Bile Derivative T3K Ameliorates Colitis by Regulating the Intestinal Microbiota-Bile Acid Axis
Abstract
1. Introduction
2. Materials and Methods
2.1. Compounds
2.2. Animals and Cells
2.3. Bioinformatics Database Search and Analysis
2.4. Establishment of DSS-Induced Colitis Mouse Model
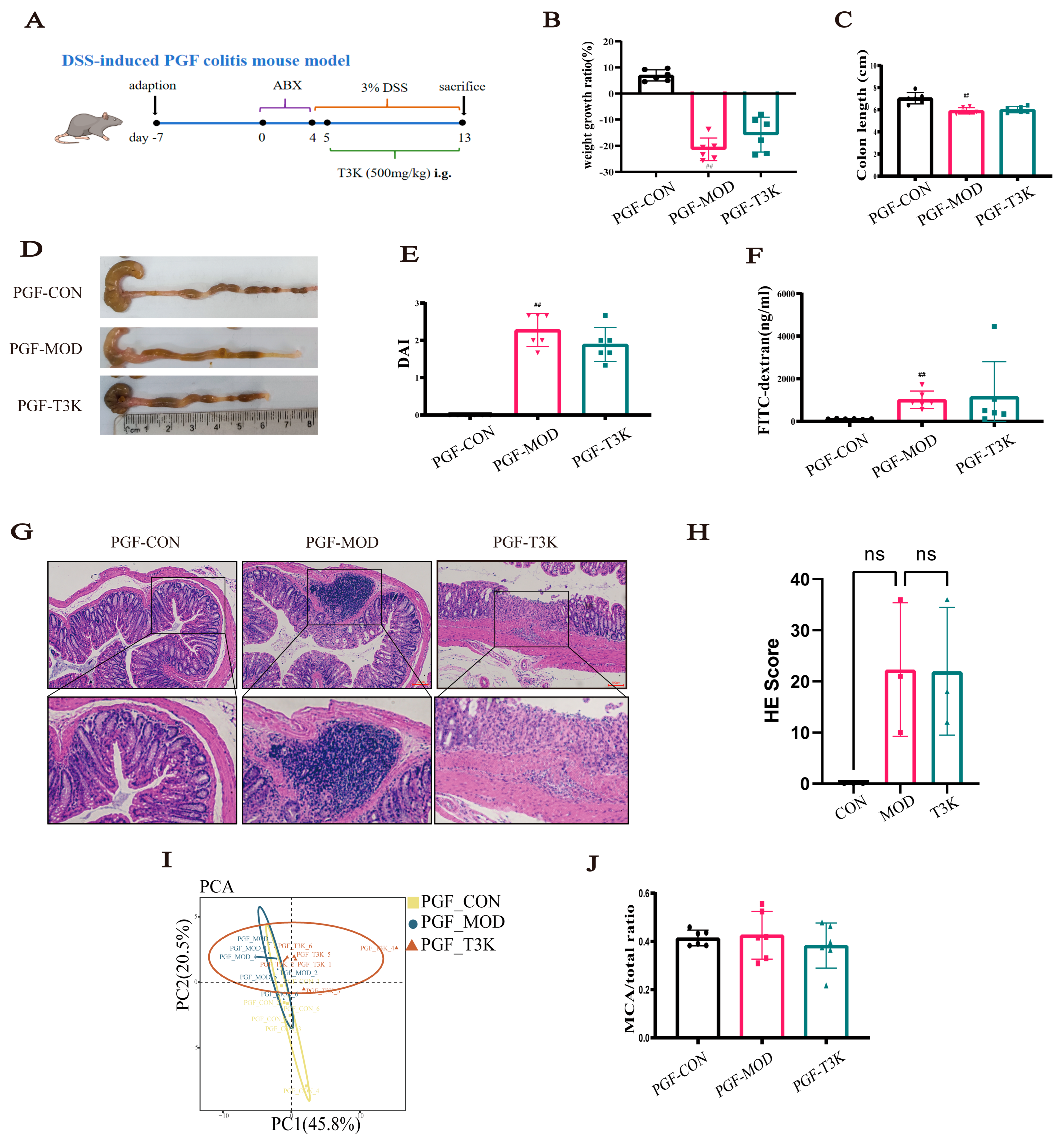
2.5. Establishment of DSS-Induced PGF Colitis Mouse Model
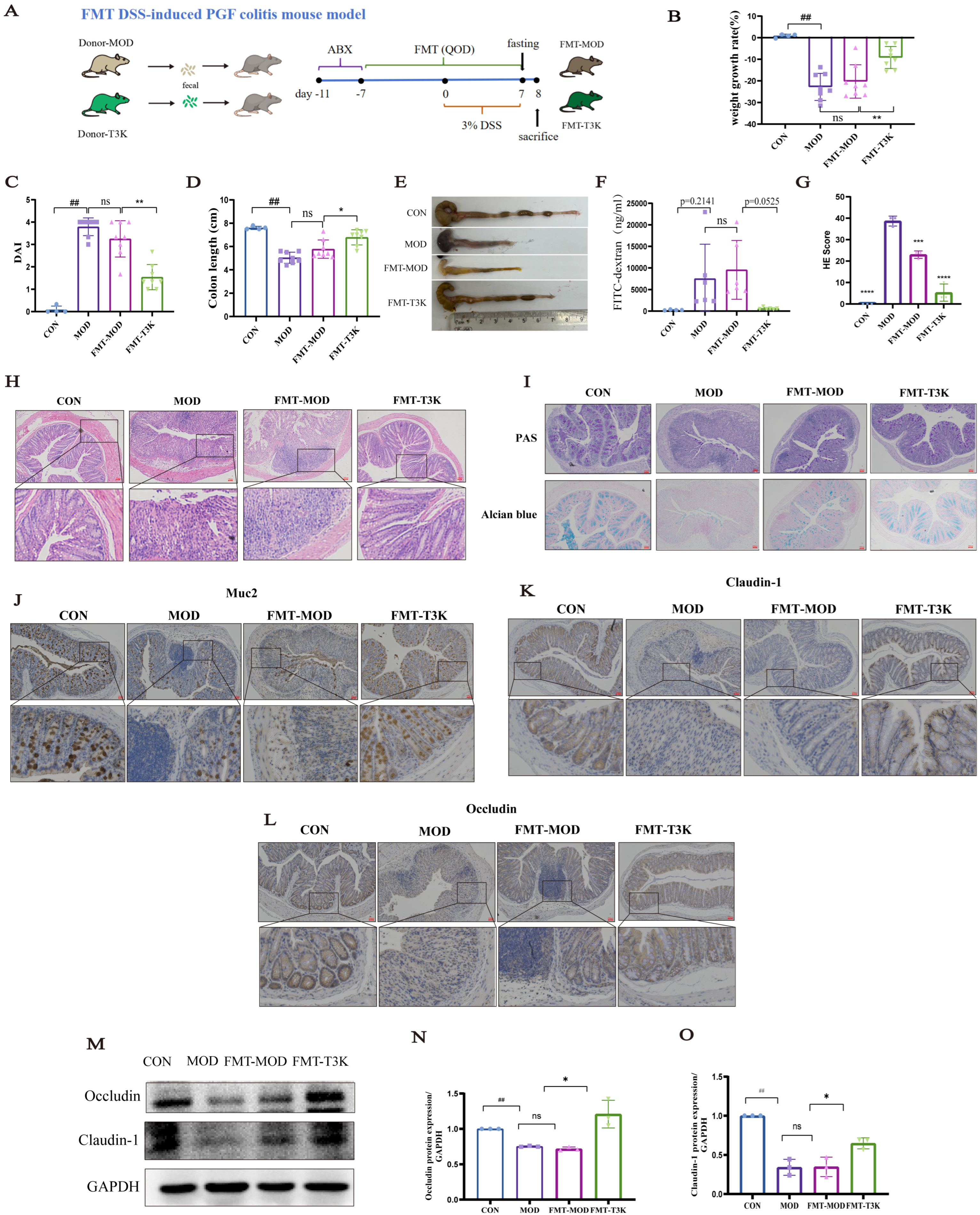
2.6. Establishment of FMT DSS-Induced PGF Colitis Mouse Model
2.7. Intestinal Permeability Assay
2.8. Histopathological Examination
2.9. 16S rRNA Sequencing Analysis
2.10. Bile Acid Analysis
2.11. Western Blot Assay
2.12. Immunohistochemical Staining
2.13. Alcian Blue Staining
2.14. Periodic Acid-Schiff (PAS) Staining
2.15. Immunofluorescence
2.16. Transwell Assays
2.17. Immunofluorescence (IF)
2.18. Data Availability
2.19. Statistics
3. Results
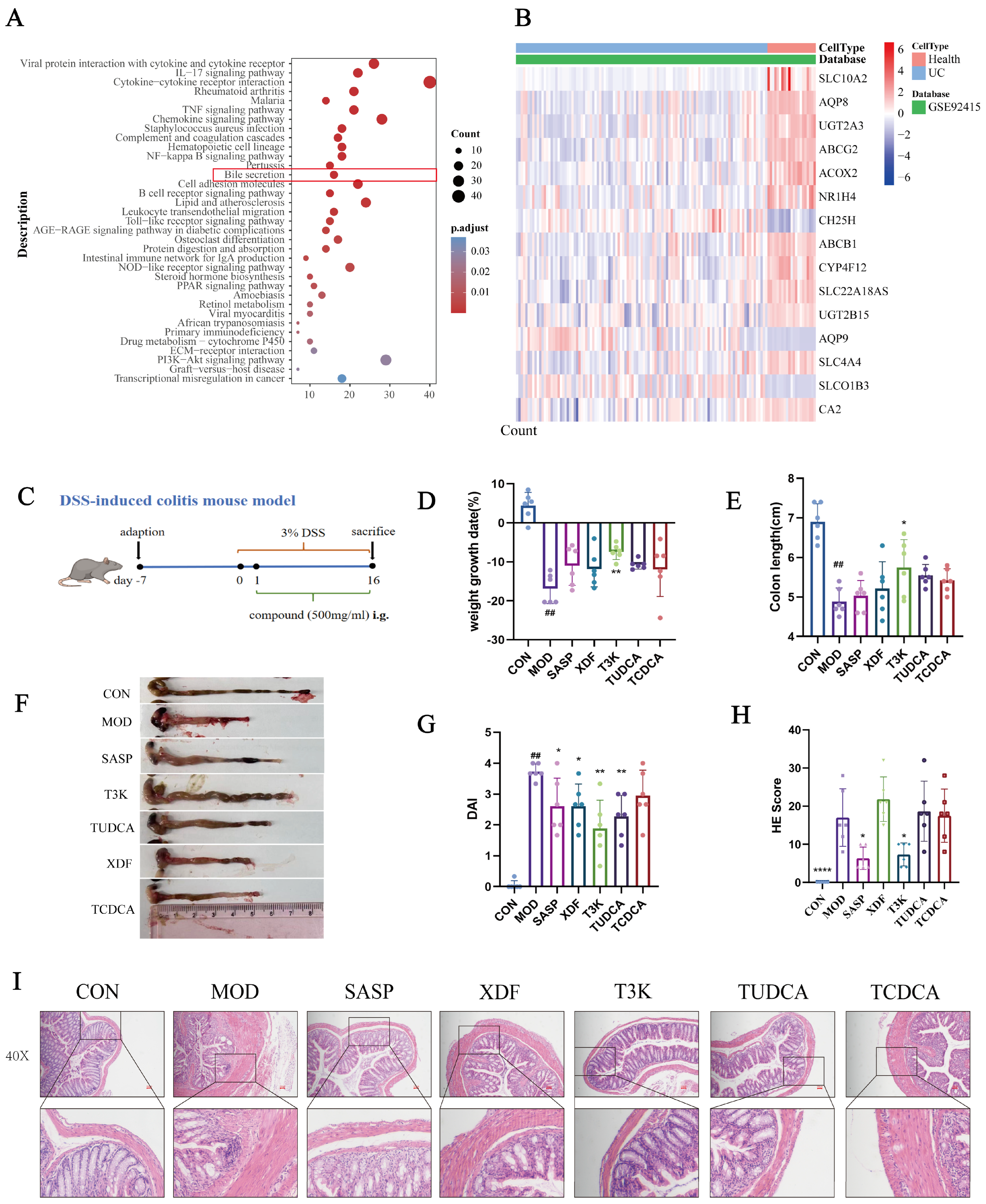
3.1. Bile Acid Metabolism Is Implicated in UC and T3K Alleviates DSS-Induced Colitis
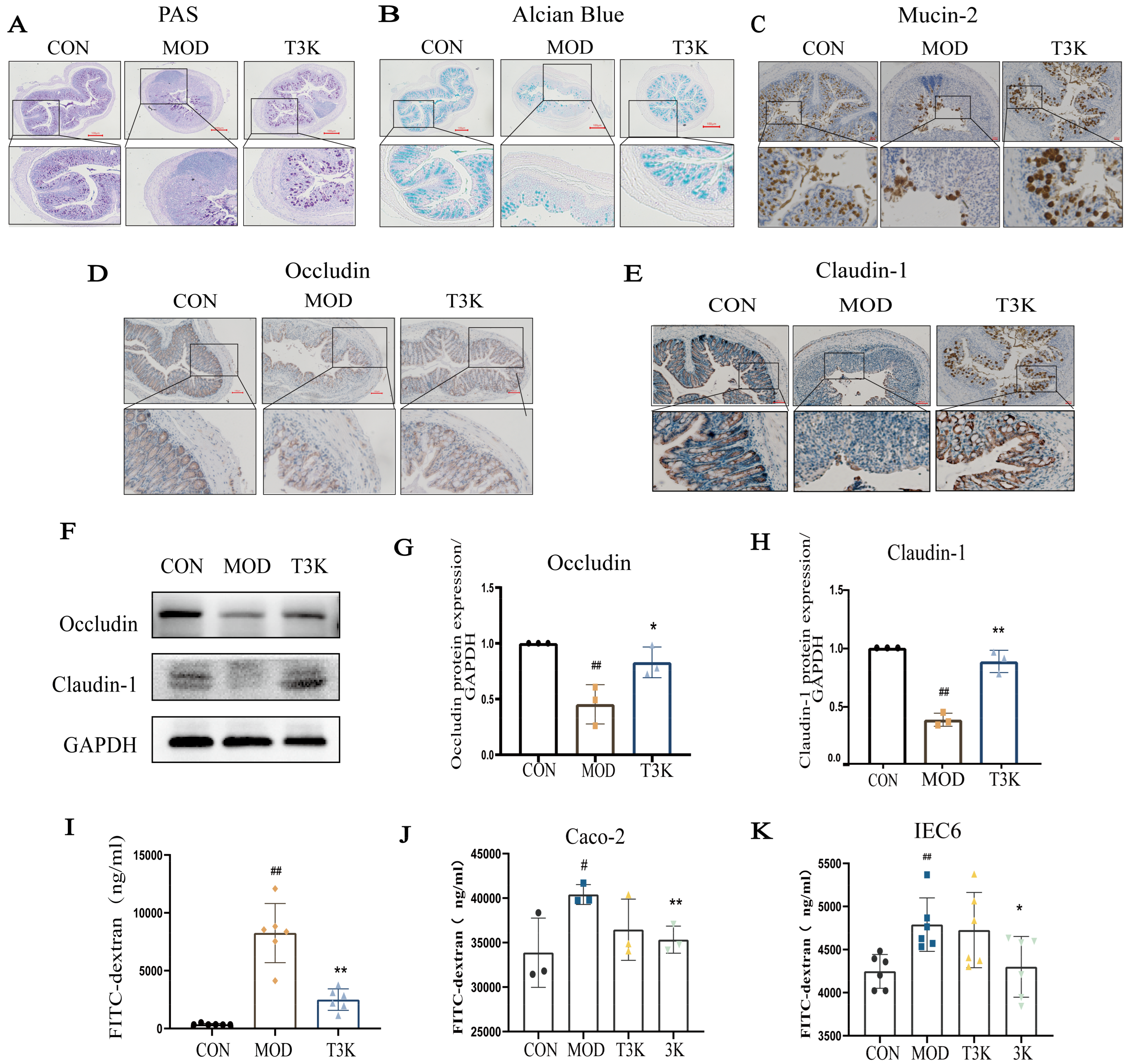
3.2. T3K Restores Intestinal Barrier Integrity and Reduces Epithelial Permeability
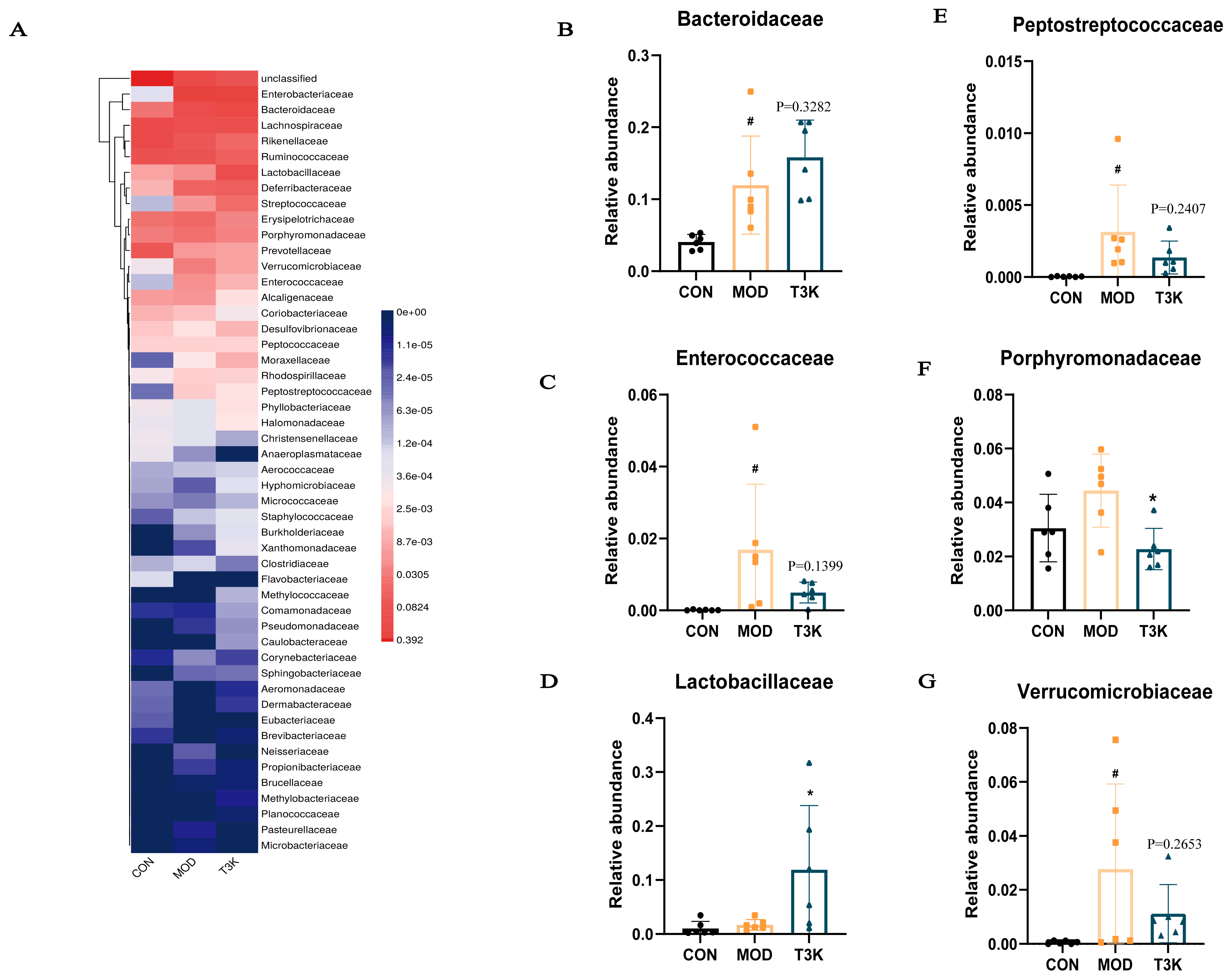
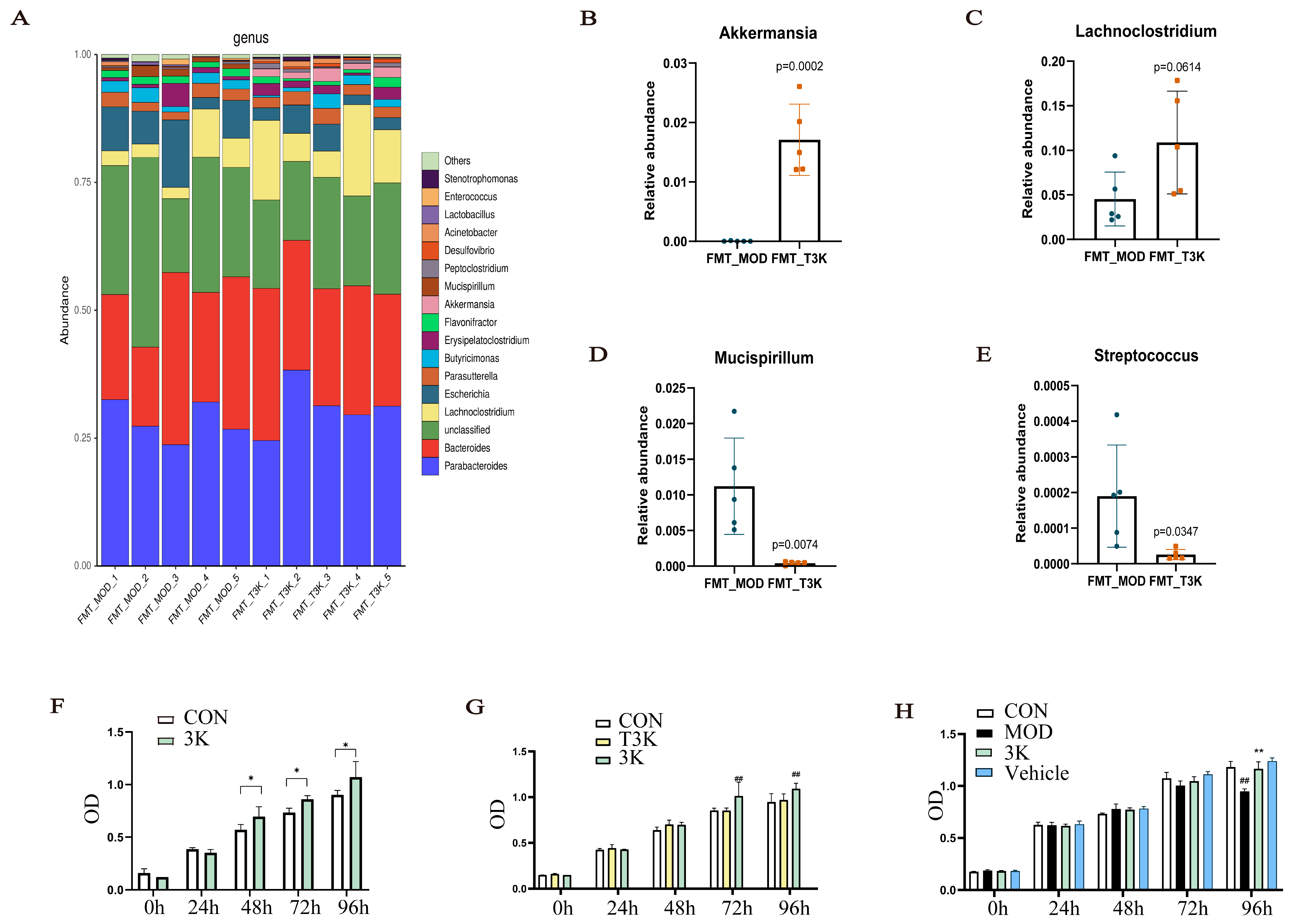
3.3. T3K Rectifies Gut Microbial Dysbiosis in Colitis Mice
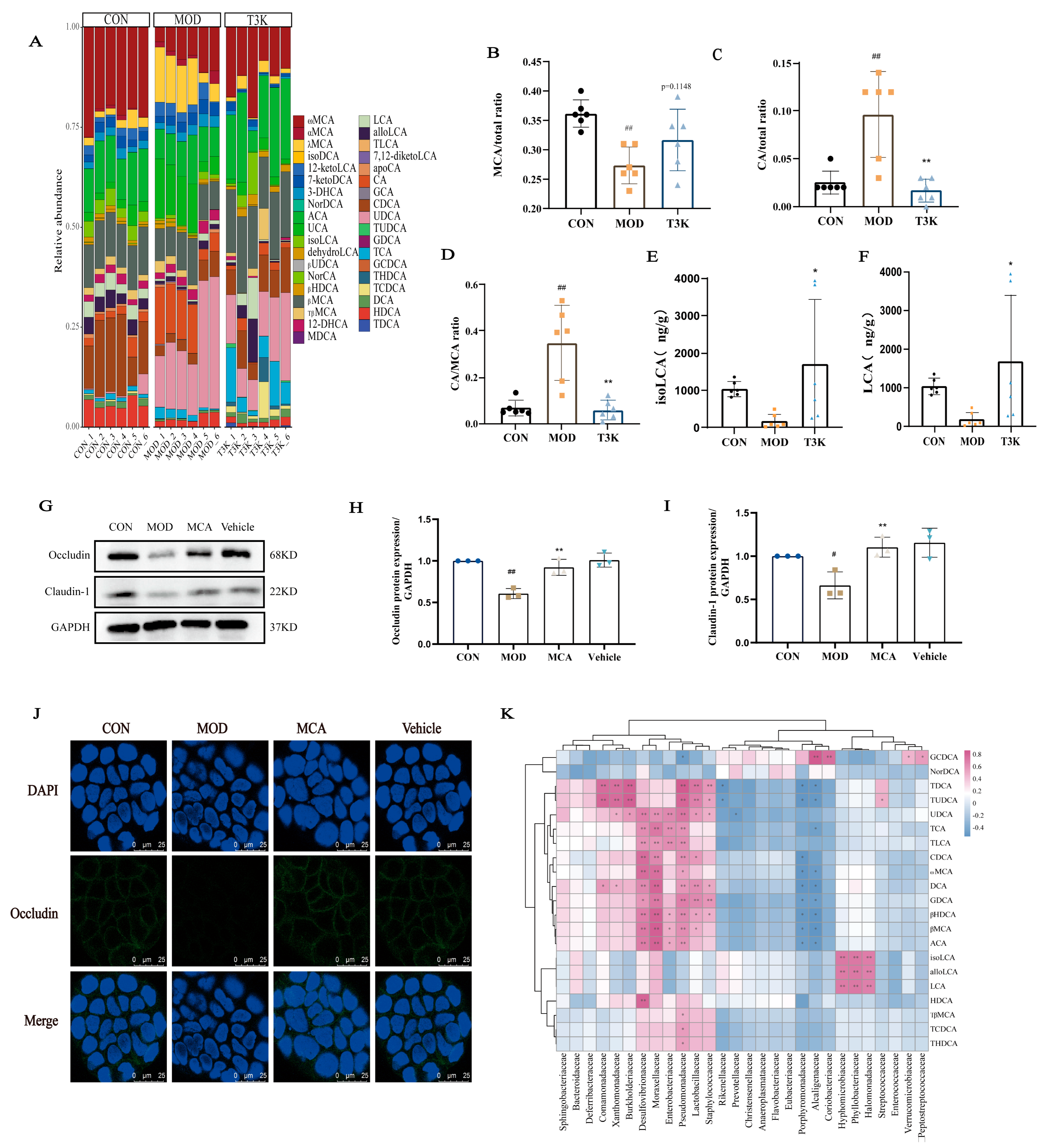
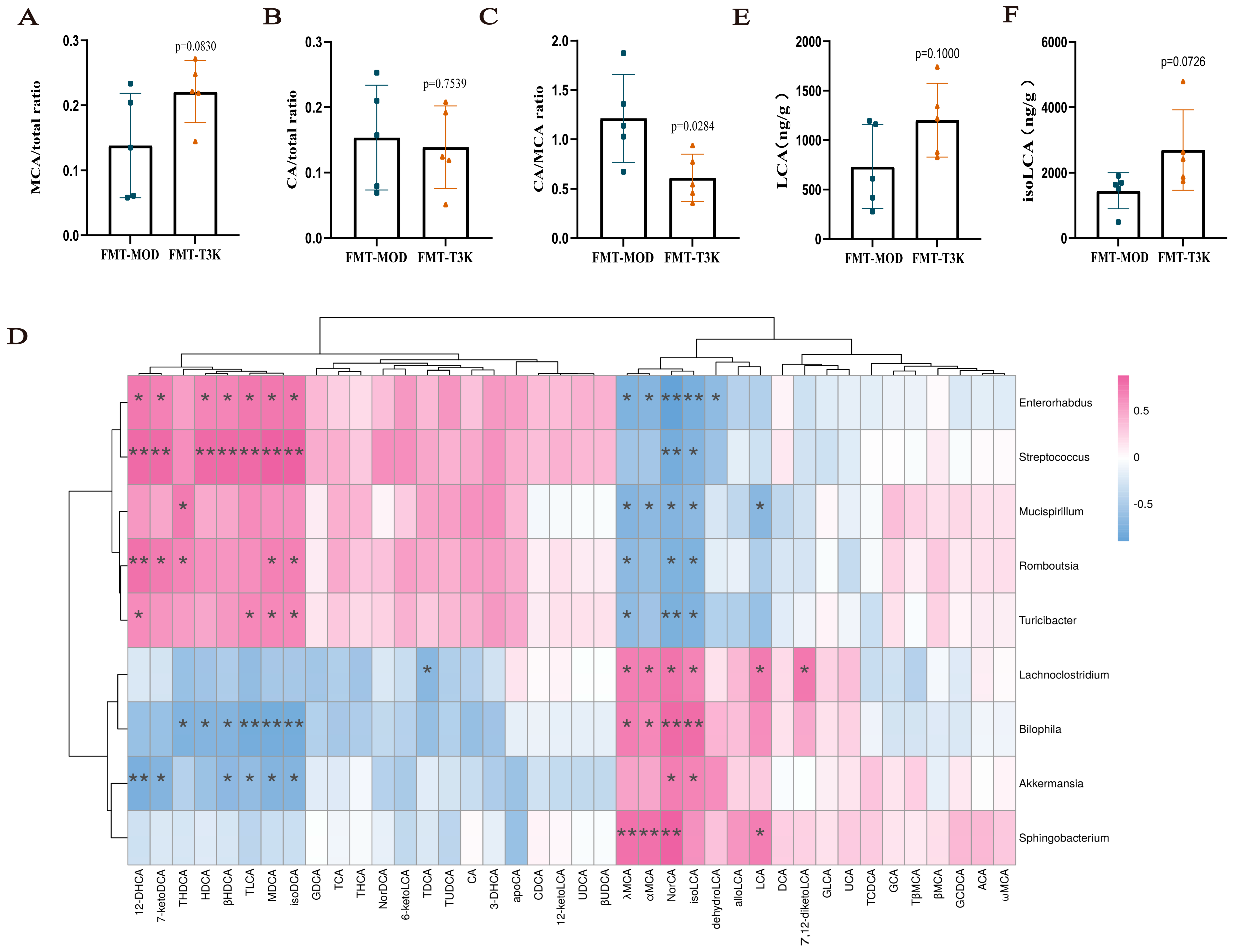
3.4. T3K Normalizes Bile Acid Metabolism and Increases Hydrophilic Bile Acids
3.5. T3K Loses Efficacy in Alleviating Symptoms of DSS-Induced Colitis in PGF-Mice
3.6. T3K Attenuates DSS-Induced Colitis Through Intestinal Microbiota Modulation
3.7. T3K Restores Gut Flora and BA Metabolism via Fecal Microbiota Transplantation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BA | Bile acid |
| DAI | Disease activity index |
| XDF | Drained bear bile powder |
| FMT | Fecal microbiota transplantation |
| FITC | Fluorescein isothiocyanate |
| GEO | Gene Expression Omnibus |
| IBD | Inflammatory bowel disease |
| LCA | Lithocholic acid |
| PAS | Periodic Acid-Schiff |
| PGF | Pseudo-germ-free |
| SASP | Sulfasalazine |
| TUDCA | Tauroursodeoxycholate |
| TCDCA | Taurochenodeoxycholate |
| T3KDCA | Tauro-7α-hydroxy-3-oxo-5β-cholanoate |
| UC | Ulcerative colitis |
References
- Le Berre, C.; Honap, S.; Peyrin-Biroulet, L. Ulcerative colitis. Lancet 2023, 402, 571–584. [Google Scholar] [CrossRef]
