, E.; Terlecki, G.; Wolańczyk-Mędrala, A.; Mędrala, W. Alternaria alternata and its allergens: A comprehensive review. Clin. Rev. Allergy Immunol. 2014, 47, 354–365. [Google Scholar] [CrossRef]
- Jones, E.B.G.; Pang, K.L.; Abdel-Wahab, M.A.; Scholz, B.; Hyde, K.D.; Boekhout, T.; Ebel, R.; Rateb, M.E.; Henderson, L.; Sakayaroj, J.; et al. An online resource for marine fungi. Fungal Divers. 2019, 96, 347–433. [Google Scholar] [CrossRef]
- Li, F.L.; Lin, S.; Zhang, S.T.; Pan, L.F.; Chai, C.W.; Su, J.C.; Yang, B.Y.; Liu, J.J.; Wang, J.P.; Hu, Z.X.; et al. Modified fusicoccane-type diterpenoids from Alternaria brassicicola. J. Nat. Prod. 2020, 83, 1931–1938. [Google Scholar] [CrossRef]
- Yang, C.L.; Wu, H.M.; Liu, C.L.; Zhang, X.; Guo, Z.K.; Chen, Y.; Liu, F.; Liang, Y.; Jiao, R.H.; Tan, R.X.; et al. Bialternacins A-F, aromatic polyketide dimers from an endophytic Alternaria sp. J. Nat. Prod. 2019, 82, 792–797. [Google Scholar] [CrossRef]
- Wu, J.C.; Hou, Y.N.; Xu, Q.H.; Jin, X.J.; Chen, Y.X.; Fang, J.G.; Hu, B.R.; Wu, Q.X. (±)-Alternamgin, a pair of enantiomeric polyketides, from the endophytic fungi Alternaria sp. MG1. Org. Lett. 2019, 21, 1551–1554. [Google Scholar] [CrossRef]
- Zhang, G.J.; Wu, G.W.; Zhu, T.J.; Kurtán, T.; Mándi, A.; Jiao, J.Y.; Li, J.; Qi, X.; Gu, Q.Q.; Li, D.H. Meroterpenoids with diverse ring systems from the sponge-associated fungus Alternaria sp. JJY-32. J. Nat. Prod. 2013, 76, 1946–1957. [Google Scholar] [CrossRef]
- Pang, X.Y.; Lin, X.P.; Wang, P.; Zhou, X.F.; Yang, B.; Wang, J.F.; Liu, Y.H. Perylenequione derivatives with anticancer activities Isolated from the marine sponge-derived fungus, Alternaria sp. SCSIO41014. Mar. Drugs 2018, 16, 280. [Google Scholar] [CrossRef]
- Kim, M.Y.; Sohn, J.H.; Ahn, J.S.; Oh, H. Alternaramide, a cyclic depsipeptide from the marine-derived fungus Alternaria sp. SF-5016. J. Nat. Prod. 2009, 72, 2065–2068. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.R.; Huo, R.Y.; Niu, S.B.; Song, F.H.; Liu, L. Two new cytotoxic decalin derivatives from marine-derived fungus Talaromyces sp. Chem. Biodivers. 2022, 19, e202100990. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.H.; Zhang, J.X.; Huo, R.Y.; Xue, Y.X.; Hong, K.; Liu, L. Sulfur-containing benzofurans and α-pyrones from the mangrove-derived fungus Talaromyces sp. WHUF0341. Front. Mar. Sci. 2022, 9, 1034945. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the "One Strain Many Compounds" (OSMAC) principle to marine microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed]
- Frelek, J.; Geiger, M.; Voelter, W. Transition-metal complexes as auxiliary chromophores in chiroptical studies on carbohydrates. Curr. Org. Chem. 1999, 3, 117–146. [Google Scholar] [CrossRef]
- Di Bari, L.; Pescitelli, G.; Pratelli, C.; Pini, D.; Salvadori, P. Determination of absolute configuration of acyclic 1,2-diols with Mo2(OAc)4. 1. snatzke’s method revisited. J. Org. Chem. 2001, 66, 4819–4825. [Google Scholar] [CrossRef]
- Naganuma, M.; Nishida, M.; Kuramochi, K.; Sugawara, F.; Yoshida, H.; Mizushina, Y. 1-Deoxyrubralactone, a novel specific inhibitor of families X and Y of eukaryotic DNA polymerases from a fungal strain derived from sea algae. Bioorg. Med. Chem. 2008, 16, 2939–2944. [Google Scholar] [CrossRef]
