Emerging Approaches for the Discovery of Lipid-Based RNA Delivery Systems
Abstract
1. Introduction
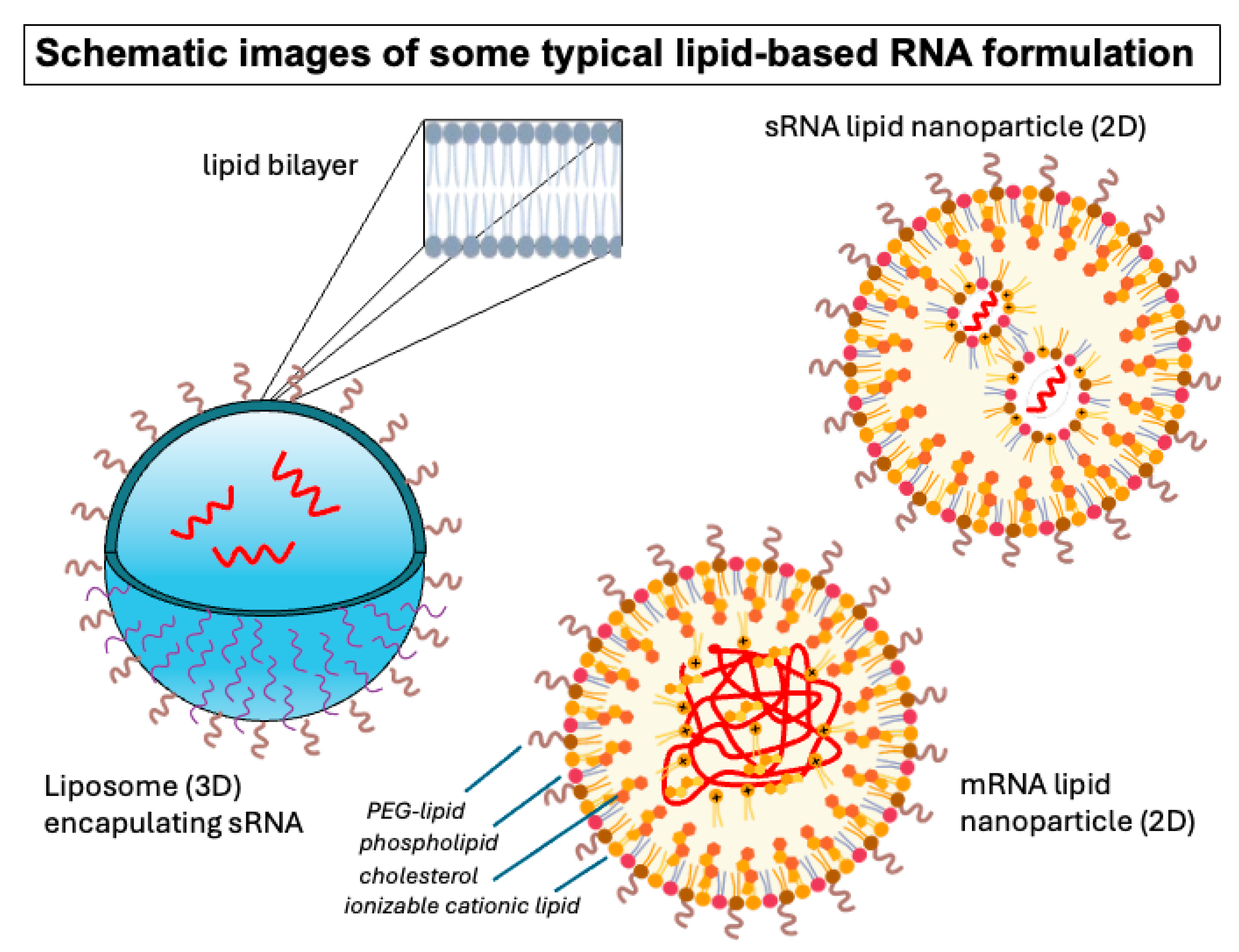
2. Historical Evolution of Lipid-Based Nucleic Acid Formulations
- High (nearly 100%) complexation efficiency (w/cationic or ionizable cationic lipids);
- Rapid formulation methods (including microfluidics);
- Combinatorial synthesis of alternative (less toxic) lipids;
- Better DNA/RNA synthesis methods for stabilization;
- Easier multiplexing of formulation and testing.
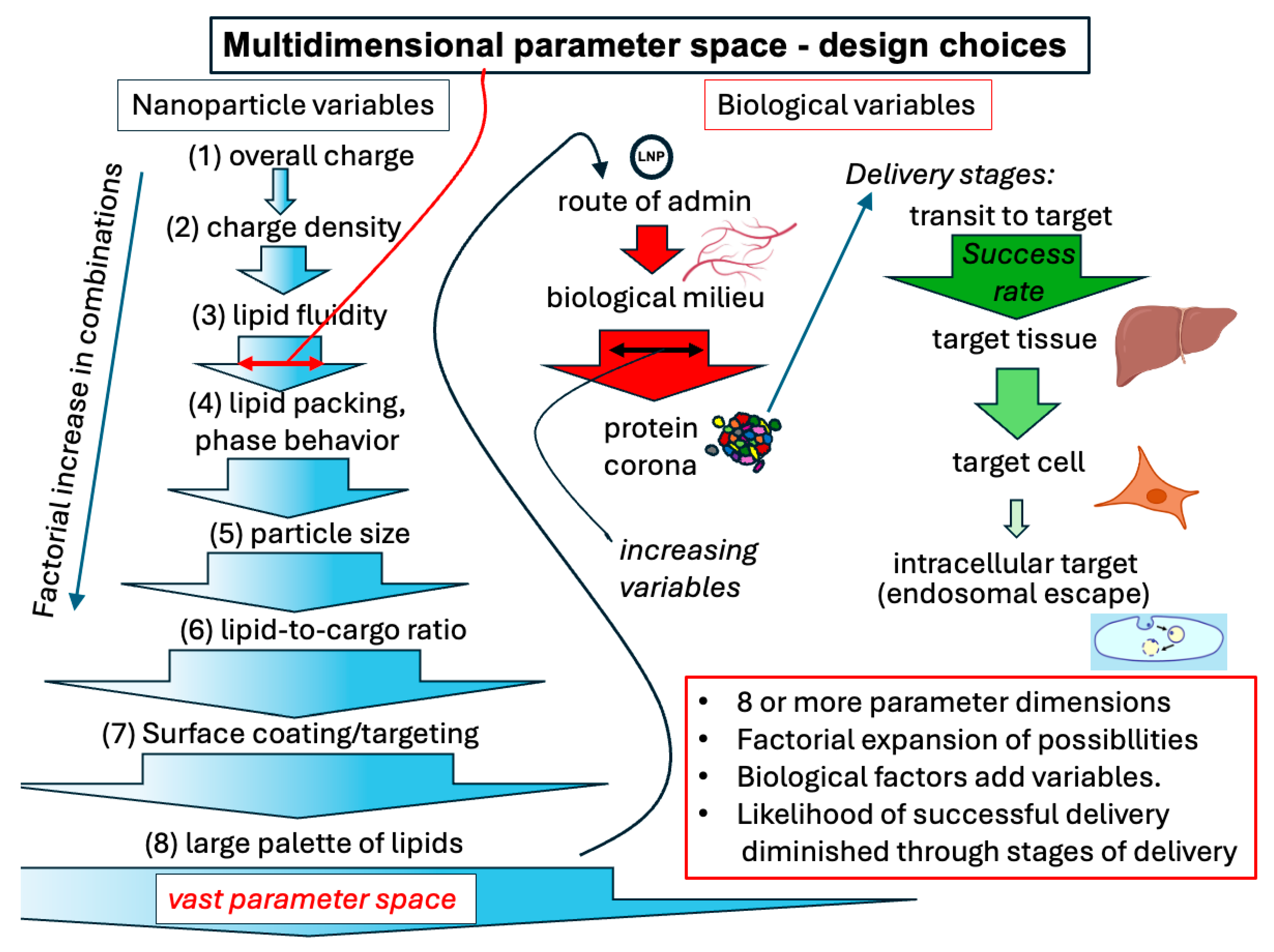
3. Emerging Technologies for Formulation Design
3.1. Top-Down Approach—Screening Formulations
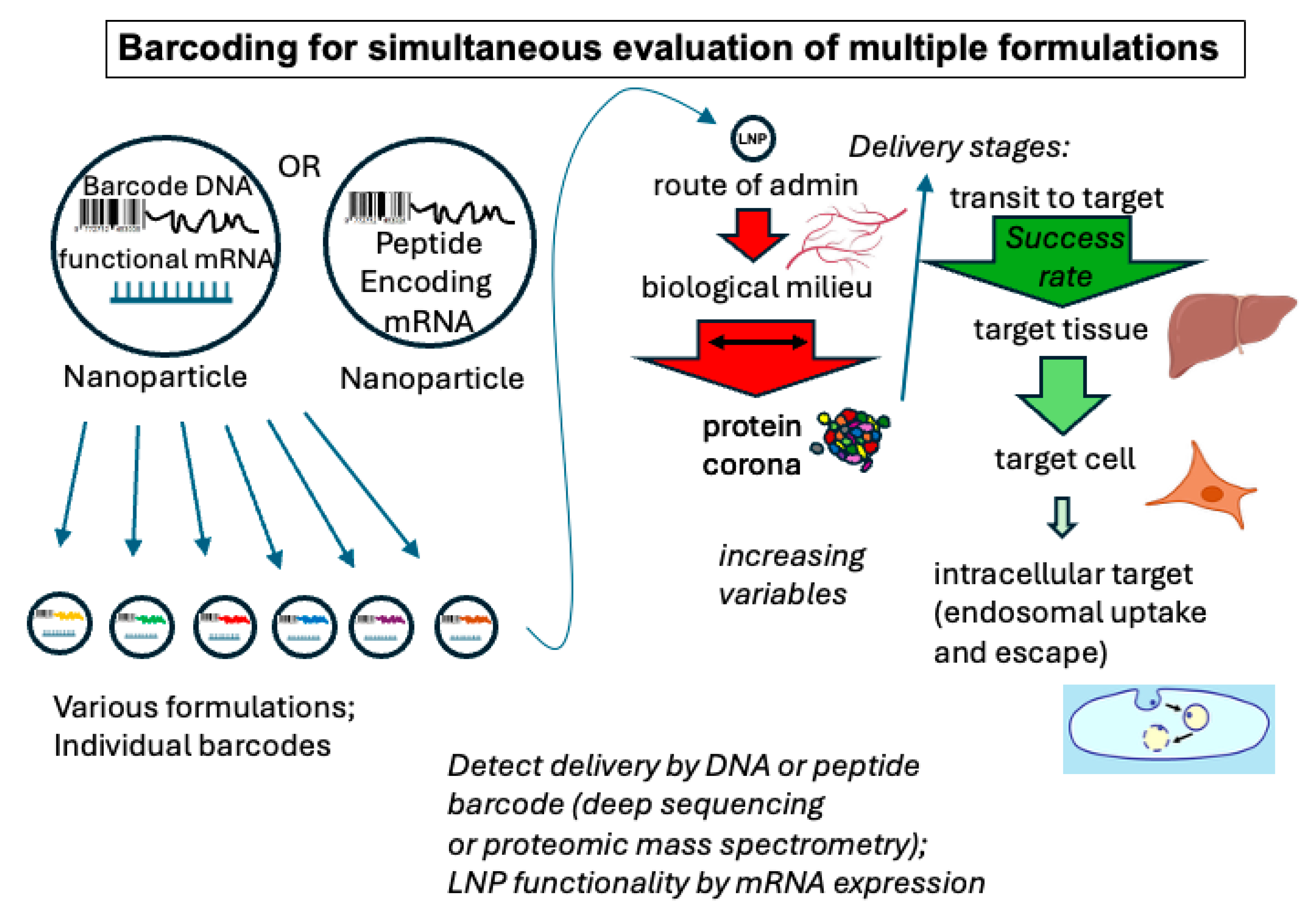
3.1.1. Multiplexed Formulation
3.1.2. Multiplexed In Vivo Testing
3.2. Bottom-Up: Understanding Biologically Dictated Design Principles
3.2.1. The Importance of the Protein Corona
3.2.2. Imaging, Spectroscopic and Computational Methods to Understand LNP/Liposome Interactions with Cells
3.2.3. Machine-Learning-Based LNP Design
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LNP | Lipid nanoparticle |
| LCMS | Liquid chromatography/mass spectrometry |
| EM | Electron microscopy |
References
- Wei, P.S.; Thota, N.; John, G.; Chang, E.; Lee, S.; Wang, Y.; Ma, Z.; Tsai, Y.H.; Mei, K.C. Enhancing RNA-lipid nanoparticle delivery: Organ- and cell-specificity and barcoding strategies. J. Control. Release 2024, 375, 366–388. [Google Scholar] [CrossRef]
- Hou, X.; Zaks, T.; Langer, R.; Dong, Y. Lipid Nanoparticles for mRNA Delivery. Nat. Rev. Mater. 2021, 6, 1078–1094. [Google Scholar] [CrossRef]
- Cullis, P.R.; Felgner, P.L. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat. Rev. Drug Discov. 2024, 23, 709–722. [Google Scholar] [CrossRef]
- Haque, M.A.; Shrestha, A.; Mikelis, C.M.; Mattheolabakis, G. Comprehensive analysis of lipid nanoparticle formulation and preparation for RNA delivery. Int. J. Pharm. X 2024, 8, 100283. [Google Scholar] [CrossRef]
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475. [Google Scholar] [CrossRef]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154–155, 37–63. [Google Scholar] [CrossRef] [PubMed]
- Mukai, H.; Ogawa, K.; Kato, N.; Kawakami, S. Recent advances in lipid nanoparticles for delivery of nucleic acid, mRNA, and gene editing-based therapeutics. Drug Metab. Pharmacokinet. 2022, 44, 100450. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Li, S.; Lin, X.; Chen, Y. A perspective of lipid nanoparticles for RNA delivery. Exploration 2024, 4, 20230147. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, X.; Liu, Y.; Yang, G.; Falconer, R.J.; Zhao, C.-X. Lipid Nanoparticles for Drug Delivery. Adv. NanoBiomed Res. 2022, 2, 2100109. [Google Scholar] [CrossRef]
- Zhang, H.; Vandesompele, J.; Braeckmans, K.; De Smedt, S.C.; Remaut, K. Nucleic acid degradation as barrier to gene delivery: A guide to understand and overcome nuclease activity. Chem. Soc. Rev. 2024, 53, 317–360. [Google Scholar] [CrossRef]
- Lee, Y.; Jeong, M.; Park, J.; Jung, H.; Lee, H. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med. 2023, 55, 2085–2096. [Google Scholar] [CrossRef]
- Bangham, A.D.; Horne, R.W. Negative staining of phospholipids and their structural modification by surface-active agents as observed in the electron microscope. J. Mol. Biol. 1964, 8, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Fraley, R.; Subramani, S.; Berg, P.; Papahadjopoulos, D. Introduction of liposome-encapsulated SV40 DNA into cells. J. Biol. Chem. 1980, 255, 10431–10435. [Google Scholar] [CrossRef] [PubMed]
- Felgner, P.L.; Gadek, T.R.; Holm, M.; Roman, R.; Chan, H.W.; Wenz, M.; Northrop, J.P.; Ringold, G.M.; Danielsen, M. Lipofection: A highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA 1987, 84, 7413–7417. [Google Scholar] [CrossRef]
- Felgner, P.L.; Ringold, G.M. Cationic liposome-mediated transfection. Nature 1989, 337, 387–388. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, B.; Sorgi, F.L.; Huang, L. New structures in complex formation between DNA and cationic liposomes visualized by freeze—Fracture electron microscopy. FEBS Lett. 1994, 356, 361–366. [Google Scholar] [CrossRef]
- Xu, Y.; Hui, S.-W.; Frederik, P.; Szoka, F.C. Physicochemical Characterization and Purification of Cationic Lipoplexes. Biophys. J. 1999, 77, 341–353. [Google Scholar] [CrossRef]
