Abstract
The second wave of COVID-19 occurred in South America in early 2021 and was mainly driven by Gamma and Lambda variants. In this study, we aimed to describe the emergence and local genomic diversity of the SARS-CoV-2 Lambda variant in Argentina, from its initial entry into the country until its detection ceased. Molecular surveillance was conducted on 9356 samples from Argentina between October 2020 and April 2022, and sequencing, phylogenetic, and phylogeographic analyses were performed. Our findings revealed that the Lambda variant was first detected in Argentina in January 2021 and steadily increased in frequency until it peaked in April 2021, with continued detection throughout the year. Phylodynamic analyses showed that at least 18 introductions of the Lambda variant into the country occurred, with nine of them having evidence of onward local transmission. The spatial–-temporal reconstruction showed that Argentine clades were associated with Lambda sequences from Latin America and suggested an initial diversification in the Metropolitan Area of Buenos Aires before spreading to other regions in Argentina. Genetic analyses of genome sequences allowed us to describe the mutational patterns of the Argentine Lambda sequences and detect the emergence of rare mutations in an immunocompromised patient. Our study highlights the importance of genomic surveillance in identifying the introduction and geographical distribution of the SARS-CoV-2 Lambda variant, as well as in monitoring the emergence of mutations that could be involved in the evolutionary leaps that characterize variants of concern.
1. Introduction
Since the onset of the COVID-19 pandemic, genome sequencing efforts around the world have monitored the spread and evolution of the SARS-CoV-2 virus. By this means, the introduction, circulation, and establishment of the SARS-CoV-2 lineages in each country were studied as well as the identification of specific mutations that impact virulence, pathogenesis, host range, immune evasion, as well as the effectiveness of diagnostic tests, vaccines, and therapeutic treatments [1].
Although there is an unprecedented number of genomes available, there still exists a significant disparity in the distribution of genomic data, especially in regions such as South America [2]. Until 26 April 2023, 405,692 whole-genome sequences from South America had been generated and shared in the GISAID database [3], accounting for 0.21% of all reported positive cases of SARS-CoV-2 from the continent. In comparison, regions with higher coverage include Europe, with 7,700,433 genomes (2.70% coverage), and North America, with 5,458,362 genomes (4.12% coverage) (WHO Coronavirus (COVID-19) Dashboard https://covid19.who.int/table (accessed on 26 April 2023). Several studies have highlighted the importance of genomic surveillance by using genomic data to examine the evolution and associated spread of dominant variants in specific countries or regions, which may be a key factor in the global community’s ability to contain and control infectious disease threats [4].
The detection of emerging viral variants of SARS-CoV-2 with characteristics of concern for public health has had a significant impact on the development of the pandemic [5]. A common characteristic of these variants is the presence of mutations at the Spike (S) glycoprotein [6]; particularly, the N-terminal domain (NTD), receptor-binding domain (RBD), and the Furin cleavage site have been identified as hotspots that concentrate amino acid changes. Several coincident amino acid changes in the SARS-CoV-2 Spike protein have independently emerged in different viral lineage, evidencing that these changes could confer an adaptive advantage towards the virus infectivity and easier spread in the population [7].
In late 2020, the Argentine Inter-Institutional SARS-CoV-2 Genomic Consortium (PAIS Consortium) implemented a molecular surveillance strategy that focused on monitoring viral variants of concern (VOC) and variants of interest (VOI) in Argentina. This allowed for monitoring the emergence of new local variants and facilitated genomic and evolutionary analyses to study their origin and dispersion within the country [8]. During the first half of 2021, the second wave of COVID-19 in Argentina occurred in the context of an ongoing vaccination campaign that began in December 2020, prioritizing strategic personnel and high-risk groups such as the elderly. By mid-2021, 20% of the population had received at least one dose of the COVID-19 vaccine, and only 7% had completed the full vaccination schedule [9]. Also, throughout 2021, the Argentine government continued with the implementation of a comprehensive plan for preventive social distancing as a critical strategy to mitigate the spread of COVID-19. This plan included significant restrictions on the mobility of people, primarily limiting movement to short distances, with interprovincial travel being the maximum allowed. International travel was mainly restricted to commercial activities, with air travel from Buenos Aires City being the primary mode of transportation. Land travel with neighboring countries was tightly regulated and limited to commercial activities. The government implemented these measures to reduce the risk of imported cases and local outbreaks, as well as to limit the transmission of the virus within the country [10].
During the period spanning from October 2020 to April 2022, a comprehensive surveillance program was implemented, involving the analysis of a total of 9356 respiratory samples collected from diverse regions of Argentina [11,12]. It was determined that all SARS-CoV-2 variants of global epidemiological importance, except for the Beta variant, were present and circulating within the country at some point during the study period [8].
Noteworthy, since mid-February 2021, a very rapid growth in the frequency of genomes carrying the nonsynonymous mutations S_L452Q and S_F490S in the Spike gene was observed in samples from the Metropolitan Area of Buenos Aires (MABA), which has been the epicenter of the epidemic in Argentina [11]. The complete genome analysis showed that these samples belong to the lineage C.37, later designated as a global VOI, and assigned the WHO label “Lambda” [13]. This variant was detected in several countries worldwide, but it has spread particularly rapidly and with growing frequency in South America, mainly in Peru, Argentina, and Chile. Remarkably, while the Alpha variant was responsible for most COVID-19 cases around the world during the beginning of 2021 [4], Lambda and Gamma were the most frequently detected variants in Argentina [11].