- Kobayashi, T.; Siegmund, B.; Le Berre, C.; Wei, S.C.; Ferrante, M.; Shen, B.; Bernstein, C.N.; Danese, S.; Peyrin-Biroulet, L.; Hibi, T.; et al. Ulcerative colitis. Nat. Rev. Dis. Primers 2020, 6, 74. [Google Scholar] [CrossRef] [PubMed]
- Burisch, J.; Zhao, M.; Odes, S.; De Cruz, P.; Vermeire, S.; Bernstein, C.N.; Kaplan, G.G.; Duricova, D.; Greenberg, D.; Melberg, H.O.; et al. The cost of inflammatory bowel disease in high-income settings: A Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol. Hepatol. 2023, 8, 458–492. [Google Scholar] [CrossRef]
- Chang, J.T. Pathophysiology of Inflammatory Bowel Diseases. N. Engl. J. Med. 2020, 383, 2652–2664. [Google Scholar] [CrossRef]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef]
- Norman, J.M.; Handley, S.A.; Baldridge, M.T.; Droit, L.; Liu, C.Y.; Keller, B.C.; Kambal, A.; Monaco, C.L.; Zhao, G.; Fleshner, P.; et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015, 160, 447–460. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Hoarau, G.; Mukherjee, P.K.; Gower-Rousseau, C.; Hager, C.; Chandra, J.; Retuerto, M.A.; Neut, C.; Vermeire, S.; Clemente, J.; Colombel, J.F.; et al. Bacteriome and Mycobiome Interactions Underscore Microbial Dysbiosis in Familial Crohn’s Disease. mBio 2016, 7, e01250-16. [Google Scholar] [CrossRef]
- Fiorucci, S.; Carino, A.; Baldoni, M.; Santucci, L.; Costanzi, E.; Graziosi, L.; Distrutti, E.; Biagioli, M. Bile Acid Signaling in Inflammatory Bowel Diseases. Dig. Dis. Sci. 2021, 66, 674–693. [Google Scholar] [CrossRef] [PubMed]
- Ott, S.J. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004, 53, 685–693. [Google Scholar] [CrossRef]
- Cai, J.; Rimal, B.; Jiang, C.; Chiang, J.Y. Bile acid metabolism and signaling, the microbiota, and metabolic disease. Pharmacol. Ther. 2022, 237, 108238. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G.; et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2013, 62, 531–539. [Google Scholar] [CrossRef]
- Lavelle, A.; Sokol, H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.Q.; Carey, M.C. Therapeutic uses of animal biles in traditional Chinese medicine: An ethnopharmacological, biophysical chemical and medicinal review. World J. Gastroenterol. 2014, 20, 9952–9975. [Google Scholar] [CrossRef]