- Bailey, A.L.; Cullis, P.R. Modulation of Membrane Fusion by Asymmetric Transbilayer Distributions of Amino Lipids. Biochemistry 1994, 33, 12573–12580. [Google Scholar] [CrossRef]
- Semple, S.C.; Klimuk, S.K.; Harasym, T.O.; Dos Santos, N.; Ansell, S.M.; Wong, K.F.; Maurer, N.; Stark, H.; Cullis, P.R.; Hope, M.J.; et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Biochim. Biophys. Acta 2001, 1510, 152–166. [Google Scholar] [CrossRef]
- Allen, T.M.; Chonn, A. Large unilamellar liposomes with low uptake into the reticuloendothelial system. FEBS Lett. 1987, 223, 42–46. [Google Scholar] [CrossRef]
- Allen, T.M.; Mehra, T.; Hansen, C.; Chin, Y.C. Stealth liposomes: An improved sustained release system for 1-beta-D-arabinofuranosylcytosine. Cancer Res. 1992, 52, 2431–2439. [Google Scholar]
- Gabizon, A.; Papahadjopoulos, D. Liposome formulations with prolonged circulation time in blood and enhanced uptake by tumors. Proc. Natl. Acad. Sci. USA 1988, 85, 6949–6953. [Google Scholar] [CrossRef]
- Symon, Z.; Peyser, A.; Tzemach, D.; Lyass, O.; Sucher, E.; Shezen, E.; Gabizon, A. Selective delivery of doxorubicin to patients with breast carcinoma metastases by stealth liposomes. Cancer 1999, 86, 72–78. [Google Scholar] [CrossRef]
- Torchilin, V.P. How do polymers prolong circulation time of liposomes? J. Liposome Res. 1996, 6, 99–116. [Google Scholar] [CrossRef]
- Xu, Y.; Szoka, F.C., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry 1996, 35, 5616–5623. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef]
- Stewart, M.J.; Plautz, G.E.; Del Buono, L.; Yang, Z.Y.; Xu, L.; Gao, X.; Huang, L.; Nabel, E.G.; Nabel, G.J. Gene transfer in vivo with DNA-liposome complexes: Safety and acute toxicity in mice. Hum. Gene Ther. 1992, 3, 267–275. [Google Scholar] [CrossRef]
- Liu, F.; Yang, J.; Huang, L.; Liu, D. New Cationic Lipid Formulations for Gene Transfer. Pharm. Res. 1996, 13, 1856–1860. [Google Scholar] [CrossRef] [PubMed]
- Love, K.T.; Mahon, K.P.; Levins, C.G.; Whitehead, K.A.; Querbes, W.; Dorkin, J.R.; Qin, J.; Cantley, W.; Qin, L.L.; Racie, T.; et al. Lipid-like materials for low-dose, in vivo gene silencing. Proc. Natl. Acad. Sci. USA 2010, 107, 1864–1869. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The Dawn of mRNA Vaccines: The Role of Lipid Nanoparticles. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef]
- Naidu, G.S.; Yong, S.B.; Ramishetti, S.; Rampado, R.; Sharma, P.; Ezra, A.; Goldsmith, M.; Hazan-Halevy, I.; Chatterjee, S.; Aitha, A.; et al. A Combinatorial Library of Lipid Nanoparticles for Cell Type-Specific mRNA Delivery. Adv. Sci. 2023, 10, e2301929. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, M.P.; Sago, C.D.; Dahlman, J.E. Testing thousands of nanoparticles in vivo using DNA barcodes. Curr. Opin. Biomed. Eng. 2018, 7, 1–8. [Google Scholar] [CrossRef]
- Johnson, B.K.; Prud’homme, R.K. Flash NanoPrecipitation of Organic Actives and Block Copolymers using a Confined Impinging Jets Mixer. Aust. J. Chem. 2003, 56, 1021–1024. [Google Scholar] [CrossRef]
- Markwalter, C.E.; Prud’homme, R.K. Design of a Small-Scale Multi-Inlet Vortex Mixer for Scalable Nanoparticle Production and Application to the Encapsulation of Biologics by Inverse Flash NanoPrecipitation. J. Pharm. Sci. 2018, 107, 2465–2471. [Google Scholar] [CrossRef]
- Pustulka, K.M.; Wohl, A.R.; Lee, H.S.; Michel, A.R.; Han, J.; Hoye, T.R.; McCormick, A.V.; Panyam, J.; Macosko, C.W. Flash nanoprecipitation: Particle structure and stability. Mol. Pharm. 2013, 10, 4367–4377. [Google Scholar] [CrossRef]
- Caggiano, N.J.; Nayagam, S.K.; Wang, L.Z.; Wilson, B.K.; Lewis, P.; Jahangir, S.; Priestley, R.D.; Prud’homme, R.K.; Ristroph, K.D. Sequential Flash NanoPrecipitation for the scalable formulation of stable core-shell nanoparticles with core loadings up to 90. Int. J. Pharm. 2023, 640, 122985. [Google Scholar] [CrossRef]
- Misra, B.; Hughes, K.A.; Pentz, W.H.; Samart, P.; Geldenhuys, W.J.; Bobbala, S. Flash nanoprecipitation assisted self-assembly of ionizable lipid nanoparticles for nucleic acid delivery. Nanoscale 2024, 16, 6939–6948. [Google Scholar] [CrossRef]
- Strelkova Petersen, D.M.; Chaudhary, N.; Arral, M.L.; Weiss, R.M.; Whitehead, K.A. The mixing method used to formulate lipid nanoparticles affects mRNA delivery efficacy and organ tropism. Eur. J. Pharm. Biopharm. 2023, 192, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Love, K.T.; Chen, Y.; Eltoukhy, A.A.; Kastrup, C.; Sahay, G.; Jeon, A.; Dong, Y.; Whitehead, K.A.; Anderson, D.G. Rapid Discovery of Potent siRNA-Containing Lipid Nanoparticles Enabled by Controlled Microfluidic Formulation. J. Am. Chem. Soc. 2012, 134, 6948–6951. [Google Scholar] [CrossRef] [PubMed]