Here, we describe the emergence, spread, predominance, and decline of the Lambda SARS-CoV-2 variant in Argentina, and the relationship with global Lambda isolates. This study provides insights into the understanding of the evolutionary dynamics of a SARS-CoV-2 viral variant in a geographic region during social distancing measures and ongoing vaccination strategies.
2. Materials and Methods
2.1. SARS-CoV-2 Sample Collection and Sequencing
Since the start of the pandemic, molecular surveillance of SARS-CoV-2 virus has been implemented in Argentina by the PAIS Consortium. Fifteen sequencing nodes throughout the country have sequenced and analyzed the SARS-CoV-2 virus using two strategies: next-generation sequencing (NGS) to obtain complete genomes and Sanger sequencing to obtain a fragment of the Spike protein gene spanning amino acids (aa) 428 to 750, as previously reported in Torres et al. [11]. These approaches together provide valuable epidemiological information, allowing lineage distribution and virus genetic diversity to be characterized in near-real time.
During the second wave of COVID-19, an intensive sampling strategy for sequencing was implemented, which involved randomly selecting a 2.5–10% fraction of the total positive cases detected weekly in different healthcare centers. This approach enabled the detection of emerging variants and the tracking of their frequency in various regions of the country. Regular sampling was conducted at sentinel laboratories, in addition to sporadic sampling in some locations. These samples were selected for sequencing after positive tests for SARS-CoV-2 were reported, considering both the Ct value (<30) and the availability of epidemiological metadata (date of collection and the place of residence of the patients). It is worth noting that all samples come from individuals who acquired the infection in the community, since those with a travel history abroad or related to travelers were excluded from the analyzed cohort. Depending on the sequencing node, libraries preparation and sequencing were performed either on Illumina and/or Oxford Nanopore Platforms. The preparation of SARS-CoV-2 genomic libraries was performed using two different strategies: (1) the Quick protocol [14] with Illumina platform and (2) the Oxford Nanopore sequencing using the ARTIC Network or Midnight primer scheme [15]. The libraries were sequenced on the Illumina MiSeq or NextSeq instruments (Illumina, San Diego, CA, USA) and in R9 flow cells on a MinIon device (Oxford Nanopore Technologies, Oxford, UK).
The reads obtained from sequencing on the Illumina platform were initially filtered by quality, both at the base level and the read level. Subsequently, PCR duplicates, spurious primer sequences, and reads potentially contaminated from other organisms, particularly human contamination (using the DeconSeq program [16]), were removed using BBMap [17]. Finally, the quality of the resulting reads was assessed using the FastQC program [18]. The resulting reads were aligned to the SARS-CoV-2 reference genome (ID EPI_ISL_402124, hCoV-19/Wuhan/WIV04/2019) using the BWA-MEM software [19]. Finally, the consensus sequence for each sample was generated in FASTA format by the pile-up command of the samtools software and the consensus command of bcftools [20].
After sequencing on Oxford Nanopore Technologies (ONT) sequencing platforms, the raw sequencing data (files in FAST5 format) were converted into DNA sequences, and the index sequences were trimmed using the Guppy program [21]. The workflow known as “ARTIC SARS-CoV-2”, developed on the Oxford Nanopore EPI2ME platform [22], was used to obtain the complete viral genome as a FASTA-formatted sequence. In this protocol, DNA sequences in FASTQ format were filtered based on sequence length and quality and then aligned to the reference SARS-CoV-2 genome using minimap2. A specific bed file from the primer scheme was used to identify regions of the mapped sequences corresponding to synthetic sequences (primers), and these regions were trimmed to ensure that the sequences were entirely of biological origin. The retained sequences were used to generate a consensus sequence, which was further polished using Medaka [23].
A total of 9356 sequences were obtained by the PAIS Consortium, with 3531 obtained through NGS (complete genomes) and 5825 obtained through Sanger sequencing (fragment of the Spike protein gene).
2.2. SARS-CoV-2 Genomic Datasets
Different datasets were generated depending on the analyses to be performed. A schematic representation of the datasets is shown in Figure 1.
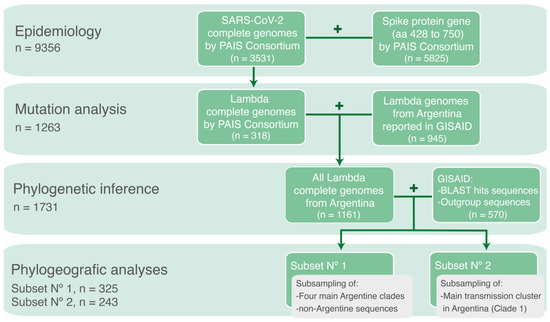
Figure 1.
Schematic representation of the datasets used according to the specific objective of each analysis.