- Liu, Z.-B.; Ye, X.; Wu, C.-J.; Wei, D.-N. Bear Bile Powder Improves Ulcerative Colitis by Protecting the Intestinal Mechanical Barrier and Regulating Intestinal Flora. Curr. Pharm. Des. 2024, 30, 1530–1540. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Huang, Y.; Feng, N.; Zhang, H.; Qu, J.; Ma, S.; Liu, Y.; Jiang, L.; Xu, S.; Wang, L.; et al. Artificial Bear Bile: A Novel Approach to Balancing Medical Requirements and Animal Welfare. Engineering 2023, 38, 100–112. [Google Scholar] [CrossRef]
- Choucair, I.; Nemet, I.; Li, L.; Cole, M.A.; Skye, S.M.; Kirsop, J.; Fischbach, M.A.; Gogonea, V.; Mark Brown, J.; Wilson Tang, W.H.; et al. Quantification of bile acids: A mass spectrometry platform for studying gut microbe connection to metabolic diseases. J. Lipid Res. 2020, 61, 159–177. [Google Scholar] [CrossRef]
- Cao, S.S.; Zimmermann, E.M.; Chuang, B.M.; Song, B.; Nwokoye, A.; Wilkinson, J.E.; Eaton, K.A.; Kaufman, R.J. The unfolded protein response and chemical chaperones reduce protein misfolding and colitis in mice. Gastroenterology 2013, 144, 989–1000.e6. [Google Scholar] [CrossRef]
- Haller, D.; Jobin, C. Interaction between resident luminal bacteria and the host: Can a healthy relationship turn sour? J. Pediatr. Gastroenterol. Nutr. 2004, 38, 123–136. [Google Scholar] [PubMed]
- He, Y.; Li, X.; Yu, H.; Ge, Y.; Liu, Y.; Qin, X.; Jiang, M.; Wang, X. The Functional Role of Fecal Microbiota Transplantation on Dextran Sulfate Sodium-Induced Colitis in Mice. Front. Cell Infect. Microbiol. 2019, 9, 393. [Google Scholar] [CrossRef]
- Fan, Q.; Guan, X.; Hou, Y.; Liu, Y.; Wei, W.; Cai, X.; Zhang, Y.; Wang, G.; Zheng, X.; Hao, H.; et al. Paeoniflorin modulates gut microbial production of indole-3-lactate and epithelial autophagy to alleviate colitis in mice. Phytomedicine 2020, 79, 153345. [Google Scholar] [CrossRef]
- Dong, S.; Zhu, M.; Wang, K.; Zhao, X.; Hu, L.; Jing, W.; Lu, H.; Wang, S. Dihydromyricetin improves DSS-induced colitis in mice via modulation of fecal-bacteria-related bile acid metabolism. Pharmacol. Res. 2021, 171, 105767. [Google Scholar] [CrossRef] [PubMed]
- Gupta, J.; Nebreda, A.R. Analysis of Intestinal Permeability in Mice. Bio-Protocol 2014, 4, e1289. [Google Scholar] [CrossRef]