- Stroock, A.D.; Dertinger, S.K.W.; Ajdari, A.; Mezić, I.; Stone, H.A.; Whitesides, G.M. Chaotic Mixer for Microchannels. Science 2002, 295, 647–651. [Google Scholar] [CrossRef]
- O’Brien Laramy, M.N.; Costa, A.P.; Cebrero, Y.M.; Joseph, J.; Sarode, A.; Zang, N.; Kim, L.J.; Hofmann, K.; Wang, S.; Goyon, A. Process robustness in lipid nanoparticle production: A comparison of microfluidic and turbulent jet mixing. Mol. Pharm. 2023, 20, 4285–4296. [Google Scholar] [CrossRef]
- Williams, M.S.; Longmuir, K.J.; Yager, P. A practical guide to the staggered herringbone mixer. Lab Chip 2008, 8, 1121–1129. [Google Scholar] [CrossRef]
- Rasouli, M.R.; Tabrizian, M. An ultra-rapid acoustic micromixer for synthesis of organic nanoparticles. Lab Chip 2019, 19, 3316–3325. [Google Scholar] [CrossRef] [PubMed]
- Ahl, P.L. Microfluidic and Turbulent Mixing for mRNA LNP Vaccines. Pharmaceutics 2025, 17, 1148. [Google Scholar] [CrossRef]
- Udepurkar, A.; Devos, C.; Sagmeister, P.; Destro, F.; Inguva, P.; Ahmadi, S.; Boulais, E.; Quan, Y.; Braatz, R.D.; Myerson, A.S. Structure and Morphology of Lipid Nanoparticles for Nucleic Acid Drug Delivery: A Review. ACS Nano 2025, 19, 21206–21242. [Google Scholar] [CrossRef] [PubMed]
- Mehraji, S.; DeVoe, D.L. Microfluidic synthesis of lipid-based nanoparticles for drug delivery: Recent advances and opportunities. Lab Chip 2024, 24, 1154–1174. [Google Scholar] [CrossRef]
- Vogelaar, A.; Marcotte, S.; Cheng, J.; Oluoch, B.; Zaro, J. Use of Microfluidics to Prepare Lipid-Based Nanocarriers. Pharmaceutics 2023, 15, 1053. [Google Scholar] [CrossRef]
- Maeki, M.; Uno, S.; Niwa, A.; Okada, Y.; Tokeshi, M. Microfluidic technologies and devices for lipid nanoparticle-based RNA delivery. J. Control. Release 2022, 344, 80–96. [Google Scholar] [CrossRef]
- Jürgens, D.C.; Deßloch, L.; Porras-Gonzalez, D.; Winkeljann, J.; Zielinski, S.; Munschauer, M.; Hörner, A.L.; Burgstaller, G.; Winkeljann, B.; Merkel, O.M. Lab-scale siRNA and mRNA LNP manufacturing by various microfluidic mixing techniques—An evaluation of particle properties and efficiency. OpenNano 2023, 12, 100161. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, J.; Sun, R.; Han, S.; Yang, Z.; Teng, L. Microfluidics for nano-drug delivery systems: From fundamentals to industrialization. Acta Pharm. Sin. B 2023, 13, 3277–3299. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Issadore, D.; Mitchell, M.J. Microfluidic formulation of nanoparticles for biomedical applications. Biomaterials 2021, 274, 120826. [Google Scholar] [CrossRef]
- Lu, M.; Ozcelik, A.; Grigsby, C.L.; Zhao, Y.; Guo, F.; Leong, K.W.; Huang, T.J. Microfluidic Hydrodynamic Focusing for Synthesis of Nanomaterials. Nano Today 2016, 11, 778–792. [Google Scholar] [CrossRef]
- Prakash, G.; Shokr, A.; Willemen, N.; Bashir, S.M.; Shin, S.R.; Hassan, S. Microfluidic fabrication of lipid nanoparticles for the delivery of nucleic acids. Adv. Drug Deliv. Rev. 2022, 184, 114197. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, G.; Hui, Y.; Ranaweera, S.; Zhao, C.-X. Microfluidic Nanoparticles for Drug Delivery. Small 2022, 18, 2106580. [Google Scholar] [CrossRef]
- Belliveau, N.M.; Huft, J.; Lin, P.J.; Chen, S.; Leung, A.K.; Leaver, T.J.; Wild, A.W.; Lee, J.B.; Taylor, R.J.; Tam, Y.K.; et al. Microfluidic Synthesis of Highly Potent Limit-size Lipid Nanoparticles for In Vivo Delivery of siRNA. Mol. Ther. Nucleic Acids 2012, 1, e37. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.S.; Bostic, W.K.V.; Malmstadt, N. 3D-printed microfluidic device for high-throughput production of lipid nanoparticles incorporating SARS-CoV-2 spike protein mRNA. Lab Chip 2024, 24, 162–170. [Google Scholar] [CrossRef]
- Shepherd, S.J.; Warzecha, C.C.; Yadavali, S.; El-Mayta, R.; Alameh, M.-G.; Wang, L.; Weissman, D.; Wilson, J.M.; Issadore, D.; Mitchell, M.J. Scalable mRNA and siRNA Lipid Nanoparticle Production Using a Parallelized Microfluidic Device. Nano Lett. 2021, 21, 5671–5680. [Google Scholar] [CrossRef] [PubMed]