To analyze the genomic epidemiology of SARS-CoV-2 variants in Argentina during the circulation period of the Lambda variant, complete- and partial-genome sequences obtained by the PAIS Consortium between epidemiological week (EW)-44/2020 and EW-17/2022 were used. A total of 9356 sequences were analyzed, with 3531 obtained through NGS (complete genomes) and 5825 obtained through Sanger sequencing (fragment of the Spike protein gene).
To analyze the mutational patterns within Lambda whole-genome sequences, all complete genomes of the Lambda variant obtained by the PAIS Consortium (n = 318) were combined with SARS-CoV-2 genomes reported by other groups in the GISAID database [3]. The search criteria were set to include genomes with a collection date between EW-44/2020 and EW-17/2022, located in Argentina, and classified as the Lambda variant (lineage C.37) using the Pangolin COVID-19 Lineage Assigner (accessed on 30 April 2022) [24]. This dataset comprised all Lambda genome sequences from Argentina throughout the studied period (n = 1263).
To study the introductions and diversification of Lambda in Argentina, a phylogenetic analysis was conducted with the 1263 genome sequences of the Lambda variant from Argentina, along with their most similar sequences (the best ten hits for each sequence) from a BLAST analysis conducted against the GISAID database (accessed on 30 April 2022), plus reference sequences from other lineages as outgroup. Sequences <29 Kb were excluded, except for isolates with 27–29 Kb in length that were kept due to the scarce genomic data available from some regions. The final dataset included 1731 sequences, with 1688 Lambda sequences.
Finally, to reconstruct the spatiotemporal distribution of the Lambda variant in Argentina, two subsets were generated by subsampling sequences previously used in the phylogenetic analysis. This approach was carried out to optimize the computational process required for the Markov chain runs and ensure convergence of the analysis. The first analysis comprised a subset of the sequences (n = 325) used in the previous phylogenetic analysis of the four main phylogenetic clades of Lambda in Argentina. The selection process considered the geographic distribution and collection dates of the sequences. As a result, redundant sequences were removed if they originated from the same location, had very similar collection dates, and clustered together within a phylogenetic clade. This subset was used to estimate the time and location of the most recent common ancestors, rates of evolution and demographic reconstruction. The geographic distribution of the Argentine sequences included in this analysis is described in the phylogeographic analysis (Section 3.4). The second analysis consisted in a different subset of sequences used in the phylogenetic analysis, focusing on the main transmission cluster in Argentina (n = 243). This subset was also chosen with consideration for geographic representation and collection dates to conduct a Bayesian phylogeographic diffusion analysis in discrete space.
We gratefully acknowledge the authors from the originating laboratories responsible for obtaining the specimens and the submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which part of this research is based. All genome sequences and associated metadata in these datasets are published in GISAID’s EpiCoV database. To view the contributors of each individual sequence with details such as accession number, Virus name, Collection date, Originating Lab and Submitting Lab and the list of Authors, visit 10.55876/gis8.230503du.
2.3. Phylogenetic Inference
Sequences were aligned using MAFFT v7.475 [25] with the default parameters and manually edited to exclude the 5′ and 3′ ends (first 54 nt and the last 67 nt, in reference to isolate hCoV-19/Wuhan/WIV04/2019, EPI_ISL_402124). The best-fitted evolutionary model was determined using ModelFinder [26] according to the Bayesian Information Criterion (BIC). Maximum-likelihood phylogenetic reconstruction was performed with the IQ-TREE COVID-19 release 2.2.0 [27] and clustering confidence was evaluated using the Ultrafast Bootstrap approximation method (UFBoot, 10,000 replicates) [28] and Shimodaira–Hasegawa approximate likelihood ratio test (SH-aLRT, 1000 replicates) [29].
To verify the lineage assignment made by Pangolin COVID-19 Lineage Assigner, maximum-likelihood phylogenetic trees were performed (data not shown), including all Argentine sequences and reference sequences for several lineages and sublineages obtained from the PANGO designation list v1.9 [30]).
2.4. Phylodynamic Analysis
A Bayesian coalescent approach was used to co-estimate the temporal and spatial history of the SARS-CoV-2 C.37 lineage in Argentina. In addition, a discrete phylogeographic analysis was performed and used to visualize the spatiotemporal spread of the main transmission cluster in Argentina.
The temporal signal of the datasets was examined by root-to-tip regression using Tempest v1.5.1 [31] software. Sequences whose genetic divergence and sampling date were incongruent (according to visual inspection) with the general pattern of the datasets were discarded, as they could have had sequencing errors, contamination, or misassigned collection date.
The analyses were carried out using an appropriate substitution model according to the BIC estimated with ModelFinder in IQ-TREE v2.1 [26,27]. The uncorrelated lognormal (UCLN) molecular clock model [32], and the Bayesian Skyline coalescent model [33] implemented in the BEAST v1.10.4 software package [34], were used. A discrete phylogeographic model with an asymmetric substitution matrix over the sampling locations was set, with all transitions equally probable as a prior.