- Dieleman, L.A.; Palmen, M.; Akol, H.; Bloemena, E.; Peña, A.S.; Meuwissen, S.G.M.; Van Rees, E.P. Chronic experimental colitis induced by dextran sulphate sodium (DSS) is char-acterized by Th1 and Th2 cytokines. Clin. Exp. Immunol. 1998, 114, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Sandborn, W.J.; Feagan, B.G.; Marano, C.; Zhang, H.; Strauss, R.; Johanns, J.; Adedokun, O.J.; Guzzo, C.; Colombel, J.F.; Reinisch, W.; et al. Subcutaneous golimumab induces clinical response and remission in patients with moderate-to-severe ulcerative colitis. Gastroenterology 2014, 146, 85–95. [Google Scholar] [CrossRef]
- Liu, Y.; Yu, X.; Zhao, J.; Zhang, H.; Zhai, Q.; Chen, W. The role of MUC2 mucin in intestinal homeostasis and the impact of dietary components on MUC2 expression. Int. J. Biol. Macromol. 2020, 164, 884–891. [Google Scholar] [CrossRef] [PubMed]
- Jing, W.; Dong, S.; Luo, X.; Liu, J.; Wei, B.; Du, W.; Yang, L.; Luo, H.; Wang, Y.; Wang, S.; et al. Berberine improves colitis by triggering AhR activation by microbial tryptophan catabolites. Pharmacol. Res. 2021, 164, 105358. [Google Scholar] [CrossRef]
- Yao, D.; Dai, W.; Dong, M.; Dai, C.; Wu, S. MUC2 and related bacterial factors: Therapeutic targets for ulcerative colitis. EBioMedicine 2021, 74, 103751. [Google Scholar] [CrossRef]
- Hang, S.; Paik, D.; Yao, L.; Kim, E.; Trinath, J.; Lu, J.; Ha, S.; Nelson, B.N.; Kelly, S.P.; Wu, L.; et al. Bile acid metabolites control T(H)17 and T(reg) cell differentiation. Nature 2019, 576, 143–148. [Google Scholar] [CrossRef]
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorsky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ(+) regulatory T cell homeostasis. Nature 2020, 577, 410–415. [Google Scholar] [CrossRef]
- O’Connor, C.J.; Wallace, R.G.; Iwamoto, K.; Taguchi, T.; Sunamoto, J. Bile salt damage of egg phosphatidylcholine liposomes. Biochim. Biophys. Acta 1985, 817, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Jiao, T.; Liu, W.; Luo, Y.; Wang, J.; Guo, X.; Tong, X.; Lin, Z.; Sun, C.; Wang, K.; et al. Hepatic cytochrome P450 8B1 and cholic acid potentiate intestinal epithelial injury in colitis by suppressing intestinal stem cell renewal. Cell Stem Cell 2022, 29, 1366–1381.e9. [Google Scholar] [CrossRef]