- Ripoll, M.; Martin, E.; Enot, M.; Robbe, O.; Rapisarda, C.; Nicolai, M.-C.; Deliot, A.; Tabeling, P.; Authelin, J.-R.; Nakach, M.; et al. Optimal self-assembly of lipid nanoparticles (LNP) in a ring micromixer. Sci. Rep. 2022, 12, 9483. [Google Scholar] [CrossRef]
- Jung, D.; Jang, S.; Park, D.; Bae, N.H.; Han, C.S.; Ryu, S.; Lim, E.-K.; Lee, K.G. Automated Microfluidic Systems Facilitating the Scalable and Reliable Production of Lipid Nanoparticles for Gene Delivery. BioChip J. 2025, 19, 79–90. [Google Scholar] [CrossRef]
- Han, J.Y.; La Fiandra, J.N.; DeVoe, D.L. Microfluidic vortex focusing for high throughput synthesis of size-tunable liposomes. Nat. Commun. 2022, 13, 6997. [Google Scholar] [CrossRef]
- Seder, I.; Zheng, T.; Zhang, J.; Rojas, C.C.; Helalat, S.H.; Téllez, R.C.; Sun, Y. A Scalable Microfluidic Platform for Nanoparticle Formulation: For Exploratory- and Industrial-Level Scales. Nano Lett. 2024, 24, 5132–5138. [Google Scholar] [CrossRef]
- Hanna, A.R.; Shepherd, S.J.; Datto, G.A.; Navarro, I.B.; Ricciardi, A.S.; Padilla, M.S.; Srikumar, N.; Zhang, S.; Yamagata, H.M.; Meng, N.Y.; et al. Automated and parallelized microfluidic generation of large and precisely-defined lipid nanoparticle libraries. bioRxiv 2025. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yen, C.-W.; Lin, H.-C.; Hou, W.; Estevez, A.; Sarode, A.; Goyon, A.; Bian, J.; Lin, J.; Koenig, S.G.; et al. Automated high-throughput preparation and characterization of oligonucleotide-loaded lipid nanoparticles. Int. J. Pharm. 2021, 599, 120392. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, M.P.; Vanover, D.; Beyersdorf, J.; Hatit, M.Z.C.; Rotolo, L.; Echeverri, E.S.; Peck, H.E.; Ni, H.; Yoon, J.-K.; Kim, Y.; et al. Optimization of lipid nanoparticles for the delivery of nebulized therapeutic mRNA to the lungs. Nat. Biomed. Eng. 2021, 5, 1059–1068. [Google Scholar] [CrossRef]
- Sago, C.D.; Lokugamage, M.P.; Paunovska, K.; Vanover, D.A.; Monaco, C.M.; Shah, N.N.; Gamboa Castro, M.; Anderson, S.E.; Rudoltz, T.G.; Lando, G.N.; et al. High-throughput in vivo screen of functional mRNA delivery identifies nanoparticles for endothelial cell gene editing. Proc. Natl. Acad. Sci. USA 2018, 115, E9944–E9952. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, J.E.; Kauffman, K.J.; Xing, Y.; Shaw, T.E.; Mir, F.F.; Dlott, C.C.; Langer, R.; Anderson, D.G.; Wang, E.T. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl. Acad. Sci. USA 2017, 114, 2060–2065. [Google Scholar] [CrossRef]
- Xue, L.; Hamilton, A.G.; Zhao, G.; Xiao, Z.; El-Mayta, R.; Han, X.; Gong, N.; Xiong, X.; Xu, J.; Figueroa-Espada, C.G.; et al. High-throughput barcoding of nanoparticles identifies cationic, degradable lipid-like materials for mRNA delivery to the lungs in female preclinical models. Nat. Commun. 2024, 15, 1884. [Google Scholar] [CrossRef]
- Hatit, M.Z.C.; Dobrowolski, C.N.; Lokugamage, M.P.; Loughrey, D.; Ni, H.; Zurla, C.; Da Silva Sanchez, A.J.; Radmand, A.; Huayamares, S.G.; Zenhausern, R.; et al. Nanoparticle stereochemistry-dependent endocytic processing improves in vivo mRNA delivery. Nat. Chem. 2023, 15, 508–515. [Google Scholar] [CrossRef]
- Rhym, L.H.; Manan, R.S.; Koller, A.; Stephanie, G.; Anderson, D.G. Peptide-encoding mRNA barcodes for the high-throughput in vivo screening of libraries of lipid nanoparticles for mRNA delivery. Nat. Biomed. Eng. 2023, 7, 901–910. [Google Scholar] [CrossRef]
- Odunze, U.; Rustogi, N.; Devine, P.; Miller, L.; Pereira, S.; Vashist, S.; Snijder, H.J.; Corkill, D.; Sabirsh, A.; Douthwaite, J.; et al. RNA encoded peptide barcodes enable efficient in vivo screening of RNA delivery systems. Nucleic Acids Res. 2024, 52, 9384–9396. [Google Scholar] [CrossRef]
- Guimaraes, P.P.G.; Zhang, R.; Spektor, R.; Tan, M.; Chung, A.; Billingsley, M.M.; El-Mayta, R.; Riley, R.S.; Wang, L.; Wilson, J.M.; et al. Ionizable lipid nanoparticles encapsulating barcoded mRNA for accelerated in vivo delivery screening. J. Control. Release 2019, 316, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E. Rational Design of Cationic Lipids for siRNA Delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Brimacombe, C.A.; Kulkarni, J.A.; Cheng, M.H.Y.; An, K.; Witzigmann, D.; Cullis, P.R. Rational design of lipid nanoparticles for enabling gene therapies. Mol. Ther. Methods Clin. Dev. 2025, 33, 101518. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Scherphof, L.G.; Kamps, J.A.A.M. Liposome Opsonization. J. Liposome Res. 2005, 15, 109–139. [Google Scholar] [CrossRef]