- Zhang, J.C.; Chen, G.Y.; Li, X.Z.; Hu, M.; Wang, B.Y.; Ruan, B.H.; Zhou, H.; Zhao, L.X.; Zhou, J.; Ding, Z.T.; et al. Phytotoxic, antibacterial, and antioxidant activities of mycotoxins and other metabolites from Trichoderma sp. Nat. Prod. Res. 2017, 31, 2745–2752. [Google Scholar] [CrossRef]
- Pero, R.W.; Harvan, D.; Blois, M.C. Isolation of the toxin, altenuisol, from the fungus, Alternaria tenuis auct. Tetrahedron Lett. 1973, 14, 945–948. [Google Scholar] [CrossRef]
- Ye, F.; Chen, G.D.; He, J.W.; Li, X.X.; Sun, X.; Guo, L.D.; Li, Y.; Gao, H. Xinshengin, the first sltenusin with tetracyclic skeleton core from Phialophora spp. Tetrahedron Lett. 2013, 54, 4551–4554. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; revision C 01; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Guo, L.F.; Lin, J.; Niu, S.B.; Liu, S.C.; Liu, L. Pestalotiones A-D: Four new secondary metabolites from the plant endophytic fungus Pestalotiopsis theae. Molecules 2020, 25, 470. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.J.; Tian, D.M.; Wei, J.H.; Li, C.; Ma, Y.H.; Gou, X.S.; Shen, Y.R.; Chen, M.; Zhang, S.H.; Li, J.; et al. Citrinin derivatives from Penicillium citrinum Y34 that inhibit α-glucosidase and ATP-citrate lyase. Front. Mar. Sci. 2022, 9, 961356. [Google Scholar] [CrossRef]



| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δH (J in Hz) | δC, mult. | δH (J in Hz) | δC, mult. | δH (J in Hz) | δC, mult. | |
| 1 | 145.0, C | 140.7, C | 141.1, C | |||
| 2 | 101.6, C | 100.6, C | 100.6, C | |||
| 3 | 165.6, C | 165.2, C | 165.2, C | |||
| 4 | 6.21 (s) | 101.8, CH | 6.28 (d, 2.2) | 103.6, CH | 6.30 (d, 2.2) | 103.5, CH |
| 5 | 166.6, C | 167.1, C | 166.7, C | |||
| 6 | 6.27 (s) | 105.1, CH | 6.51 (d, 2.2) | 104.5, CH | 6.52 (d, 2.2) | 104.5, CH |
| 7 | 170.6, C | 170.4, C | 170.5, C | |||
| 1′ | 3.14 (d, 12.4) | 43.5, CH | 135.2, C | 135.0, C | ||
| 2′ | 84.7, C | 82.4, C | 82.3, C | |||
| 3′α | 2.06 (dd, 12.4, 3.1) | 43.4, CH2 | 2.21 (dd, 14.0, 2.8) | 40.8, CH2 | 2.40 (dd, 14.4, 3.9) | 40.9, CH2 |
| 3′β | 2.23 (dd, 12.4, 3.3) | 2.38 (dd, 14.0, 6.3) | 1.97 (dd, 14.4, 9.4) | |||
| 4′ | 4.11 (m) | 70.0, CH | 4.12 (m) | 68.2, CH | 3.78 (ddd, 9.4, 5.9, 3.9) | 70.7, CH |
| 5′ | 3.86 (m) | 72.2, CH | 4.37 (t, 3.3) | 68.4, CH | 4.07 (dd, 5.9, 2.8) | 72.3, CH |
| 6′α | 2.23 (dd, 12.4, 3.3) | 28.5, CH2 | 6.16 (d, 3.3) | 129.7, CH | 6.16 (d, 2.8) | 131.0, CH |
| 6′β | 1.71 (q, 12.4) | |||||
| 7′ | 1.36 (s) | 20.9, CH3 | 1.61 (s) | 27.9, CH3 | 1.50 (s) | 28.0, CH3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Zhang, B.; Cai, L.; Liu, L. New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata. Mar. Drugs 2022, 20, 778. https://doi.org/10.3390/md20120778
Zhang J, Zhang B, Cai L, Liu L. New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata. Marine Drugs. 2022; 20(12):778. https://doi.org/10.3390/md20120778
Chicago/Turabian StyleZhang, Jinxin, Baodan Zhang, Lei Cai, and Ling Liu. 2022. "New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata" Marine Drugs 20, no. 12: 778. https://doi.org/10.3390/md20120778
APA StyleZhang, J., Zhang, B., Cai, L., & Liu, L. (2022). New Dibenzo-α-pyrone Derivatives with α-Glucosidase Inhibitory Activities from the Marine-Derived Fungus Alternaria alternata. Marine Drugs, 20(12), 778. https://doi.org/10.3390/md20120778