Three independent Markov chain Monte Carlo (MCMC) chains were run for each dataset. Results were examined with Tracer v1.7.1 [35] to evaluate the convergence of parameters (effective sample size ≥200, acceptable mixing without tendencies in traces, with a burn-in of 10%) and concatenated with LogCombiner [34]. Uncertainty in parameter estimate was evaluated in the 95% highest posterior density (HPD95%) interval. The maximum clade credibility tree (MCCT) was summarized using Tree Annotator v1.10.5 [34], visualized with FigTree v1.4.4 [36], and analyzed further in the SPREAD3 program [37].
2.5. Mutation Analysis
To study the emergence of mutations in Argentina, we traced the profiles of nonsynonymous mutations and compared their frequency in the studied population. The mutational profile of each sequence from lineage C.37 was investigated using the CoVsurver tool available at the GISAID EpiCoV platform [3] to identify nonsynonymous changes in comparison with the Wuhan-Hu-4 reference sequence (GISAID: EPI_ISL_402124). All frequencies were calculated based on the coverage of each position in the genome sequence, and those positions with less than 50% coverage were excluded from the analysis.
2.6. Statistical Analysis
The frequencies of detection of VOI, VOC, or mutations and the 95% CIs were estimated with the Wilson/Brown method, implemented in the GraphPad Prism v.9.2 program (San Diego, CA, USA).
2.7. Data Visualization
The plots were generated using graphical visualization tools at covdb.stanford.edu [38] (Philip L. Tzou et al., 2022) and/or Microsoft Excel v16.47.1 and edited in Adobe Illustrator 23.1.1.
3. Results
3.1. The Molecular Epidemiology of Lineage C.37 in Argentina
From October 2020 to April 2022, 9356 respiratory samples positive for SARS-CoV-2 from Argentina were subjected to sequencing, out of which 3531 were obtained through NGS and the remainder were obtained through Sanger sequencing. In this study, we analyzed genomic sequences obtained from the six regions of the country [38], but with a heterogeneous distribution. A high number of genomes from the MABA and Pampeana regions were sequenced, while in regions such as Cuyo, Northeast, and Northwest, a lower number of sequences were obtained due to their lower population and, as a consequence, a lower total number of cases (Figure 2A) [39].
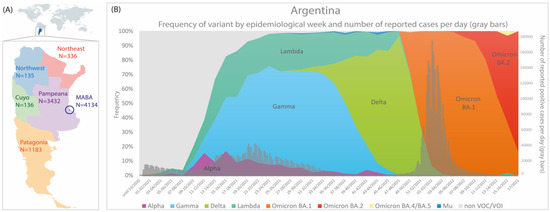
Figure 2.
(A) Number of partial/complete genomes by Argentine regions. (B) Frequency of SARS-CoV-2 variants and number of reported cases per day by epidemiological week (n = 9356); only cases that did not present a history of travel or close contact. The colors in (B) represent the cumulative abundance of each SARS-CoV-2 variant per epidemiological week.
Of note, 82.04% of the samples corresponded to a VOC/VOI: 26.96% Gamma, 21.37% Omicron, 16.10% Delta, 13.24% Lambda, 4.19% Alpha, and 0.18% Mu (Figure 2). In previous works, we have reported the emergence and evolution of SARS-CoV-2 lineages during the first and third waves of COVID-19 in Argentina [11,12,40]. In this study, we have focused on the analysis of the Lambda variant during the second wave of COVID-19 in Argentina that took place during the first half of 2021.
The Lambda variant was first detected in Argentina on 30 January 2021, through the detection of its characteristic mutations in the Sanger-sequenced region of Spike protein gene (S_L452Q and S_F490S). Since then, it was detected in 1239 out of 9356 genome sequences from Argentina with a frequency that increased steadily since February (EW-5/2021) and reached a value of 31.1% (EW-17/2021, end of April) (Figure 2B and Table S1). However, the impact of the Lambda variant was heterogenous in each region of the country, and the highest frequencies were found in the MABA (44.7%, SE 17-18/2021) and Pampeana regions (27.4%, SE 33-34/2021).
By the end of October 2021, the Lambda frequency decreased to values below 10% in accordance with the end of the second wave of COVID-19 and the introduction of the Delta variant in Argentina. Since then, Lambda was detected only in sporadic cases until April 2022. Notably, the last detected case occurred during the third wave of COVID-19, when the Omicron variant had a prevalence above 99% since EW 03-04/2022. The clinical–epidemiological investigation revealed that this single case was an immunocompromised individual with an advanced HIV infection. Indeed, a particular set of mutations was found in the genome sequence, which indicates a highly divergent Lambda variant that has not yet been reported in databases.
3.2. Phylogenetic Analysis
Argentine sequences were analyzed along with the most similar sequences in the GISAID database to infer common transmission chains and establish the presence of circulating viruses. The Argentine sequences included in this analysis were distributed across regions as follows: 35% Pampeana, 26% Northwest, 15% MABA, 11% Patagonia, 9% Northeast, and 4% Cuyo.