- Tavella, T.; Rampelli, S.; Guidarelli, G.; Bazzocchi, A.; Gasperini, C.; Pujos-Guillot, E.; Comte, B.; Barone, M.; Biagi, E.; Candela, M.; et al. Elevated gut microbiome abundance of Christensenellaceae, Porphyromonadaceae and Rikenellaceae is associated with reduced visceral adipose tissue and healthier metabolic profile in Italian elderly. Gut Microbes 2021, 13, 1880221. [Google Scholar] [CrossRef]
- Hu, S.; Bourgonje, A.R.; Gacesa, R.; Jansen, B.H.; Björk, J.R.; Bangma, A.; Hidding, I.J.; van Dullemen, H.M.; Visschedijk, M.C.; Faber, K.N.; et al. Mucosal host-microbe interactions associate with clinical phenotypes in inflammatory bowel disease. Nat. Commun. 2024, 15, 1470. [Google Scholar] [CrossRef]
- Cani, P.D.; Depommier, C.; Derrien, M.; Everard, A.; de Vos, W.M. Akkermansia muciniphila: Paradigm for next-generation beneficial microorganisms. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 682. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.R.; Hwang, J.; Koh, H.J.; Jang, K.; Lee, J.D.; Choi, J.; Yang, C.S. The targeted delivery of the c-Src peptide complexed with schizophyllan to macrophages inhibits polymicrobial sepsis and ulcerative colitis in mice. Biomaterials 2016, 89, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, E.; Guzmán-Jofre, L.; Moore-Carrasco, R.; Palomo, I. Role of PPARs in inflammatory processes associated with metabolic syndrome (Review). Mol. Med. Rep. 2013, 8, 1611–1616. [Google Scholar] [CrossRef] [PubMed]
- Bang, B.; Lichtenberger, L.M. Methods of Inducing Inflammatory Bowel Disease in Mice. Curr. Protoc. Pharmacol. 2016, 72, 5.58.1–5.58.42. [Google Scholar] [CrossRef]
- Gadaleta, R.M.; Garcia-Irigoyen, O.; Moschetta, A. Exploration of Inflammatory Bowel Disease in Mice: Chemically Induced Murine Models of Inflammatory Bowel Disease (IBD). Curr. Protoc. Mouse Biol. 2017, 7, 13–28. [Google Scholar] [CrossRef]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016–6029. [Google Scholar] [CrossRef]
- Martel, J.; Chang, S.H.; Ko, Y.F.; Hwang, T.L.; Young, J.D.; Ojcius, D.M. Gut barrier disruption and chronic disease. Trends Endocrinol. Metab. 2022, 33, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Knoop, K.A.; Newberry, R.D. Goblet cells: Multifaceted players in immunity at mucosal surfaces. Mucosal Immunol. 2018, 11, 1551–1557. [Google Scholar] [CrossRef]