- Roerdink, F.; Wassef, N.M.; Richardson, E.C.; Alving, C.R. Effects of negatively charged lipids on phagocytosis of liposomes opsonized by complement. Biochim. Et. Biophys. Acta (BBA)—Biomembr. 1983, 734, 33–39. [Google Scholar] [CrossRef]
- Rampado, R.; Crotti, S.; Caliceti, P.; Pucciarelli, S.; Agostini, M. Recent Advances in Understanding the Protein Corona of Nanoparticles and in the Formulation of “Stealthy” Nanomaterials. Front. Bioeng. Biotechnol. 2020, 8, 166. [Google Scholar] [CrossRef]
- Francia, V.; Schiffelers, R.M.; Cullis, P.R.; Witzigmann, D. The Biomolecular Corona of Lipid Nanoparticles for Gene Therapy. Bioconjug Chem. 2020, 31, 2046–2059. [Google Scholar] [CrossRef]
- Mahmoudi, M.; Landry, M.P.; Moore, A.; Coreas, R. The protein corona from nanomedicine to environmental science. Nat. Rev. Mater. 2023, 8, 422–438. [Google Scholar] [CrossRef]
- Huzar, J.; Coreas, R.; Landry, M.P.; Tikhomirov, G. AI-Based Prediction of Protein Corona Composition on DNA Nanostructures. ACS Nano 2025, 19, 4333–4345. [Google Scholar] [CrossRef]
- van Straten, D.; Sork, H.; van de Schepop, L.; Frunt, R.; Ezzat, K.; Schiffelers, R.M. Biofluid specific protein coronas affect lipid nanoparticle behavior in vitro. J. Control. Release 2024, 373, 481–492. [Google Scholar] [CrossRef] [PubMed]
- Blume, J.E.; Manning, W.C.; Troiano, G.; Hornburg, D.; Figa, M.; Hesterberg, L.; Platt, T.L.; Zhao, X.; Cuaresma, R.A.; Everley, P.A.; et al. Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat. Commun. 2020, 11, 3662. [Google Scholar] [CrossRef]
- Ashkarran, A.A.; Gharibi, H.; Sadeghi, S.A.; Modaresi, S.M.; Wang, Q.; Lin, T.J.; Yerima, G.; Tamadon, A.; Sayadi, M.; Jafari, M.; et al. Small molecule modulation of protein corona for deep plasma proteome profiling. Nat. Commun. 2024, 15, 9638. [Google Scholar] [CrossRef]
- Papi, M.; Caputo, D.; Palmieri, V.; Coppola, R.; Palchetti, S.; Bugli, F.; Martini, C.; Digiacomo, L.; Pozzi, D.; Caracciolo, G. Clinically approved PEGylated nanoparticles are covered by a protein corona that boosts the uptake by cancer cells. Nanoscale 2017, 9, 10327–10334. [Google Scholar] [CrossRef] [PubMed]
- Navarro-Marchal, S.A.; Martín-Contreras, M.; Castro-Santiago, D.; del Castillo-Santaella, T.; Graván, P.; Jódar-Reyes, A.B.; Marchal, J.A.; Peula-García, J.M. Effect of the Protein Corona Formation on Antibody Functionalized Liquid Lipid Nanocarriers. Int. J. Mol. Sci. 2023, 24, 16759. [Google Scholar] [CrossRef]
- Chou, W.-C.; Lin, Z. Impact of protein coronas on nanoparticle interactions with tissues and targeted delivery. Curr. Opin. Biotechnol. 2024, 85, 103046. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. The role of surface chemistry in serum protein corona-mediated cellular delivery and gene silencing with lipid nanoparticles. Nanoscale 2019, 11, 8760–8775. [Google Scholar] [CrossRef]
- Amici, A.; Caracciolo, G.; Digiacomo, L.; Gambini, V.; Marchini, C.; Tilio, M.; Capriotti, A.L.; Colapicchioni, V.; Matassa, R.; Familiari, G.; et al. In vivo protein corona patterns of lipid nanoparticles. RSC Adv. 2017, 7, 1137–1145. [Google Scholar] [CrossRef]
- Giulimondi, F.; Vulpis, E.; Digiacomo, L.; Giuli, M.V.; Mancusi, A.; Capriotti, A.L.; Laganà, A.; Cerrato, A.; Zenezini Chiozzi, R.; Nicoletti, C.; et al. Opsonin-Deficient Nucleoproteic Corona Endows UnPEGylated Liposomes with Stealth Properties In Vivo. ACS Nano 2022, 16, 2088–2100. [Google Scholar] [CrossRef]
- Chen, D.; Ganesh, S.; Wang, W.; Amiji, M. Protein Corona-Enabled Systemic Delivery and Targeting of Nanoparticles. Aaps J. 2020, 22, 83. [Google Scholar] [CrossRef]
- Kim, W.; Ly, N.K.; He, Y.; Li, Y.; Yuan, Z.; Yeo, Y. Protein corona: Friend or foe? Co-opting serum proteins for nanoparticle delivery. Adv. Drug Deliv. Rev. 2023, 192, 114635. [Google Scholar] [CrossRef]
- Chen, D.; Parayath, N.; Ganesh, S.; Wang, W.; Amiji, M. The role of apolipoprotein- and vitronectin-enriched protein corona on lipid nanoparticles for in vivo targeted delivery and transfection of oligonucleotides in murine tumor models. Nanoscale 2019, 11, 18806–18824. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiao, W.; He, S.; Xia, X.; Yang, W.; Yang, Z.; Hu, H.; Wang, Y.; Wang, X.; Li, H.; et al. Lipid-mediated protein corona regulation with increased apolipoprotein A-I recruitment for glioma targeting. J. Control. Release 2024, 368, 42–51. [Google Scholar] [CrossRef]