The introduction and diversification study performed on the Argentine Lambda genomes revealed a minimum of 18 independent introduction events, 9 of them with evidence of onward transmission. Most Lambda sequences from Argentina clustered into four main clades, labeled as 1, 5, 6, and 8, where samples from all Argentine regions were found (Figure 3). Clades 5, 6, and 8 were associated with specific regions: Clade 5 circulated mainly in the Northwest region, Clade 6 mostly in the Pampeana region, and Clade 8 between the Patagonia and Pampeana regions. Regarding Clade 1, the largest one, the 861 Argentine sequences were split into several subclades, with few sequences collected from other countries (mostly the USA, Chile, Spain, and Mexico). Although the basal relationship between subclades could not be resolved with confidence, highly supported local groups with viral diversification to different cities could be identified (complete full-detailed phylogenetic tree is in the Supplementary Materials).
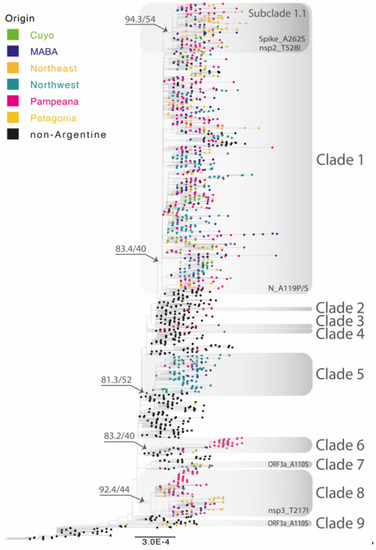
Figure 3.
Maximum-likelihood phylogenetic tree of SARS-CoV-2 whole-genome sequences of Lambda (lineage C.37). Tips are colored by region and clades with Argentine genomes are highlighted by grey rectangles. Nonsynonymous mutations associated with one or more of the Argentine clades are shown at the bottom of the rectangles. B.1.1.1 sequences were used as the outgroup. The SH-like/UFB values for the relevant groups are indicated for some groups. UFB: ultrafast bootstrap. The scale indicates the number of substitutions per site.
In addition to these major clades, we also identified a number of smaller clades (2, 3, 4, 7, and 9), as well as 7 singletons which were randomly interspersed with Lambda sequences mostly from Peru, Chile, and the United States.
Notably, Clade 4 contained the latest Lambda sequence reported in Argentina in April 2022, PAIS-G1123 (EPI_ISL_13466784). This clade included 5 other sequences from Argentina, Chile, and the United States, all reported between February and July 2021. As expected, the branch containing PAIS-G1123 shows a large number of substitutions per site compared to the rest of the group.
3.3. Mutational Patterns in Argentine SARS-CoV-2 Whole-Genome Sequences
Compared to the reference sequence hCoV-19/Wuhan/WIV04/2019 (GISAID accession number EPI_ISL_402124), the 1263 genome sequences displayed a total of 1631 amino acid changes in different viral genes (912 in ORF1a/1b, 268 in S, 121 in N, 114 in ORF3a, 94 in ORF7a, 47 in ORF8, 28 in M, 18 in ORF6, 17 in ORF7b, and 12 in E). In the Argentine sequences, all the C.37 lineage defining amino acid changes [41], shown in grey in Figure 4A, were found at a frequency higher than 95.9%.
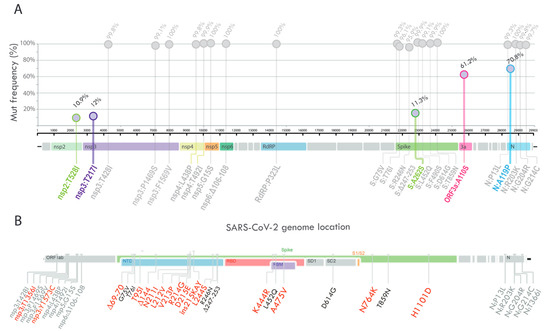
Figure 4.
(A) Lollipop plot summarizing the frequency of SARS-CoV-2 nonsynonymous mutations and deletions with >10% prevalence. The bubbles’ y-coordinates indicate mutation frequencies, which are also shown above the bubbles. The five amino acid substitutions with frequency values between 10% and 90% are shown in colors. (B) Schematic summary of the changes found in the PAIS-G1123 sequence. Text in grey indicates the constellation of mutations characteristic of the Lambda variant, and red text indicates amino acid positions with changes that are unique to the PAIS-G1123 sequence compared to the most-related Argentine genome sequences of the Lambda variant.
Five amino acid substitutions, with frequency values between 10 and 90% (shown with colors in Figure 4A), were further analyzed to determine if they could be assigned to one or more of the Argentine clades identified by phylogenetic inference (in Figure 3, the nonsynonymous mutations related to each clade are shown). While the amino acid substitution ORF3a_A110S was observed in several clades with Argentine sequences (clades 1, 7, and 9), the remaining mutations were found to be associated with specific Argentine clades. For instance, the amino acid substitution nsp3_T217I was only found in Clade 8, which includes genome sequences mainly from two regions, Pampeana and Patagonia. In relation to Clade 1, an amino acid replacement was found throughout the clade at position N_A119, of which more than 70% corresponded to the N_A119P substitution. In addition, the Spike_A262S and nsp2_T528I mutations were found as signatures of Subclade 1.1.
