Postnatal Epigenetic Alterations in Calves Persistently Infected with Bovine Viral Diarrhea Virus
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and PI Identification
2.2. Blood Collection and Complete Blood Counts (CBCs)
2.3. Isolation of White Blood Cells
2.4. Confirmation of PI Status by RT-PCR
2.5. Viral Neutralization Test
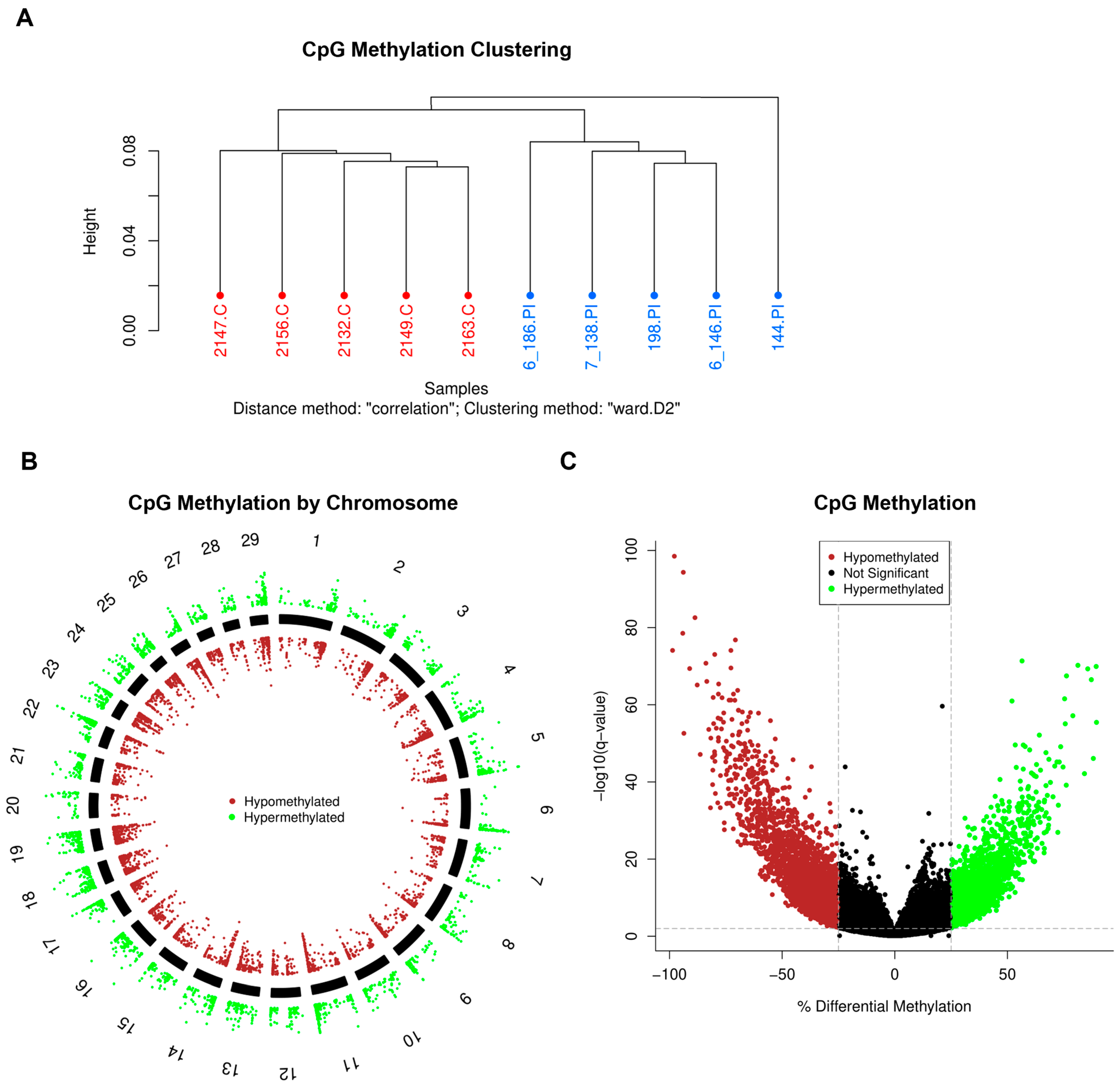
2.6. Reduced Representation Bisulfite Sequencing (RRBS)
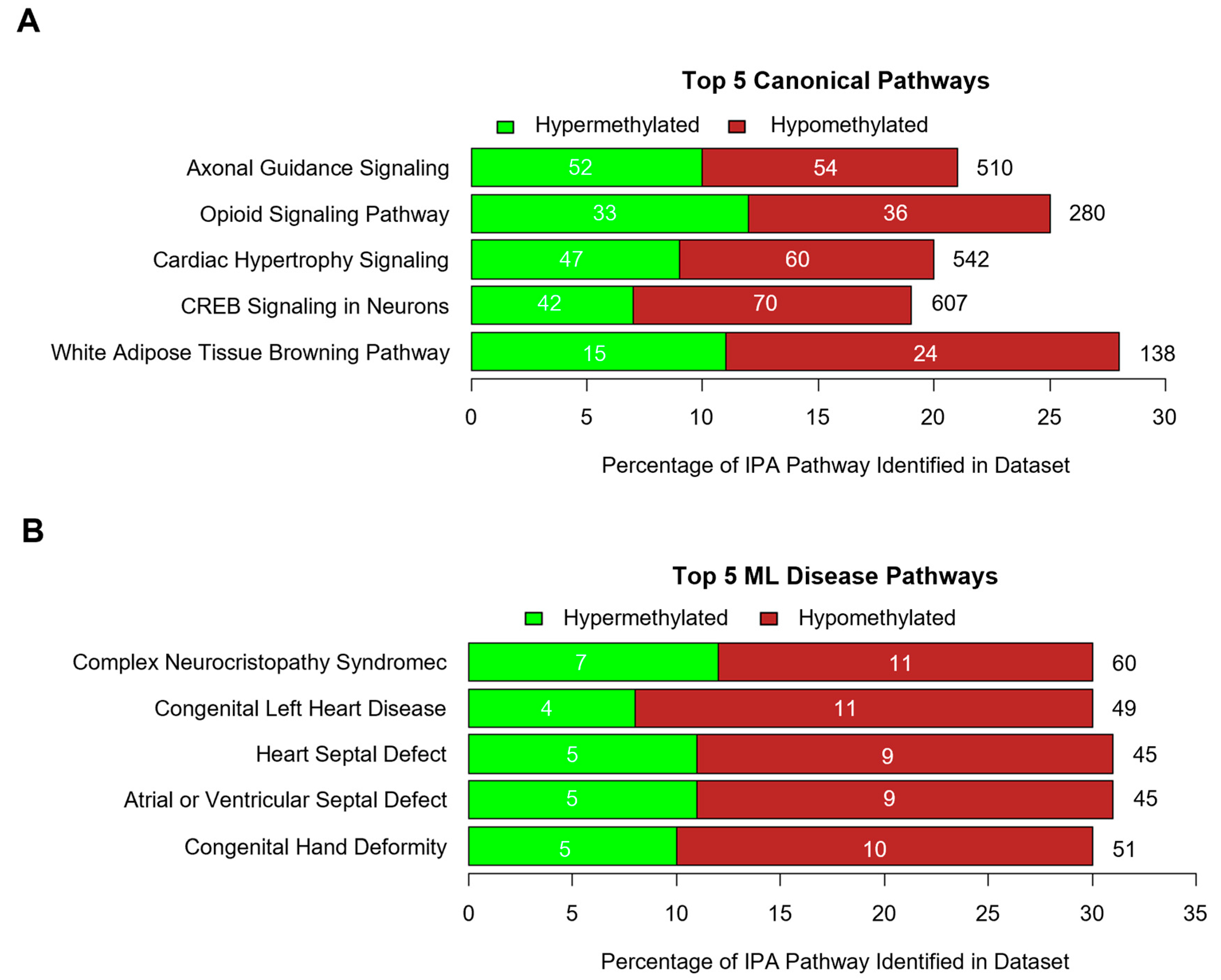
2.7. Methylation Bioinformatics and Pathway Analysis
2.8. Spectral Cytometry
2.9. Statistical Analysis
3. Results
3.1. BVDV Identification and Serology
3.2. DNA Methylation
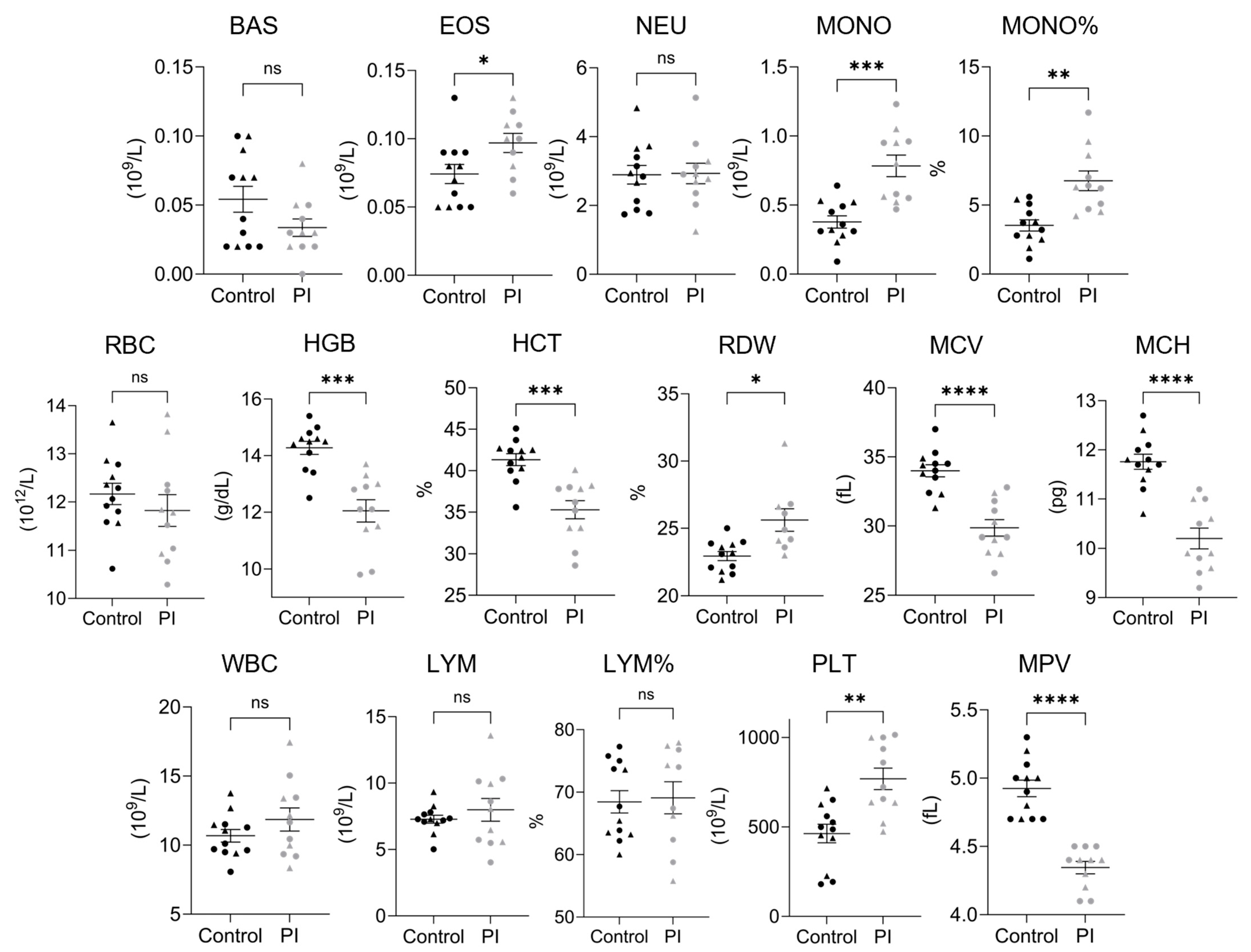
3.3. Complete Blood Counts
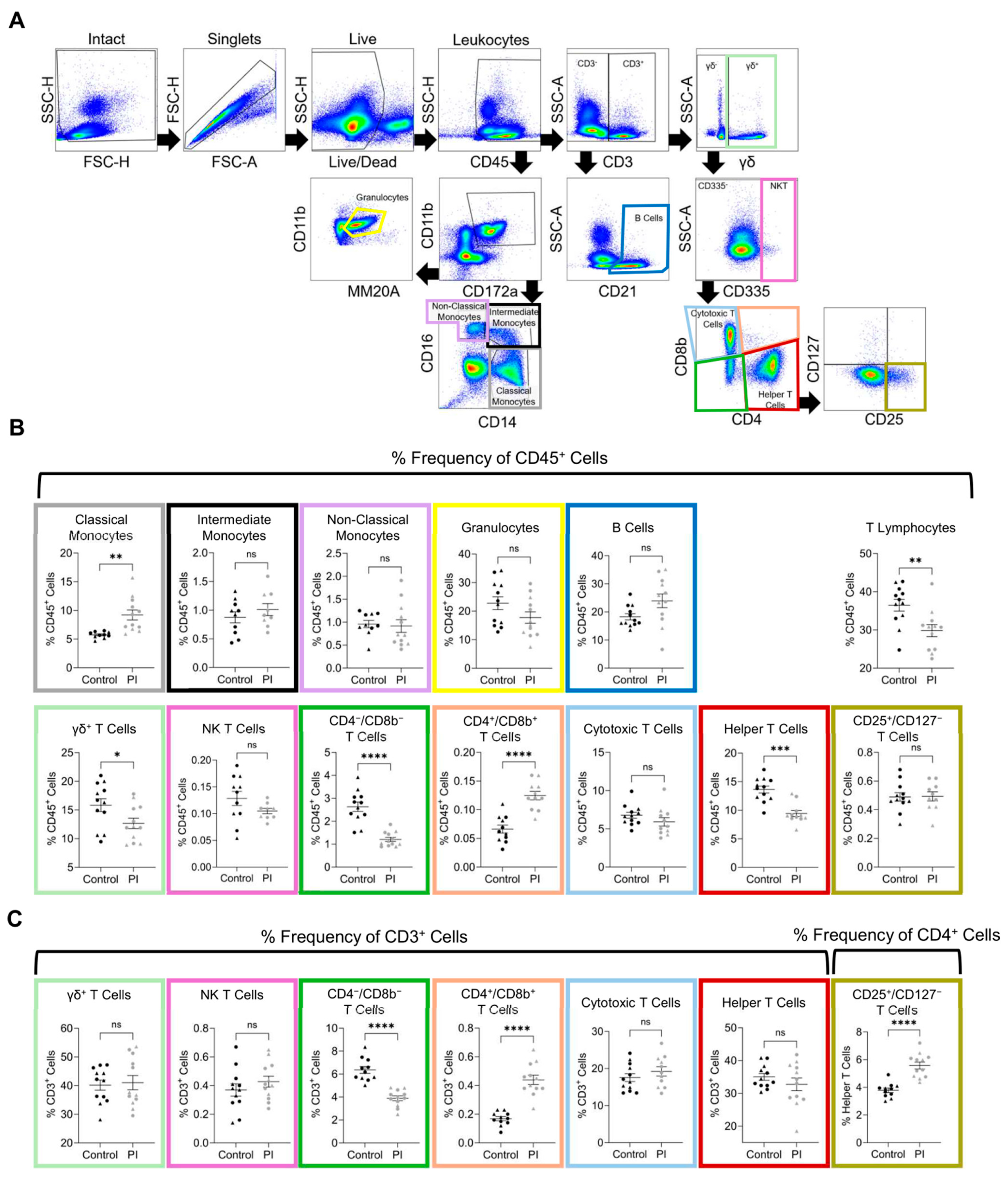
3.4. Flow Cytometry
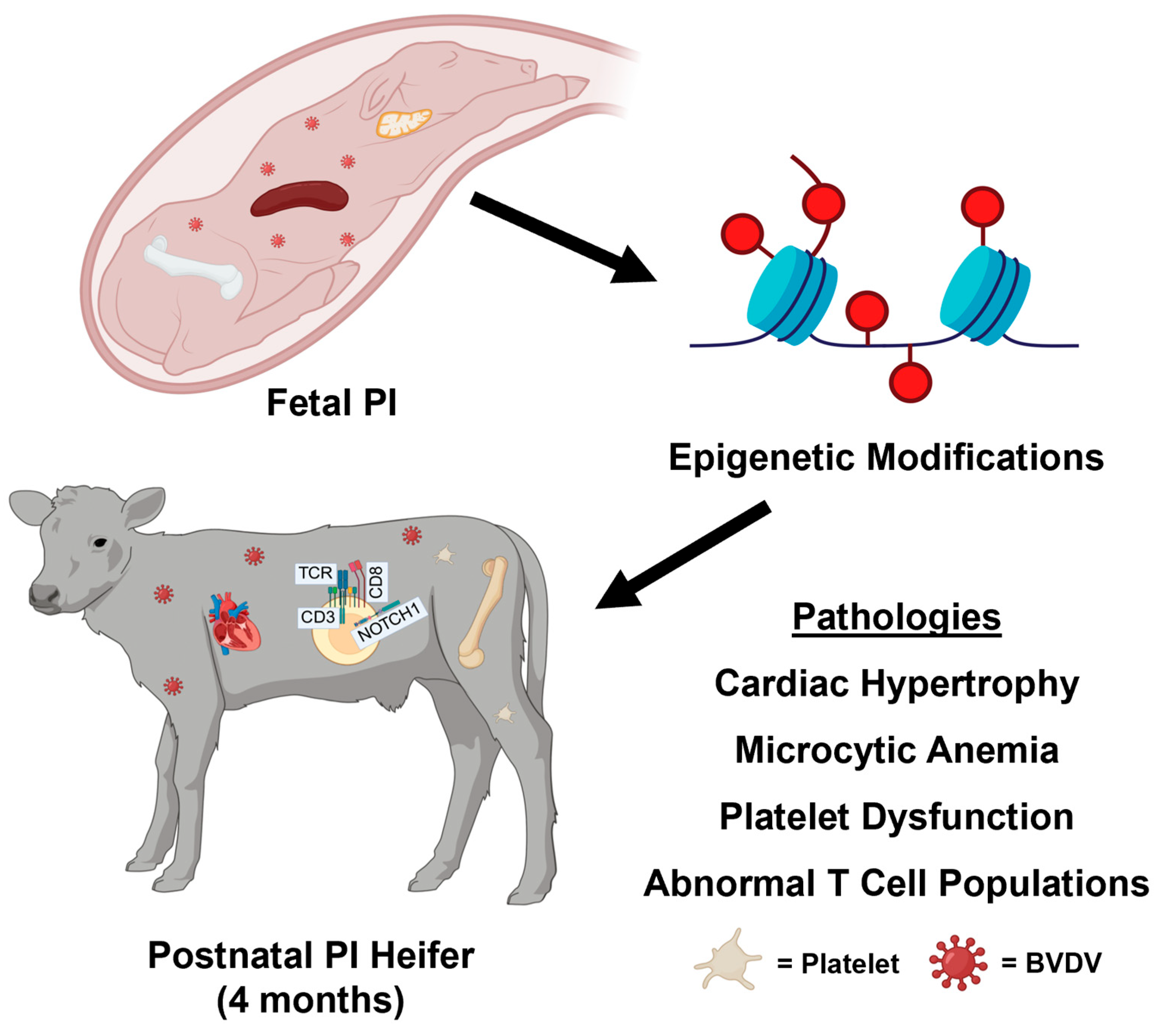
4. Discussion
4.1. Relationships Between PI CBCs, Flow Cytometry, and DNA Methylation
4.2. Epigenetic Findings in PI Calves Are Supported by Previous Research
4.3. Potential Confounders and Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pinior, B.; Firth, C.L.; Richter, V.; Lebl, K.; Trauffler, M.; Dzieciol, M.; Hutter, S.E.; Burgstaller, J.; Obritzhauser, W.; Winter, P.; et al. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. Prev. Vet. Med. 2017, 137, 77–92. [Google Scholar] [CrossRef]
- Gillespie, J.H.; Baker, J.A.; McEntee, K. A cytopathogenic strain of virus diarrhea virus. Cornell Vet. 1960, 50, 73–79. [Google Scholar]
- Underdahl, N.R.; Grace, O.D.; Hoerlein, A.B. Cultivation in tissue-culture of cytopathogenic agent from bovine mucosal disease. Proc. Soc. Exp. Biol. Med. 1957, 94, 795–797. [Google Scholar] [CrossRef]
- Tautz, N.; Tews, B.A.; Meyers, G. Chapter Two—The Molecular Biology of Pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [CrossRef]
- Moennig, V.; Liess, B. Pathogenesis of intrauterine infections with bovine viral diarrhea virus. Vet. Clin. N. Am. Food Anim. Pract. 1995, 11, 477–487. [Google Scholar] [CrossRef]
- Moerman, A.; Straver, P.J.; de Jong, M.C.; Quak, J.; Baanvinger, T.; van Oirschot, J.T. Clinical consequences of a bovine virus diarrhoea virus infection in a dairy herd: A longitudinal study. Vet. Q. 1994, 16, 115–119. [Google Scholar] [CrossRef]
- Dubovi, E.J. Impact of bovine viral diarrhea virus on reproductive performance in cattle. Vet. Clin. N. Am. Food Anim. Pract. 1994, 10, 503–514. [Google Scholar] [CrossRef]
- Brownlie, J.; Clarke, M.C.; Howard, C.J.; Pocock, D.H. Pathogenesis and epidemiology of bovine virus diarrhoea virus infection of cattle. Ann. Rech. Vet. 1987, 18, 157–166. [Google Scholar]
- Hessman, B.E.; Fulton, R.W.; Sjeklocha, D.B.; Murphy, T.A.; Ridpath, J.F.; Payton, M.E. Evaluation of economic effects and the health and performance of the general cattle population after exposure to cattle persistently infected with bovine viral diarrhea virus in a starter feedlot. Am. J. Vet. Res. 2009, 70, 73–85. [Google Scholar] [CrossRef]
- Baker, J.C. The clinical manifestations of bovine viral diarrhea infection. Vet. Clin. N. Am. Food Anim. Pract. 1995, 11, 425–445. [Google Scholar] [CrossRef]
- Bielefeldt-Ohmann, H. The pathologies of bovine viral diarrhea virus infection. A window on the pathogenesis. Vet. Clin. N. Am. Food Anim. Pract. 1995, 11, 447–476. [Google Scholar] [CrossRef] [PubMed]