- Velcich, A.; Yang, W.; Heyer, J.; Fragale, A.; Nicholas, C.; Viani, S.; Kucherlapati, R.; Lipkin, M.; Yang, K.; Augenlicht, L. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 2002, 295, 1726–1729. [Google Scholar] [CrossRef]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.P.; Van Goudoever, J.B.; Büller, H.A.; Dekker, J.; Van Seuningen, I.; Renes, I.B.; et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Kuo, W.T.; Odenwald, M.A.; Turner, J.R.; Zuo, L. Tight junction proteins occludin and ZO-1 as regulators of epithelial proliferation and survival. Ann. N. Y. Acad. Sci. 2022, 1514, 21–33. [Google Scholar] [CrossRef]
- Kim, B.; Breton, S. The MAPK/ERK-Signaling Pathway Regulates the Expression and Distribution of Tight Junction Proteins in the Mouse Proximal Epididymis. Biol. Reprod. 2016, 94, 22. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.H.; Zhu, C.X.; Quan, Y.S.; Yang, Z.Y.; Wu, S.; Luo, W.W.; Tan, B.; Wang, X.Y. Relationship between intestinal microbiota and ulcerative colitis: Mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018, 24, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Wallace, K.L.; Zheng, L.B.; Kanazawa, Y.; Shih, D.Q. Immunopathology of inflammatory bowel disease. World J. Gastroenterol. 2014, 20, 6–21. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Hansen, J.; Gulati, A.; Sartor, R.B. The role of mucosal immunity and host genetics in defining intestinal commensal bacteria. Curr. Opin. Gastroenterol. 2010, 26, 564–571. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Sang, L.X.; Chang, B.; Zhang, W.L.; Wu, X.M.; Li, X.H.; Jiang, M. Remission induction and maintenance effect of probiotics on ulcerative colitis: A meta-analysis. World J. Gastroenterol. 2010, 16, 1908–1915. [Google Scholar] [CrossRef]
- Sun, H.; Guo, Y.; Wang, H.; Yin, A.; Hu, J.; Yuan, T.; Zhou, S.; Xu, W.; Wei, P.; Yin, S.; et al. Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis. Gut 2023, 72, 1664–1677. [Google Scholar] [CrossRef]
- Hegyi, P.; Maléth, J.; Walters, J.R.; Hofmann, A.F.; Keely, S.J. Guts and Gall: Bile Acids in Regulation of Intestinal Epithelial Function in Health and Disease. Physiol. Rev. 2018, 98, 1983–2023. [Google Scholar] [CrossRef]
- Annese, V. A Review of Extraintestinal Manifestations and Complications of Inflammatory Bowel Disease. Saudi. J. Med. Med. Sci. 2019, 7, 66–73. [Google Scholar] [CrossRef]