The analysis of the complete SARS-CoV-2 genome PAIS-G1123 (EPI_ISL_13466784) from the patient detected in April 2022 showed a total of 30 nucleotide substitutions, 4 deletions, and 1 insertion compared to the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402124). Of these, 19 mutations correspond to the constellation of mutations characteristic of the Lambda variant. Interestingly, this case occurred at a moment of no circulation of Lambda in our country, suggesting a prolonged infection time that started months before their detection. Remarkably, compared to the most related genome sequences of the Lambda variant, this sequence displayed 13 nonsynonymous mutations, 2 deletions, and 1 insertion distributed in 2 genes: ORF1ab (replicase polyprotein) and Spike glycoprotein gene (Figure 4B). Of the 13 changes in the Spike protein, 2 were in the receptor-binding domain, 10 were in the N-terminal domain, and 2 were in the intergenic region (shown in red text, Figure 4B).
3.4. Phylogeographic Analyses
A discrete phylogeographic analysis was performed on a subset of Lambda sequences (n = 325), including the four main Argentine clades that showed local transmission (clades 1, 5, 6, and 8). The Argentine sequences included in this analysis were distributed across regions as follows: 26% Pampeana, 24% Northwest, 23% MABA, 11% Patagonia, 10% Northeast, and 6% Cuyo.
The Lambda variant displayed a rate of evolution of 3.88×10−4 substitutions per site per year (s/s/y) (HPD95% = 3.54 × 10−4 − 4.22 × 10−4), with an estimated date for its most recent common ancestor on 13 November 2020 (HPD95% = 15 October to 9 December 2020), placed in Peru (posterior probability (pp) 1.0).
The demographic reconstruction of the Lambda isolates, represented by the Bayesian skyline plot, reveals a notable increase in the effective number of infections during the period spanning from December 2020 to mid-April 2021, when it reached a plateau continuing until the last sampling time (Figure 5).
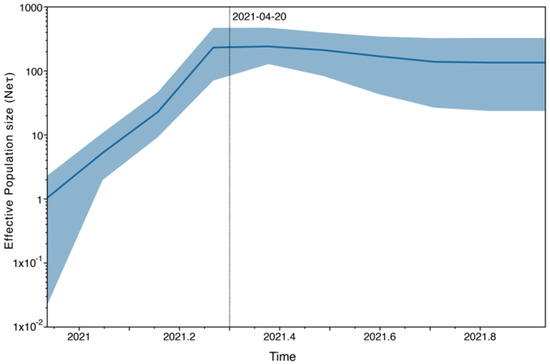
Figure 5.
Coalescent Bayesian Skyline analysis. The black line is the mean estimate of the estimated effective population size. The two blue lines are the upper and lower bounds of the 95% HPD interval. The x-axis is the time in years, and the y-axis is on a log scale.
To explore the time/date of the introduction of the viruses that gave rise to the four main clades that circulated in Argentina, we estimated the time of the most recent common ancestor (TMRCA) and its location for each clade (Figure 6A).
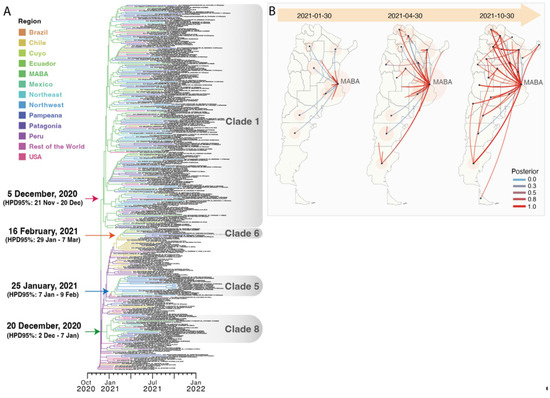
Figure 6.
(A) The Bayesian discrete phylogeographic analysis. Maximum clade credibility tree for the Lambda variant. The branches’ colors represent the MRCA’s location (described in the legend). The time scale in years is detailed at the bottom. (B) Different stages of phylogeographic history of Clade 1 under a discrete diffusion model. The lines colors represent the posterior probability support for each transition rate between locations calculated by the BEAST program and summarized by the SPREAD3 program (described in the legend). The size of the polygons around a sampling location is proportional to the number of lineages that maintain that location.
The TMRCA of Clade 1 was estimated on 5 December 2020 and was in MABA (pp, posterior probability, 0.84). To evaluate the regional distribution of this clade, which was the main clade of Lambda in Argentina, a second discrete phylogeographic analysis was conducted. The reconstruction of the diffusion history of Clade 1 is shown in Figure 6B. The result suggested an early spread of Lambda from MABA to other provinces that persisted throughout 2021, with a more limited onward spread among provinces since early May.
The TMRCA of Clade 5 was estimated to be on 26 January 2021, and was located in Peru (pp 0.84). An Argentine sequence with a travel history to Peru was placed basally to this clade on the tree, so this sequence is likely to share a common ancestor with the probable source of introduction. Viruses within this transmission cluster were diversified predominantly in the Northwest region.
The TMRCA of Clade 6 was estimated to be on 16 February 2021, and was located in Chile (pp 0.92). Viruses in this cluster were distributed among all Argentine regions but were particularly widespread in the Pampeana region, where transmission was persistent until December 2021.