- Done, J.T.; Terlecki, S.; Richardson, C.; Harkness, J.W.; Sands, J.J.; Patterson, D.S.; Sweasey, D.; Shaw, I.G.; Winkler, C.E.; Duffell, S.J. Bovine virus diarrhoea-mucosal disease virus: Pathogenicity for the fetal calf following maternal infection. Vet. Rec. 1980, 106, 473–479. [Google Scholar] [CrossRef] [PubMed]
- Brownlie, J.; Clarke, M.C.; Howard, C.J. Experimental production of fatal mucosal disease in cattle. Vet. Rec. 1984, 114, 535–536. [Google Scholar] [CrossRef] [PubMed]
- McClurkin, A.W.; Littledike, E.T.; Cutlip, R.C.; Frank, G.H.; Coria, M.F.; Bolin, S.R. Production of cattle immunotolerant to bovine viral diarrhea virus. Can. J. Comp. Med. 1984, 48, 156–161. [Google Scholar]
- Knapek, K.J.; Georges, H.M.; Van Campen, H.; Bishop, J.V.; Bielefeldt-Ohmann, H.; Smirnova, N.P.; Hansen, T.R. Fetal Lymphoid Organ Immune Responses to Transient and Persistent Infection with Bovine Viral Diarrhea Virus. Viruses 2020, 12, 816. [Google Scholar] [CrossRef]
- Georges, H.M.; Knapek, K.J.; Bielefeldt-Ohmann, H.; Van Campen, H.; Hansen, T.R. Attenuated lymphocyte activation leads to the development of immunotolerance in bovine fetuses persistently infected with bovine viral diarrhea virusdagger. Biol. Reprod. 2020, 103, 560–571. [Google Scholar] [CrossRef]
- Smirnova, N.P.; Ptitsyn, A.A.; Austin, K.J.; Bielefeldt-Ohmann, H.; Van Campen, H.; Han, H.; van Olphen, A.L.; Hansen, T.R. Persistent fetal infection with bovine viral diarrhea virus differentially affects maternal blood cell signal transduction pathways. Physiol. Genom. 2009, 36, 129–139. [Google Scholar] [CrossRef]
- Smirnova, N.P.; Webb, B.T.; McGill, J.L.; Schaut, R.G.; Bielefeldt-Ohmann, H.; Van Campen, H.; Sacco, R.E.; Hansen, T.R. Induction of interferon-gamma and downstream pathways during establishment of fetal persistent infection with bovine viral diarrhea virus. Virus Res. 2014, 183, 95–106. [Google Scholar] [CrossRef]
- Scruggs, D.W.; Fleming, S.A.; Maslin, W.R.; Groce, A.W. Osteopetrosis, anemia, thrombocytopenia, and marrow necrosis in beef calves naturally infected with bovine virus diarrhea virus. J. Vet. Diagn. Investig. 1995, 7, 555–559. [Google Scholar] [CrossRef]
- Webb, B.T.; Norrdin, R.W.; Smirnova, N.P.; Van Campen, H.; Weiner, C.M.; Antoniazzi, A.Q.; Bielefeldt-Ohmann, H.; Hansen, T.R. Bovine viral diarrhea virus cyclically impairs long bone trabecular modeling in experimental persistently infected fetuses. Vet. Pathol. 2012, 49, 930–940. [Google Scholar] [CrossRef]
- Nuss, K.; Spiess, A.; Hilbe, M.; Sterr, K.; Reiser, M.; Matis, U. Transient benign osteopetrosis in a calf persistently infected with bovine virus diarrhoea virus. Vet. Comp. Orthop. Traumatol. 2005, 18, 100–104. [Google Scholar]
- Brodersen, B.W. Bovine viral diarrhea virus infections: Manifestations of infection and recent advances in understanding pathogenesis and control. Vet. Pathol. 2014, 51, 453–464. [Google Scholar] [CrossRef]
- Fulton, R.W.; Hessman, B.E.; Ridpath, J.F.; Johnson, B.J.; Burge, L.J.; Kapil, S.; Braziel, B.; Kautz, K.; Reck, A. Multiple diagnostic tests to identify cattle with Bovine viral diarrhea virus and duration of positive test results in persistently infected cattle. Can. J. Vet. Res. 2009, 73, 117–124. [Google Scholar]