- Miao, L.; Lin, J.; Huang, Y.; Li, L.; Delcassian, D.; Ge, Y.; Shi, Y.; Anderson, D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020, 11, 2424. [Google Scholar] [CrossRef] [PubMed]
- Gray, N. Knowing the limit. Nat. Cell Biol. 2009, 11, S8. [Google Scholar] [CrossRef]
- Crawford, R.; Dogdas, B.; Keough, E.; Haas, R.M.; Wepukhulu, W.; Krotzer, S.; Burke, P.A.; Sepp-Lorenzino, L.; Bagchi, A.; Howell, B.J. Analysis of lipid nanoparticles by Cryo-EM for characterizing siRNA delivery vehicles. Int. J. Pharm. 2011, 403, 237–244. [Google Scholar] [CrossRef]
- Pattipeiluhu, R.; Zeng, Y.; Hendrix, M.M.R.M.; Voets, I.K.; Kros, A.; Sharp, T.H. Liquid crystalline inverted lipid phases encapsulating siRNA enhance lipid nanoparticle mediated transfection. Nat. Commun. 2024, 15, 1303. [Google Scholar] [CrossRef]
- Leung, A.K.K.; Hafez, I.M.; Baoukina, S.; Belliveau, N.M.; Zhigaltsev, I.V.; Afshinmanesh, E.; Tieleman, D.P.; Hansen, C.L.; Hope, M.J.; Cullis, P.R. Lipid Nanoparticles Containing siRNA Synthesized by Microfluidic Mixing Exhibit an Electron-Dense Nanostructured Core. J. Phys. Chem. C 2012, 116, 18440–18450. [Google Scholar] [CrossRef] [PubMed]
- Hammel, M.; Fan, Y.; Sarode, A.; Byrnes, A.E.; Zang, N.; Kou, P.; Nagapudi, K.; Leung, D.; Hoogenraad, C.C.; Chen, T.; et al. Correlating the Structure and Gene Silencing Activity of Oligonucleotide-Loaded Lipid Nanoparticles Using Small-Angle X-ray Scattering. ACS Nano 2023, 17, 11454–11465. [Google Scholar] [CrossRef] [PubMed]
- Yap, S.L.; Yu, H.; Li, S.; Drummond, C.J.; Conn, C.E.; Tran, N. Cell interactions with lipid nanoparticles possessing different internal nanostructures: Liposomes, bicontinuous cubosomes, hexosomes, and discontinuous micellar cubosomes. J. Colloid. Interface Sci. 2024, 656, 409–423. [Google Scholar] [CrossRef]
- Dao, H.M.; AboulFotouh, K.; Hussain, A.F.; Marras, A.E.; Johnston, K.P.; Cui, Z.; Williams, R.O. Characterization of mRNA Lipid Nanoparticles by Electron Density Mapping Reconstruction: X-ray Scattering with Density from Solution Scattering (DENSS) Algorithm. Pharm. Res. 2024, 41, 501–512. [Google Scholar] [CrossRef]
- Szebeni, J.; Kiss, B.; Bozó, T.; Turjeman, K.; Levi-Kalisman, Y.; Barenholz, Y.; Kellermayer, M. Insights into the Structure of Comirnaty Covid-19 Vaccine: A Theory on Soft, Partially Bilayer-Covered Nanoparticles with Hydrogen Bond-Stabilized mRNA–Lipid Complexes. ACS Nano 2023, 17, 13147–13157. [Google Scholar] [CrossRef] [PubMed]
- Unruh, T.; Götz, K.; Vogel, C.; Fröhlich, E.; Scheurer, A.; Porcar, L.; Steiniger, F. Mesoscopic Structure of Lipid Nanoparticle Formulations for mRNA Drug Delivery: Comirnaty and Drug-Free Dispersions. ACS Nano 2024, 18, 9746–9764. [Google Scholar] [CrossRef]
- Zheng, L.; Bandara, S.R.; Tan, Z.; Leal, C. Lipid nanoparticle topology regulates endosomal escape and delivery of RNA to the cytoplasm. Proc. Natl. Acad. Sci. USA 2023, 120, e2301067120. [Google Scholar] [CrossRef]
- Eygeris, Y.; Patel, S.; Jozic, A.; Sahay, G. Deconvoluting Lipid Nanoparticle Structure for Messenger RNA Delivery. Nano Lett. 2020, 20, 4543–4549. [Google Scholar] [CrossRef]
- Ellens, H.; Siegel, D.P.; Alford, D.; Yeagle, P.L.; Boni, L.; Lis, L.J.; Quinn, P.J.; Bentz, J. Membrane fusion and inverted phases. Biochemistry 1989, 28, 3692–3703. [Google Scholar] [CrossRef]
- Verkleij, A.J.; van Echteld, C.J.; Gerritsen, W.J.; Cullis, P.R.; de Kruijff, B. The lipidic particle as an intermediate structure in membrane fusion processes and bilayer to hexagonal HII transitions. Biochim. Biophys. Acta 1980, 600, 620–624. [Google Scholar] [CrossRef]
- Cheng, M.H.Y.; Leung, J.; Zhang, Y.; Strong, C.; Basha, G.; Momeni, A.; Chen, Y.; Jan, E.; Abdolahzadeh, A.; Wang, X.; et al. Induction of Bleb Structures in Lipid Nanoparticle Formulations of mRNA Leads to Improved Transfection Potency. Adv. Mater. 2023, 35, 2303370. [Google Scholar] [CrossRef]
- Grzetic, D.J.; Hamilton, N.B.; Shelley, J.C. Coarse-Grained Simulation of mRNA-Loaded Lipid Nanoparticle Self-Assembly. Mol. Pharm. 2024, 21, 4747–4753. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, N.B.; Arns, S.; Shelley, M.; Bechis, I.; Shelley, J.C. Calculating Apparent pKa Values of Ionizable Lipids in Lipid Nanoparticles. Mol. Pharm. 2025, 22, 588–593. [Google Scholar] [CrossRef] [PubMed]
- Trollmann, M.F.W.; Böckmann, R.A. mRNA lipid nanoparticle phase transition. Biophys. J. 2022, 121, 3927–3939. [Google Scholar] [CrossRef]