The TMRCA of Clade 8 was estimated to be on 20 December 2020. The location of the MRCA (Most Recent Common Ancestor) was estimated to be in MABA (pp 0.99), from where viruses within this cluster were mainly distributed between two Argentine regions (Pampeana and Patagonia), resulting in two highly supported chains of transmission.
4. Discussion
Since the C.37 lineage of SARS-CoV-2 was initially reported in Peru in December 2020 [42], considerable research has been conducted, primarily in Peru, to elucidate the dispersion and evolutionary dynamics of the Lambda variant during the early stages of viral circulation [43,44]. However, the transmission and evolutionary dynamics of this variant in other affected regions remain incompletely characterized. To address this knowledge gap, our study aimed to comprehensively characterize the emergence and spread of the SARS-CoV-2 Lambda variant throughout Argentina, from its initial entry into the country until its detection ceased. Our investigation provides insights into the genetic and epidemiological features of the Lambda variant, contributing to a more comprehensive understanding of one of the SARS-CoV-2 variants of interest that had a major impact on South America, a region that has been less studied during the COVID-19 pandemic in comparison with higher-income countries.
Our analysis of 9356 SARS-CoV-2-positive respiratory samples revealed a consistent increase in the frequency of the Lambda variant from February 2021 (EW-5/2021) through the end of April (EW-17/2021) in Argentina. These findings were consistent with the earlier emergence of the Lambda variant in Peru, where the first case was detected in November 2020, and its frequency increased until April 2021 [45]. Lambda became the predominant variant in the Coastal and Andean regions of Peru during the first half of 2021, surpassing Gamma in almost the entire country for several months, likely because the Lambda variant held off Gamma expansion initially due to a “founder effect” (having been detected one month earlier) [44]. In contrast, in Brazil, a low level of Lambda VOI circulation was found, and Gamma was the dominant variant during early 2021 [46]. Argentina was the region where these two scenarios converged, as Lambda and Gamma cocirculated during the first half of 2021, with Gamma having a higher detection frequency. Given that Argentina shares large borders with other Latin-American countries, commercial contact with neighboring countries via terrestrial transportation may have led to the introduction and dissemination of both variants in the country. This phenomenon was also observed at the borders of the Amazon region of Peru with Brazil, where the circulation of the Gamma variant predominated in contrast to the rest of Peru [44]. Further information on the circulation of these variants in neighboring countries of Argentina, such as Bolivia or Paraguay, may confirm this hypothesis. In contrast, Alpha, which was the prevalent variant in Europe, had a minimum impact in Argentina, possibly because the closure of air borders possibly mitigated the epidemiological impact [47,48].
The demographic reconstruction of the Lambda variant showed an increase in the effective number of infections that corresponded with the increase in Lambda frequency in our dataset and with the reported cases in the National Surveillance System during the second wave of COVID-19 in Argentina [39]. In early April 2021, the national government decided to increase movement restrictions within the country and to cancel all flights to Latin-American countries due to the epidemiological situation in South America [49]. This may be reflected in the demographic reconstruction of the Lambda variant, where the plateau observed at the end of April could indicate a stabilization in the diversification of the Lambda variant due to increased social distancing and restrictions on foreign travel entry.
The persistence of the Lambda variant’s detection continued until October 2021, subsequently resulting in a decrease in its frequency to less than 10%. This decline coincided with the end of Argentina’s second wave of COVID-19 and the emergence of the Delta variant. The spread of Delta throughout Argentina started in August 2021, later than in other countries [50,51,52], and, like neighboring countries, the emergence of Delta did not lead to an increase in the incidence of COVID-19 cases and deaths (Figure 2). This could be a consequence of the strict limitations imposed on air travel, which also enabled higher vaccination rates before the entry and dominance of Delta in the country [53].
Our phylogenetic analyses of all reported Lambda sequences from Argentina indicated that there were at least 18 introductions into the country. The analyses showed the national spread of Lambda, as indicated by several large clades of closely related SARS-CoV-2 lineages, which were defined by mixtures of samples from patients living in different regions of Argentina. Also, smaller clades clustering within specific regions were observed, providing evidence of local transmission chains of the virus. These findings were consistent with interprovincial community transmission within the same region, such as that observed in the Northwest region (Clade 5) and Pampeana region (Clade 6), as movement to other regions was increasingly restricted since the emergence of Lambda in the country [49]. Clades with highly moderate support, including sequences from Argentina, were observed. However, the basal relationship between the clades could not be confidently resolved.
The phylogeographic analyses showed that the four main Argentine clades had an estimated date for their most recent common ancestors between December 2020 and February 2021; this finding is consistent with the detection of the first sample in the country for each clade occurring two months later. In clades 5, 6, and 8, the estimated location was in the neighboring countries, Peru and Chile. Interestingly, sequences from MABA were observed in all clades, often in basal positions, and given that the Lambda variant was first detected in MABA, this observation could suggest the significant role played by the pandemic epicenter of Argentina as the point of entry and subsequent transmission of the virus across the country. Moreover, in Clade 1, our phylodynamic analysis suggested an initial diversification in MABA, which subsequently spread to other regions within Argentina, leading to widespread transmission of the Lambda variant within the country, as shown in the spatiotemporal reconstruction of the dispersal history of Lambda in Figure 6B.