- Cai, J.; Sun, L.; Gonzalez, F.J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell Host Microbe 2022, 30, 289–300. [Google Scholar] [CrossRef]
- Jean-Louis, S.; Akare, S.; Ali, M.A.; Mash, E.A.; Meuillet, E.; Martinez, J.D. Deoxycholic acid induces intracellular signaling through membrane perturbations. J. Biol. Chem. 2006, 281, 14948–14960. [Google Scholar] [CrossRef]
- Sorrentino, G.; Perino, A.; Yildiz, E.; El Alam, G.; Sleiman, M.B.; Gioiello, A.; Pellicciari, R.; Schoonjans, K. Bile acids signalvia TGR5 to activate intestinal stem cells and epithelial regeneration. Gastroenterology 2020, 159, 956–968.e8. [Google Scholar] [CrossRef] [PubMed]
- Paik, D.; Yao, L.; Zhang, Y.; Bae, S.; D’Agostino, G.D.; Zhang, M.; Kim, E.; Franzosa, E.A.; Avila-Pacheco, J.; Bisanz, J.E.; et al. Human gut bacteria produce ΤH17-modulating bile acid metabolites. Nature 2022, 603, 907–912. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Gao, J.; Chen, L.; Chen, Y.; Jiang, J.; Chen, H.; Ma, L. Lithocholic acid ameliorates ulcerative colitis via the PXR/TLR4/NF-κB/NLRP3 signaling pathway and gut microbiota modulation. Cell Mol. Life Sci. 2025, 82, 336. [Google Scholar] [CrossRef]
- Zhang, X.; Cai, J.; Chen, L.; Tai, J.; Liu, J.; Cao, Y. Gastrodin ameliorates DSS-induced ulcerative colitis via enhancing the abundance of Akkermansia muciniphila and modulating the metabolites. Phytomedicine 2025, 146, 157138. [Google Scholar] [CrossRef] [PubMed]
- Herrera-deGuise, C.; Varela, E.; Sarrabayrouse, G.; Pozuelo Del Río, M.; Alonso, V.R.; Sainz, N.B.; Casellas, F.; Mayorga, L.F.; Manichanh, C.; Vidaur, F.A.; et al. Gut Microbiota Composition in Long-Remission Ulcerative Colitis is Close to a Healthy Gut Microbiota. Inflamm. Bowel Dis. 2023, 29, 1362–1369. [Google Scholar] [CrossRef]
- Ghavami, S.B.; Moshiri, A.; Bonaretti, C.; Farmani, M.; Squillario, M.; Di Marco, E.; Shahrokh, S.; Balaii, H.; Corrias, M.V.; Ponzoni, M.; et al. Predictive Microbial Markers Distinguish Responders and Non-Responders to Adalimumab: A Step Toward Precision Medicine in Ulcerative Colitis. Microorganisms 2025, 13, 1941. [Google Scholar] [CrossRef] [PubMed]