- Antos, A.; Miroslaw, P.; Rola, J.; Polak, M.P. Vaccination Failure in Eradication and Control Programs for Bovine Viral Diarrhea Infection. Front. Vet. Sci. 2021, 8, 688911. [Google Scholar] [CrossRef]
- Kelling, C.L.; Steffen, D.J.; Topliff, C.L.; Eskridge, K.M.; Donis, R.O.; Higuchi, D.S. Comparative virulence of isolates of bovine viral diarrhea virus type II in experimentally inoculated six- to nine-month-old calves. Am. J. Vet. Res. 2002, 63, 1379–1384. [Google Scholar] [CrossRef]
- Barker, D.J. The origins of the developmental origins theory. J. Intern. Med. 2007, 261, 412–417. [Google Scholar] [CrossRef]
- McRae, A.F.; Powell, J.E.; Henders, A.K.; Bowdler, L.; Hemani, G.; Shah, S.; Painter, J.N.; Martin, N.G.; Visscher, P.M.; Montgomery, G.W. Contribution of genetic variation to transgenerational inheritance of DNA methylation. Genome Biol. 2014, 15, R73. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Bird, A.P. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980, 8, 1499–1504. [Google Scholar] [CrossRef]
- Coulondre, C.; Miller, J.H.; Farabaugh, P.J.; Gilbert, W. Molecular basis of base substitution hotspots in Escherichia coli. Nature 1978, 274, 775–780. [Google Scholar] [CrossRef]
- Mohn, F.; Weber, M.; Rebhan, M.; Roloff, T.C.; Richter, J.; Stadler, M.B.; Bibel, M.; Schubeler, D. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol. Cell 2008, 30, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Georges, H.M.; Van Campen, H.; Bielefeldt-Ohmann, H.; Hansen, T.R. Epigenomic and Proteomic Changes in Fetal Spleens Persistently Infected with Bovine Viral Diarrhea Virus: Repercussions for the Developing Immune System, Bone, Brain, and Heart. Viruses 2022, 14, 506. [Google Scholar] [CrossRef] [PubMed]
- Madejon, A.; Sheldon, J.; Francisco-Recuero, I.; Perales, C.; Dominguez-Beato, M.; Lasa, M.; Sanchez-Perez, I.; Muntane, J.; Domingo, E.; Garcia-Samaniego, J.; et al. Hepatitis C virus-mediated Aurora B kinase inhibition modulates inflammatory pathway and viral infectivity. J. Hepatol. 2015, 63, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Claycombe, K.J.; Brissette, C.A.; Ghribi, O. Epigenetics of inflammation, maternal infection, and nutrition. J. Nutr. 2015, 145, 1109S–1115S. [Google Scholar] [CrossRef]
- Ridpath, J.F.; Bolin, S.R. Differentiation of types 1a, 1b and 2 bovine viral diarrhoea virus (BVDV) by PCR. Mol. Cell Probes 1998, 12, 101–106. [Google Scholar] [CrossRef]
- Akalin, A.; Kormaksson, M.; Li, S.; Garrett-Bakelman, F.E.; Figueroa, M.E.; Melnick, A.; Mason, C.E. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012, 13, R87. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Carlson, M. org.Bt.eg.db: Genome Wide Annotation for Bovine, version 3.18. In BioCore; Bioconductor: Seattle, WA, USA, 2019. [Google Scholar]
- Akalin, A.; Franke, V.; Vlahovicek, K.; Mason, C.E.; Schubeler, D. Genomation: A toolkit to summarize, annotate and visualize genomic intervals. Bioinformatics 2015, 31, 1127–1129. [Google Scholar] [CrossRef]
- Pinchuk, G.V.; Lee, S.R.; Nanduri, B.; Honsinger, K.L.; Stokes, J.V.; Pinchuk, L.M. Bovine viral diarrhea viruses differentially alter the expression of the protein kinases and related proteins affecting the development of infection and anti-viral mechanisms in bovine monocytes. Biochim. Biophys. Acta 2008, 1784, 1234–1247. [Google Scholar] [CrossRef]