- Parot, J.; Mehn, D.; Jankevics, H.; Markova, N.; Carboni, M.; Olaisen, C.; Hoel, A.D.; Sigfúsdóttir, M.S.; Meier, F.; Drexel, R.; et al. Quality assessment of LNP-RNA therapeutics with orthogonal analytical techniques. J. Control. Release 2024, 367, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Kon, E.; Sharma, P.; Peer, D. Endosomal escape: A bottleneck for LNP-mediated therapeutics. Proc. Natl. Acad. Sci. USA 2024, 121, e2307800120. [Google Scholar] [CrossRef] [PubMed]
- Sahay, G.; Querbes, W.; Alabi, C.; Eltoukhy, A.; Sarkar, S.; Zurenko, C.; Karagiannis, E.; Love, K.; Chen, D.; Zoncu, R.; et al. Efficiency of siRNA delivery by lipid nanoparticles is limited by endocytic recycling. Nat. Biotechnol. 2013, 31, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Wittrup, A.; Ai, A.; Liu, X.; Hamar, P.; Trifonova, R.; Charisse, K.; Manoharan, M.; Kirchhausen, T.; Lieberman, J. Visualizing lipid-formulated siRNA release from endosomes and target gene knockdown. Nat. Biotechnol. 2015, 33, 870–876. [Google Scholar] [CrossRef]
- Gilleron, J.; Querbes, W.; Zeigerer, A.; Borodovsky, A.; Marsico, G.; Schubert, U.; Manygoats, K.; Seifert, S.; Andree, C.; Stöter, M. Image-Based Analysis of Lipid Nanoparticle-Mediated siRNA Delivery, Intracellular Trafficking and Endosomal Escape. Nat. Biotechnol. 2013, 31, 638–646. [Google Scholar] [CrossRef]
- Paramasivam, P.; Franke, C.; Stöter, M.; Höijer, A.; Bartesaghi, S.; Sabirsh, A.; Lindfors, L.; Arteta, M.Y.; Dahlén, A.; Bak, A.; et al. Endosomal escape of delivered mRNA from endosomal recycling tubules visualized at the nanoscale. J. Cell Biol. 2021, 221, e202110137. [Google Scholar] [CrossRef]
- Aliakbarinodehi, N.; Niederkofler, S.; Emilsson, G.; Parkkila, P.; Olsén, E.; Jing, Y.; Sjöberg, M.; Agnarsson, B.; Lindfors, L.; Höök, F. Time-Resolved Inspection of Ionizable Lipid-Facilitated Lipid Nanoparticle Disintegration and Cargo Release at an Early Endosomal Membrane Mimic. ACS Nano 2024, 18, 22989–23000. [Google Scholar] [CrossRef]
- Lam, Y.Y.; Hawley, A.; Tan, A.; Boyd, B.J. Coupling in vitro cell culture with synchrotron SAXS to understand the bio-interaction of lipid-based liquid crystalline nanoparticles with vascular endothelial cells. Drug Deliv. Transl. Res. 2020, 10, 610–620. [Google Scholar] [CrossRef]
- Bruininks, B.M.; Souza, P.C.; Ingolfsson, H.; Marrink, S.J. A molecular view on the escape of lipoplexed DNA from the endosome. Elife 2020, 9, e52012. [Google Scholar] [CrossRef]
- Garaizar, A.; Díaz-Oviedo, D.; Zablowsky, N.; Rissanen, S.; Köbberling, J.; Sun, J.; Steiger, C.; Steigemann, P.; Mann, F.A.; Meier, K. Toward understanding lipid reorganization in RNA lipid nanoparticles in acidic environments. Proc. Natl. Acad. Sci. USA 2024, 121, e2404555121. [Google Scholar] [CrossRef]
- Dorsey, P.J.; Lau, C.L.; Chang, T.-c.; Doerschuk, P.C.; D’Addio, S.M. Review of machine learning for lipid nanoparticle formulation and process development. J. Pharm. Sci. 2024, 113, 3413–3433. [Google Scholar] [CrossRef]
- Amoako, K.; Mokhammad, A.; Malik, A.; Yesudasan, S.; Wheba, A.; Olagunju, O.; Gu, S.X.; Yarovinsky, T.; Faustino, E.V.S.; Nguyen, J.; et al. Enhancing nucleic acid delivery by the integration of artificial intelligence into lipid nanoparticle formulation. Front. Med. Technol. 2025, 7, 1591119. [Google Scholar] [CrossRef]
- Moayedpour, S.; Broadbent, J.; Riahi, S.; Bailey, M.; Thu, H.V.; Dobchev, D.; Balsubramani, A.; Santos, R.N.D.; Kogler-Anele, L.; Corrochano-Navarro, A.; et al. Representations of lipid nanoparticles using large language models for transfection efficiency prediction. Bioinformatics 2024, 40, btae342. [Google Scholar] [CrossRef]
- Li, B.; Raji, I.O.; Gordon, A.G.R.; Sun, L.; Raimondo, T.M.; Oladimeji, F.A.; Jiang, A.Y.; Varley, A.; Langer, R.S.; Anderson, D.G. Accelerating ionizable lipid discovery for mRNA delivery using machine learning and combinatorial chemistry. Nat. Mater. 2024, 23, 1002–1008. [Google Scholar] [CrossRef]
- Shan, X.; Cai, Y.; Zhu, B.; Zhou, L.; Sun, X.; Xu, X.; Yin, Q.; Wang, D.; Li, Y. Rational strategies for improving the efficiency of design and discovery of nanomedicines. Nat. Commun. 2024, 15, 9990. [Google Scholar] [CrossRef]