- Tsifintaris, M.; Kiousi, D.E.; Repanas, P.; Kamarinou, C.S.; Kavakiotis, I.; Galanis, A. Probio-Ichnos: A Database of Microorganisms with In Vitro Probiotic Properties. Microorganisms 2024, 12, 1955. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Li, S.; Zhang, J.; Wu, F.; Li, X.; Wu, D.; Zhang, M.; Ou, Z.; Jie, Z.; Yan, Q.; et al. Genome sequencing of 39 Akkermansia muciniphila isolates reveals its population structure, genomic and functional diverisity, and global distribution in mammalian gut microbiotas. BMC Genom. 2017, 18, 800. [Google Scholar] [CrossRef]
- Kang, E.J.; Kim, J.H.; Kim, Y.E.; Lee, H.; Jung, K.B.; Chang, D.H.; Lee, Y.; Park, S.; Lee, E.Y.; Lee, E.J.; et al. The secreted protein Amuc_1409 from Akkermansia muciniphila improves gut health through intestinal stem cell regulation. Nat. Commun. 2024, 15, 2983. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, J.; Ren, X.; Zhang, Y.; Ke, Z.; Zhou, J.; Wang, Y.; Zhang, Y.; Liu, Y. Cholecystectomy-induced secondary bile acids accumulation ameliorates colitis through inhibiting monocyte/macrophage recruitment. Gut Microbes 2022, 14, 2107387. [Google Scholar] [CrossRef]
- Zhao, Q.; Duan, Z.; Lai, R.; Ma, P. Novel microbially transformed bile acids: Biosynthesis, structure, and function. Pharmacol. Res. 2025, 216, 107775. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhou, Y.; Zhang, Y.; Li, Y.; Chen, Y.; Chi, X.; You, Z.; Zhang, H.; Li, Y.; Wu, L. Bile Derivative T3K Ameliorates Colitis by Regulating the Intestinal Microbiota-Bile Acid Axis. Pharmaceutics 2026, 18, 20. https://doi.org/10.3390/pharmaceutics18010020
Zhou Y, Zhang Y, Li Y, Chen Y, Chi X, You Z, Zhang H, Li Y, Wu L. Bile Derivative T3K Ameliorates Colitis by Regulating the Intestinal Microbiota-Bile Acid Axis. Pharmaceutics. 2026; 18(1):20. https://doi.org/10.3390/pharmaceutics18010020
Chicago/Turabian StyleZhou, Yu, Yixiang Zhang, Ying Li, Yu Chen, Xiaoqian Chi, Zhongyu You, Haijing Zhang, Yong Li, and Lianqiu Wu. 2026. "Bile Derivative T3K Ameliorates Colitis by Regulating the Intestinal Microbiota-Bile Acid Axis" Pharmaceutics 18, no. 1: 20. https://doi.org/10.3390/pharmaceutics18010020
APA StyleZhou, Y., Zhang, Y., Li, Y., Chen, Y., Chi, X., You, Z., Zhang, H., Li, Y., & Wu, L. (2026). Bile Derivative T3K Ameliorates Colitis by Regulating the Intestinal Microbiota-Bile Acid Axis. Pharmaceutics, 18(1), 20. https://doi.org/10.3390/pharmaceutics18010020