- Zhang, Z.; Bossila, E.A.; Li, L.; Hu, S.; Zhao, Y. Central gene transcriptional regulatory networks shaping monocyte development in bone marrow. Front. Immunol. 2022, 13, 1011279. [Google Scholar] [CrossRef]
- Radtke, F.; Wilson, A.; Stark, G.; Bauer, M.; van Meerwijk, J.; MacDonald, H.R.; Aguet, M. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity 1999, 10, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Avram, D.; Califano, D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: Impact on immune diseases. J. Immunol. 2014, 193, 2059–2065. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Leid, M.; Rothenberg, E.V. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science 2010, 329, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Okano, M.; Bell, D.W.; Haber, D.A.; Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999, 99, 247–257. [Google Scholar] [CrossRef]
- Di Pietro, C.; Zhang, P.X.; O'Rourke, T.K.; Murray, T.S.; Wang, L.; Britto, C.J.; Koff, J.L.; Krause, D.S.; Egan, M.E.; Bruscia, E.M. Ezrin links CFTR to TLR4 signaling to orchestrate anti-bacterial immune response in macrophages. Sci. Rep. 2017, 7, 10882. [Google Scholar] [CrossRef]
- Gavin, A.L.; Huang, D.; Huber, C.; Martensson, A.; Tardif, V.; Skog, P.D.; Blane, T.R.; Thinnes, T.C.; Osborn, K.; Chong, H.S.; et al. PLD3 and PLD4 are single-stranded acid exonucleases that regulate endosomal nucleic-acid sensing. Nat. Immunol. 2018, 19, 942–953. [Google Scholar] [CrossRef]
- Langenmayer, M.C.; Jung, S.; Fux, R.; Wittlinger, C.; Tschoner, T.; Majzoub-Altweck, M.; Knubben-Schweizer, G.; Fries, R.; Hermanns, W.; Trefz, F.M. Macrophages in dermal disease progression of phospholipase D4-deficient Fleckvieh calves. Vet. Pathol. 2022, 59, 319–327. [Google Scholar] [CrossRef]
- Murphy, K.; Weaver, C. Janeway's Immunobiology, 9th ed.; Garland Science/Taylor & Francis Group, LLC: New York, NY, USA, 2016; pp. 49–71 and 265–282. 904p. [Google Scholar]
- Elinav, E.; Strowig, T.; Kau, A.L.; Henao-Mejia, J.; Thaiss, C.A.; Booth, C.J.; Peaper, D.R.; Bertin, J.; Eisenbarth, S.C.; Gordon, J.I.; et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011, 145, 745–757. [Google Scholar] [CrossRef]
- Grenier, J.M.; Wang, L.; Manji, G.A.; Huang, W.J.; Al-Garawi, A.; Kelly, R.; Carlson, A.; Merriam, S.; Lora, J.M.; Briskin, M.; et al. Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-kappaB and caspase-1. FEBS Lett. 2002, 530, 73–78. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Gaidt, M.M.; Schmidt, T.; Ebert, T.S.; Bartok, E.; Hornung, V. Caspase-4 mediates non-canonical activation of the NLRP3 inflammasome in human myeloid cells. Eur. J. Immunol. 2015, 45, 2911–2917. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef] [PubMed]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O'Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, B.J.; Dai, Y.S.; Bueno, O.F.; Parsons, S.A.; Xu, J.; Plank, D.M.; Jones, F.; Kimball, T.R.; Molkentin, J.D. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 2004, 94, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Molkentin, J.D.; Lu, J.R.; Antos, C.L.; Markham, B.; Richardson, J.; Robbins, J.; Grant, S.R.; Olson, E.N. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell 1998, 93, 215–228. [Google Scholar] [CrossRef]