- Witten, J.; Raji, I.; Manan, R.S.; Beyer, E.; Bartlett, S.; Tang, Y.; Ebadi, M.; Lei, J.; Nguyen, D.; Oladimeji, F.; et al. Artificial intelligence-guided design of lipid nanoparticles for pulmonary gene therapy. Nat. Biotechnol. 2024. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.-H.; Choi, H.; Lee, J.; Kang, M.-H.; Ahn, S.-H.; Lee, Y.-S.; Choi, H.; Jo, S.; Lee, Y.; Park, H.-J.; et al. Rational Design of Lipid Nanoparticles for Enhanced mRNA Vaccine Delivery via Machine Learning. Small 2025, 21, 2405618. [Google Scholar] [CrossRef]
- Wang, W.; Chen, K.; Jiang, T.; Wu, Y.; Wu, Z.; Ying, H.; Yu, H.; Lu, J.; Lin, J.; Ouyang, D. Artificial intelligence-driven rational design of ionizable lipids for mRNA delivery. Nat. Commun. 2024, 15, 10804. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Ma, S.; Cui, H.; Chen, J.; Xu, S.; Gong, F.; Golubovic, A.; Zhou, M.; Wang, K.C.; Varley, A.; et al. AGILE platform: A deep learning powered approach to accelerate LNP development for mRNA delivery. Nat. Commun. 2024, 15, 6305. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Wang, Z.; Yang, X.; Chen, Y.; Mastrogiovanni, F.; Zhang, J.; Liu, L. TransMA: An explainable multi-modal deep learning model for predicting properties of ionizable lipid nanoparticles in mRNA delivery. Brief. Bioinform. 2025, 26, bbaf307. [Google Scholar] [CrossRef]
- Cheng, L.; Zhu, Y.; Ma, J.; Aggarwal, A.; Toh, W.H.; Shin, C.; Sangpachatanaruk, W.; Weng, G.; Kumar, R.; Mao, H.-Q. Machine Learning Elucidates Design Features of Plasmid Deoxyribonucleic Acid Lipid Nanoparticles for Cell Type-Preferential Transfection. ACS Nano 2024, 18, 28735–28747. [Google Scholar] [CrossRef]
- Kumar, G.; Ardekani, A.M. Machine-Learning Framework to Predict the Performance of Lipid Nanoparticles for Nucleic Acid Delivery. ACS Appl. Bio Mater. 2025, 8, 3717–3727. [Google Scholar] [CrossRef]
- Hunter, M.R.; Cui, L.; Porebski, B.T.; Pereira, S.; Sonzini, S.; Odunze, U.; Iyer, P.; Engkvist, O.; Lloyd, R.L.; Peel, S.; et al. Understanding Intracellular Biology to Improve mRNA Delivery by Lipid Nanoparticles. Small Methods 2023, 7, 2201695. [Google Scholar] [CrossRef]
- Fu, X.; Yang, C.; Su, Y.; Liu, C.; Qiu, H.; Yu, Y.; Su, G.; Zhang, Q.; Wei, L.; Cui, F.; et al. Machine Learning Enables Comprehensive Prediction of the Relative Protein Abundance of Multiple Proteins on the Protein Corona. Research 2024, 7, 0487. [Google Scholar] [CrossRef] [PubMed]
- Kopac, T. Leveraging Artificial Intelligence and Machine Learning for Characterizing Protein Corona, Nanobiological Interactions, and Advancing Drug Discovery. Bioengineering 2025, 12, 312. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Coreas, R.; Liu, Y.; Bitounis, D.; Zhang, Z.; Parviz, D.; Strano, M.; Demokritou, P.; Zhong, W. Prediction of protein corona on nanomaterials by machine learning using novel descriptors. NanoImpact 2020, 17, 100207. [Google Scholar] [CrossRef] [PubMed]


| Device | Source of Relevant Information | General Comment |
|---|---|---|
| T-junction/Y-junction | [44,45,46,47,48,49,50,51] | The type of mixer that has a low Re, long mixing time, higher polydispersity |
| Hydrodynamic flow focusing (HFF/3-inlet flow focusing) | [41,44,46,48,49,50,51,52,53,54] | faster mixing, can dilute samples |
| Staggered herringbone mixer (SHM)/ chaotic micromixers | [38,39,40,41,42,44,46,47,48,49,50,51,53,54,55,56,57] | Faster mixing, can clog |
| Ring/toroidal micromixer | [44,51,53,58,59] | Faster mixing, can clog |
| 3D/multi-layer microfluidic focusing (vertical focusing VFF, multi-inlet), vortex | [46,50,51,60,61] | Faster mixing |
| Droplet (emulsion) microfluidics → solvent removal | [50,51] | |
| Acoustic/electrokinetic micro-mixers (on-chip active mixing) | [43,46,50,54] | Faster mixing |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meers, P. Emerging Approaches for the Discovery of Lipid-Based RNA Delivery Systems. Pharmaceutics 2025, 17, 1231. https://doi.org/10.3390/pharmaceutics17091231
Meers P. Emerging Approaches for the Discovery of Lipid-Based RNA Delivery Systems. Pharmaceutics. 2025; 17(9):1231. https://doi.org/10.3390/pharmaceutics17091231
Chicago/Turabian StyleMeers, Paul. 2025. "Emerging Approaches for the Discovery of Lipid-Based RNA Delivery Systems" Pharmaceutics 17, no. 9: 1231. https://doi.org/10.3390/pharmaceutics17091231
APA StyleMeers, P. (2025). Emerging Approaches for the Discovery of Lipid-Based RNA Delivery Systems. Pharmaceutics, 17(9), 1231. https://doi.org/10.3390/pharmaceutics17091231