- Yang, T.; Roden, D.M. Extracellular potassium modulation of drug block of IKr. Implications for torsade de pointes and reverse use-dependence. Circulation 1996, 93, 407–411. [Google Scholar] [CrossRef]
- Larson, R.L. Bovine Viral Diarrhea Virus-Associated Disease in Feedlot Cattle. Vet. Clin. N. Am. Food Anim. Pract. 2015, 31, 367–380, vi. [Google Scholar] [CrossRef]
- Riley, J.M. Economic consequences of beef cow-calf disease mismanagement: Bovine viral diarrhea virus. Appl. Anim. Sci. 2019, 35, 606–614. [Google Scholar] [CrossRef]
- Fulton, R.W.; Briggs, R.E.; Ridpath, J.F.; Saliki, J.T.; Confer, A.W.; Payton, M.E.; Duff, G.C.; Step, D.L.; Walker, D.A. Transmission of bovine viral diarrhea virus 1b to susceptible and vaccinated calves by exposure to persistently infected calves. Can. J. Vet. Res. 2005, 69, 161–169. [Google Scholar]
- Bachofen, C.; Braun, U.; Hilbe, M.; Ehrensperger, F.; Stalder, H.; Peterhans, E. Clinical appearance and pathology of cattle persistently infected with bovine viral diarrhoea virus of different genetic subgroups. Vet. Microbiol. 2010, 141, 258–267. [Google Scholar] [CrossRef]
- Karpatkin, S. Heterogeneity of human platelets. II. Functional evidence suggestive of young and old platelets. J. Clin. Investig. 1969, 48, 1083–1087. [Google Scholar] [CrossRef]
- Karpatkin, S.; Strick, N. Heterogeneity of human platelets. V. Differences in glycolytic and related enzymes with possible relation to platelet age. J. Clin. Investig. 1972, 51, 1235–1243. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wood, R.D.; Goens, S.D.; Carman, P.S.; Deregt, D.; Jefferson, B.; Jacobs, R.M. Effect on hematopoietic tissue of experimental infection of calves with noncytopathic type 2 bovine viral diarrhea virus. Can. J. Vet. Res. 2004, 68, 42–48. [Google Scholar] [PubMed]
- Spagnuolo, M.; Kennedy, S.; Foster, J.C.; Moffett, D.A.; Adair, B.M. Bovine viral diarrhoea virus infection in bone marrow of experimentally infected calves. J. Comp. Pathol. 1997, 116, 97–100. [Google Scholar] [CrossRef]
- Bolin, S.R.; McClurkin, A.W.; Coria, M.F. Effects of bovine viral diarrhea virus on the percentages and absolute numbers of circulating B and T lymphocytes in cattle. Am. J. Vet. Res. 1985, 46, 884–886. [Google Scholar] [CrossRef]
- Ellis, J.A.; Davis, W.C.; Belden, E.L.; Pratt, D.L. Flow cytofluorimetric analysis of lymphocyte subset alterations in cattle infected with bovine viral diarrhea virus. Vet. Pathol. 1988, 25, 231–236. [Google Scholar] [CrossRef]
- Brewoo, J.N.; Haase, C.J.; Sharp, P.; Schultz, R.D. Leukocyte profile of cattle persistently infected with bovine viral diarrhea virus. Vet. Immunol. Immunopathol. 2007, 115, 369–374. [Google Scholar] [CrossRef]
- Kitchen, S.G.; Whitmire, J.K.; Jones, N.R.; Galic, Z.; Kitchen, C.M.; Ahmed, R.; Zack, J.A. The CD4 molecule on CD8+ T lymphocytes directly enhances the immune response to viral and cellular antigens. Proc. Natl. Acad. Sci. USA 2005, 102, 3794–3799. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, D.; Yao, Y.; Yang, J.; Yang, Z.; Yang, Y. Characterization of T-cell subsets in response to foot-and-mouth disease bivalent inactivated vaccine in Chinese Holstein cows. Microbiol. Spectr. 2023, 11, e0102923. [Google Scholar] [CrossRef]
- Hoek, A.; Rutten, V.P.; Kool, J.; Arkesteijn, G.J.; Bouwstra, R.J.; Van Rhijn, I.; Koets, A.P. Subpopulations of bovine WC1(+) gammadelta T cells rather than CD4(+)CD25(high) Foxp3(+) T cells act as immune regulatory cells ex vivo. Vet. Res. 2009, 40, 6. [Google Scholar] [CrossRef]
- Ammari, M.; McCarthy, F.M.; Nanduri, B.; Pinchuk, L.M. Analysis of Bovine Viral Diarrhea Viruses-infected monocytes: Identification of cytopathic and non-cytopathic biotype differences. BMC Bioinform. 2010, 11 (Suppl. S6), S9. [Google Scholar] [CrossRef]
- Glew, E.J.; Carr, B.V.; Brackenbury, L.S.; Hope, J.C.; Charleston, B.; Howard, C.J. Differential effects of bovine viral diarrhoea virus on monocytes and dendritic cells. J. Gen. Virol. 2003, 84, 1771–1780. [Google Scholar] [CrossRef]
- Sopp, P.; Hooper, L.B.; Clarke, M.C.; Howard, C.J.; Brownlie, J. Detection of bovine viral diarrhoea virus p80 protein in subpopulations of bovine leukocytes. J. Gen. Virol. 1994, 75 Pt 5, 1189–1194. [Google Scholar] [CrossRef]
- Baule, C.; Kulcsar, G.; Belak, K.; Albert, M.; Mittelholzer, C.; Soos, T.; Kucsera, L.; Belak, S. Pathogenesis of primary respiratory disease induced by isolates from a new genetic cluster of bovine viral diarrhea virus type I. J. Clin. Microbiol. 2001, 39, 146–153. [Google Scholar] [CrossRef]
- Liebler-Tenorio, E.M.; Kenklies, S.; Greiser-Wilke, I.; Makoschey, B.; Pohlenz, J.F. Incidence of BVDV1 and BVDV2 infections in cattle submitted for necropsy in Northern Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 2006, 53, 363–369. [Google Scholar] [CrossRef]
- Khodakaram-Tafti, A. The comparative evaluation of cellular localization of viral antigens with microscopic changes in the ileum of cattle infected with bovine viral diarrhea. Comp. Clin. Pathol. 2006, 15, 90–93. [Google Scholar] [CrossRef]
- Shahriar, F.M.; Clark, E.G.; Janzen, E.; West, K.; Wobeser, G. Coinfection with bovine viral diarrhea virus and Mycoplasma bovis in feedlot cattle with chronic pneumonia. Can. Vet. J. 2002, 43, 863–868. [Google Scholar]
- Haines, D.M.; Martin, K.M.; Clark, E.G.; Jim, G.K.; Janzen, E.D. The immunohistochemical detection of Mycoplasma bovis and bovine viral diarrhea virus in tissues of feedlot cattle with chronic, unresponsive respiratory disease and/or arthritis. Can. Vet. J. 2001, 42, 857–860. [Google Scholar]
- Kirkbride, C.A. Viral agents and associated lesions detected in a 10-year study of bovine abortions and stillbirths. J. Vet. Diagn. Investig. 1992, 4, 374–379. [Google Scholar] [CrossRef]
- Breshears, M.A.; Johnson, B.J. Systemic reactive angioendotheliomatosis-like syndrome in a steer presumed to be persistently infected with bovine viral diarrhea virus. Vet. Pathol. 2008, 45, 645–649. [Google Scholar] [CrossRef]
- Yang, J.; Rothermel, B.; Vega, R.B.; Frey, N.; McKinsey, T.A.; Olson, E.N.; Bassel-Duby, R.; Williams, R.S. Independent signals control expression of the calcineurin inhibitory proteins MCIP1 and MCIP2 in striated muscles. Circ. Res. 2000, 87, E61–E68. [Google Scholar] [CrossRef]
- University of California, Davis. Clinical Diagnostic Laboratory CBC Reference Intervals. 2011. Available online: https://www.vetmed.ucdavis.edu/sites/g/files/dgvnsk491/files/local_resources/pdfs/lab_pdfs/UC_Davis_VMTH_Hematology_Reference_Intervals.pdf (accessed on 12 May 2025).
- MacPhillamy, C.; Chen, T.; Hiendleder, S.; Williams, J.L.; Alinejad-Rokny, H.; Low, W.Y. DNA methylation analysis to differentiate reference, breed, and parent-of-origin effects in the bovine pangenome era. Gigascience 2024, 13. [Google Scholar] [CrossRef]
- Powell, J.; Talenti, A.; Fisch, A.; Hemmink, J.D.; Paxton, E.; Toye, P.; Santos, I.; Ferreira, B.R.; Connelley, T.K.; Morrison, L.J.; et al. Profiling the immune epigenome across global cattle breeds. Genome Biol. 2023, 24, 127. [Google Scholar] [CrossRef]
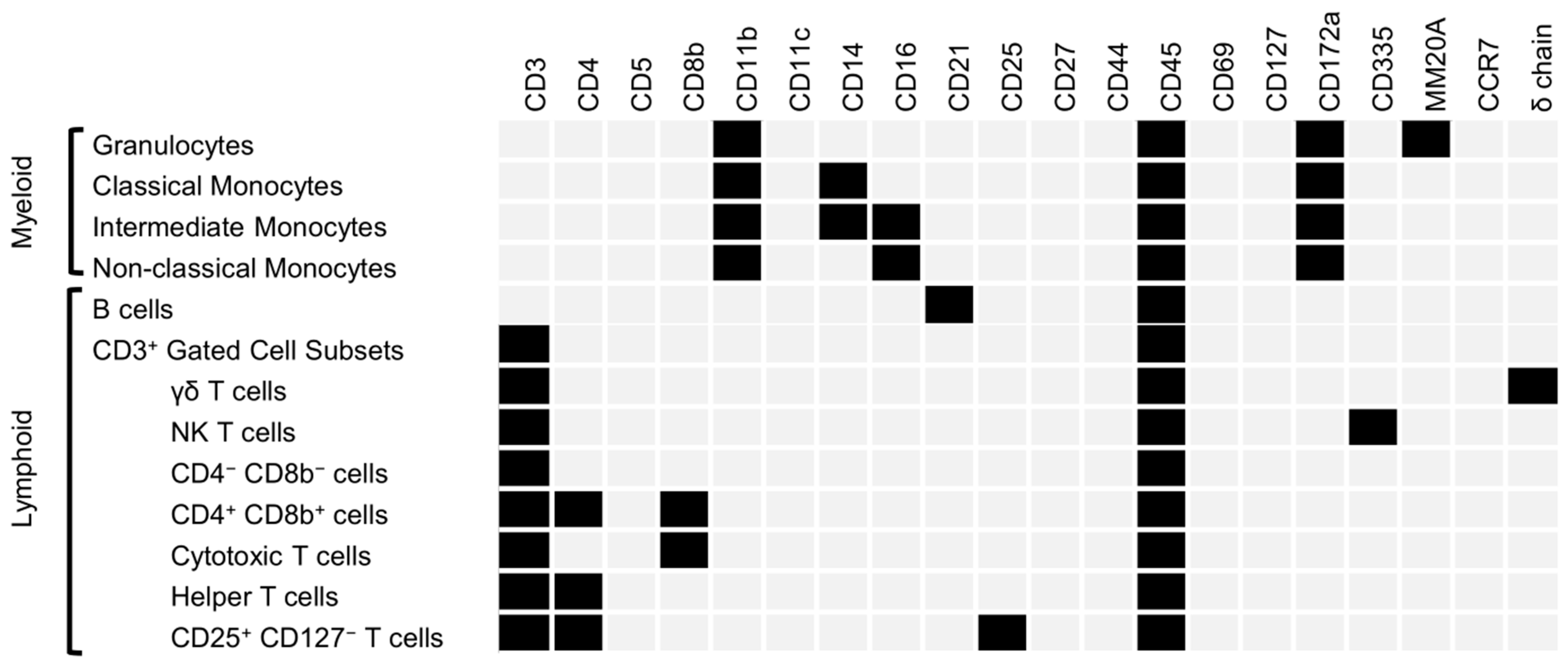
| Flow Cytometry Fluorophore Panel | |||||
|---|---|---|---|---|---|
| Laser | Marker | Species (Clone) | Manufacturer | Fluorophore (Conjugate) | Ex/Em |
| 405 | CD11b | Anti-bovine (CC126) | BioRad | DyLight405 (conjugate) | 400/420 |
| CD4 | Anti-bovine (CC8) | BioRad | CF405 (conjugate) | 408/452 | |
| CCR7 | Anti-human (3D12) | BD Horizon | BV480 | 436/478 | |
| CD25 | Anti-bovine (IL-A111) | BioRad | PacOrange | 400/551 | |
| CD44 | Anti-mouse/human (IM7) | BioLegend | BV570 | 405/570 | |
| CD127 | Anti-mouse (A7R34) | BioLegend | BV605 | 405/605 | |
| CD11c | Anti-human (HL3) | BD Horizon | BV650 | 405/645 | |
| CD27 | Anti-human (M-T271) | BD Horizon | BV711 | 405/711 | |
| CD69 | Anti-mouse (HI.2F3) | BioLegend | BV785 | 405/785 | |
| 488 | CD45 | Anti-bovine (CC1) | BioRad | FITC | 495/519 |
| CD14 | Anti-human (M5E2) | BD | BB700 | 485/693 | |
| 561 | CD5 | Anti-bovine (CC17) | BioRad | AF555 (conjugate) | 553/568 |
| CD8b | Anti-bovine (CC58) | BioRad | PE | 566/574 | |
| CD21 | Anti-bovine (CC21) | BioRad | AF568 (conjugate) | 578/603 | |
| Viability | Live/Dead | BD | FVS620 | 523/617 | |
| CD335 | Anti-bovine | BioRad | AF594 (conjugate) | 590/618 | |
| CD172a | Anti-bovine (CC149) | BioRad | PECy5 | 561/665 | |
| MM20A | Anti-bovine | WSU | PECy7 (conjugate) | 565/778 | |
| 637 | CD16 | Anti-bovine (KD1) | BioRad | AF647 | 650/671 |
| δ chain | Anti-bovine (TCR1-N24 δ) | WSU | AF680 (conjugate) | 684/707 | |
| CD3 | Anti-bovine (MM1A) | BioRad | AF750 (conjugate) | 752/776 | |
| Pathology-Related Genes Containing Differential Methylation in PI Calves | ||
|---|---|---|
| Associated Impact | Gene Names | References |
| Increased monocyte survival | STAT6, IRF7 | [40,41] |
| Decreased T cell development | GATA3, NOTCH1, CD3, CD8a, ZAP70, BCL11b | [15,42,43,44] |
| Improper de novo methylation | DNMT3A, DNMT3B | [32,45] |
| Impaired macrophage phagocytosis | EZR | [46] |
| Macrophage-induced crusting dermatitis | PLD4 | [47,48] |
| Increased inflammation | C1r, C2, C8 | [49] |
| Increased inflammasome activation | NLRP6, NEK7, GSDMD, CASP4, PANX1 | [50,51,52,53,54] |
| Cardiac hypertrophy | PPP3R2, CALM1, NFATc1, NFATc2 | [55,56,57] |
| Hypomethylated Hypermethylated | ||
| Complete Blood Counts in 4-Month-Old Heifer Calves | ||||||
|---|---|---|---|---|---|---|
| Control | PI | |||||
| Population | Average | SEM | Average | SEM | Reference | Unit |
| Basophil Count | 0.05 | ±0.01 | 0.03 | ±0.01 | 0–0.35 | 109/L |
| Eosinophil Count †* | 0.07 | ±0.01 | 0.10 | ±0.01 | 0–1.3 | 109/L |
| Neutrophil Count | 2.89 | ±0.27 | 2.93 | ±0.30 | 0.60–4.9 | 109/L |
| Monocyte Count *** | 0.38 | ±0.04 | 0.78 | ±0.08 | 0–1.02 | 109/L |
| Monocyte Percentage ** | 3.53 | ±0.41 | 6.80 | ±0.71 | 0–9.5 | % |
| Red Blood Cell Count | 12.17 | ±0.22 | 11.82 | ±0.33 | 5.0–10.1 | 1012/L |
| Hemoglobin *** | 14.28 | ±0.23 | 12.05 | ±0.39 | 8.0–14.2 | g/dL |
| Hematocrit *** | 41.33 | ±0.72 | 35.29 | ±1.10 | 23.0–42.5 | % |
| Red Cell Distribution Width †* | 22.96 | ±0.34 | 25.62 | ±0.84 | 17.5–26.5 | % |
| Mean Corpuscular Volume **** | 33.99 | ±0.44 | 29.86 | ±0.60 | 37.0–55.0 | fL |
| Mean Corpuscular Hemoglobin **** | 11.76 | ±0.15 | 10.20 | ±0.21 | 12.5–18.2 | pg |
| White Blood Cell Count | 10.68 | ±0.46 | 11.86 | ±0.84 | 4.6–15.8 | 109/L |
| Lymphocyte Count | 7.28 | ±0.30 | 7.99 | ±0.86 | 1.5–11.8 | 109/L |
| Lymphocyte Percentage † | 68.44 | ±1.79 | 69.10 | ±2.57 | 52.3–85.6 | % |
| Platelet Count ** | 462.8 | ±51.5 | 768.2 | ±60.5 | 100–720 | 109/L |
| Mean Platelet Volume **** | 4.93 | ±0.06 | 4.35 | ±0.05 | 4.8–7.6 | fL |
| Flow Cytometry in 4-Month-Old Heifer Calves | |||||
|---|---|---|---|---|---|
| Control | PI | ||||
| Population | Average (%) | SEM | Average (%) | SEM | Freq. of |
| Classical Monocytes ** | 5.73 | ±0.18 | 9.21 | ±0.86 | CD45 |
| Intermediate Monocytes | 0.88 | ±0.10 | 1.01 | ±0.10 | CD45 |
| Non-Classical Monocytes | 0.96 | ±0.08 | 0.92 | ±0.14 | CD45 |
| Granulocytes | 22.76 | ±2.24 | 17.75 | ±2.01 | CD45 |
| B Cells * | 18.28 | ±1.09 | 23.95 | ±2.44 | CD45 |
| T Lymphocytes ** | 36.48 | ±1.53 | 29.84 | ±1.57 | CD45 |
| γδ+ T Cells * | 15.85 | ±1.13 | 12.67 | ±0.09 | CD45 |
| NK T Cells | 0.13 | ±0.01 | 0.10 | ±0.01 | CD45 |
| CD4−/CD8b− T Cells **** | 2.63 | ±0.2 | 1.21 | ±0.09 | CD45 |
| CD4+/CD8b+ T Cells ** | 0.07 | ±0.1 | 0.12 | ±0.1 | CD45 |
| Cytotoxic T Cells | 6.78 | ±0.39 | 5.91 | ±0.55 | CD45 |
| Helper T Cells | 13.64 | ±0.61 | 9.43 | ±0.54 | CD45 |
| CD25+/CD127− T Cells | 0.49 | ±0.03 | 0.49 | ±0.03 | CD45 |
| γδ+ T Cells | 40.17 | ±1.79 | 41.04 | ±2.49 | CD3 |
| NK T Cells | 0.37 | ±0.04 | 0.43 | ±0.04 | CD3 |
| CD4−/CD8b− T Cells **** | 6.36 | ±0.30 | 3.9 | ±0.21 | CD3 |
| CD4+/CD8b+ T Cells **** | 0.17 | ±0.01 | 0.44 | ±0.03 | CD3 |
| Cytotoxic T Cells | 17.55 | ±1.06 | 19.22 | ±1.26 | CD3 |
| Helper T Cells | 35.02 | ±1.02 | 32.76 | ±1.93 | CD3 |
| CD25+/CD127− T Cells ** | 3.79 | ±0.16 | 5.58 | ±0.25 | CD4 |
| Differentially Expressed Genes Containing Differential Methylation | ||
|---|---|---|
| Gene Symbol | Methylation | Expression |
| SH3GL1 | Hyper | Decreased |
| MCM4 | Hyper | Decreased |
| RANBP3 | Hyper ×/Hypo ×1 | Decreased |
| IGF3R | Hypo | Increased |
| MAN2B1 | Hypo ×3 | Increased |
| PDXK | Hypo ×3/Hyper ×1 | Increased |
| ANXA2 | Hypo ×2 | Increased |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kincade, J.N.; Murtazina, D.A.; Georges, H.M.; Gonzalez-Berrios, C.L.; Bishop, J.V.; Engle, T.E.; Henao-Tamayo, M.; Eder, J.M.; McDonald, E.M.; Deines, D.M.; et al. Postnatal Epigenetic Alterations in Calves Persistently Infected with Bovine Viral Diarrhea Virus. Viruses 2025, 17, 708. https://doi.org/10.3390/v17050708
Kincade JN, Murtazina DA, Georges HM, Gonzalez-Berrios CL, Bishop JV, Engle TE, Henao-Tamayo M, Eder JM, McDonald EM, Deines DM, et al. Postnatal Epigenetic Alterations in Calves Persistently Infected with Bovine Viral Diarrhea Virus. Viruses. 2025; 17(5):708. https://doi.org/10.3390/v17050708
Chicago/Turabian StyleKincade, Jessica N., Dilyara A. Murtazina, Hanah M. Georges, Carolina L. Gonzalez-Berrios, Jeanette V. Bishop, Terry E. Engle, Marcela Henao-Tamayo, Jordan M. Eder, Erin M. McDonald, Darcy M. Deines, and et al. 2025. "Postnatal Epigenetic Alterations in Calves Persistently Infected with Bovine Viral Diarrhea Virus" Viruses 17, no. 5: 708. https://doi.org/10.3390/v17050708
APA StyleKincade, J. N., Murtazina, D. A., Georges, H. M., Gonzalez-Berrios, C. L., Bishop, J. V., Engle, T. E., Henao-Tamayo, M., Eder, J. M., McDonald, E. M., Deines, D. M., Wright, B. M., Van Campen, H., & Hansen, T. R. (2025). Postnatal Epigenetic Alterations in Calves Persistently Infected with Bovine Viral Diarrhea Virus. Viruses, 17(5), 708. https://doi.org/10.3390/v17050708