The Lambda variant is characterized by multiple lineage-specific deletions and amino acid substitutions in its viral genome, as shown in Figure 4A [41]. Specifically, the spike protein of the Lambda variant has a unique pattern of eight mutations (G75V, T76I, R246N, Δ247-253, L452Q, F490S, D614G, and T859N). Interestingly, the amino acid substitution in the L452 position had also been reported in the Delta and Omicron variants [54,55], and it had been demonstrated that the L452Q mutation conferred increased viral infectivity and resistance to vaccine-induced antiviral humoral immunity [56]. While L452Q is almost exclusive to C.37, L452R is present in Delta and former variants of interest Epsilon (B.1.427/B.1.429) and Kappa (B.1.617.1) and is associated with increased affinity for the human ACE2 receptor [57]. Furthermore, the F490S mutation had been associated with escape from convalescent sera [58]. The 247-253 deletion in the NTD of the Lambda Spike is located in an antigenic site, suggesting that this deletion might have contributed to immune escape by shortening or fully deleting neutralizing epitopes or exploiting increased glycosylation [59,60,61,62]. Lambda also displays the nsp6 106–108 deletion, found in VOCs alpha, beta, and gamma [63]. Given that the mutations present in Lambda and their genomic location are shared in many cases with the mutations found in the designated variants of concern, a greater epidemiological impact of the Lambda variant could have been observed if molecular surveillance in Latin America had been comparable to that of higher-income countries. Additionally, the government’s implementation of containment measures to prevent the spread of COVID-19 may have contributed to the decrease in Lambda transmission to other continents.
Some of the mutations found in Argentine sequences were assigned to one or more of the Argentine clades identified by phylogenetic inference, such as Clade 1 (N_A119P, Spike_A262S, and nsp2_T528I), Clade 8 (nsp3_T217I), and clades 1, 7, and 9 (ORF3a_A110S). In accordance with our results, it has been reported that the genes encoding the nonstructural protein NSP3 and the structural proteins S and N exhibit the highest number of mutations in the Lambda sequences isolated from Peru, and that the proteins with some sites under positive natural selection were ORF3a, ORF8, and S [45].
The Lambda sequence, PAIS-G1123 (EPI_ISL_13466784), was detected in April 2022, several months after the last Lambda sequence was reported in Argentina and exhibited 14 nonsynonymous mutations, two deletions, and one insertion in comparison to the most closely related genome sequences of the Lambda variant detected in Argentina. The most remarkable characteristic of this genome is the acquisition of multiple mutations within the NTD of the S glycoprotein, including deletions at positions 69–70 and 144. These deletions have been identified in the Omicron and Alpha variants of concern and have been associated with an increase in infectivity as a result of enhanced incorporation of cleaved spike into virions and evasion of antibody response [64,65]. Given that the sample belonged to an immunocompromised patient, it is likely that the changes in the amino acid composition of the viral Spike protein were a result of an ongoing interaction between the persistent virus and the patient’s adaptive immune system, as has been documented in other comparable clinical cases [66,67]. The emergence of rare mutations in chronically infected immunocompromised hosts, implying a radical difference in selective pressure compared to common cases, could be involved in the evolutionary leaps that characterize variants of concern. Therefore, understanding the evolution of SARS-CoV-2 and the emergence of new variants through studies is critical in developing effective prevention and control strategies to minimize the impact of the virus on public health.
This study has contributed to the understanding of the evolutionary trends exhibited by SARS-CoV-2 and emphasizes the role of genomic surveillance in identifying the emergence and geographical distribution of the SARS-CoV-2 Lambda variant. Furthermore, it underscores the significance of intercountry interactions, thereby highlighting the importance of implementing monitoring measures, not only at the local but also at border level. The ecological competition between two viral variants, Lambda and Gamma, within a vaccinated population raises intriguing questions, as the mutations within these variants could potentially play a critical role in determining their competitive advantage. Moreover, the study investigates the emergence of intravariant mutations, along with their occurrence in immunocompromised patients. These patients serve as reservoirs for viral diversity, amplifying the significance of mutations observed within their cases. This emphasizes the imperative need for comprehensive monitoring and a thorough understanding of viral dynamics in both the general and immunocompromised populations.
We believe that increasing data sharing and research in South America is crucial to enhance our comprehension of the COVID-19 pandemic in this region. Furthermore, revealing the virological characteristics of mutations in VOCs and VOIs can establish a baseline, which is necessary to evaluate the risk of newly emerging SARS-CoV-2 variants in the future, as well as their impact on therapeutics or vaccination effectiveness.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v15061382/s1, Table S1: Frequency of cases classified as VOC/VOI by epidemiological week (EW) in Argentina; Tree S1: Maximum-likelihood phylogenetic tree of SARS-CoV-2 whole-genome sequences of Lambda (lineage C.37).
Author Contributions
Conceptualization, M.S.N.J. and M.V.; data curation, L.M. (Laura Mojsiejczuk), D.A., L.E.V., M.N., S.L., S.A., A.A. (Ariel Amadio), M.E.A., C.A.L., A.A. (Andres Angelletti), E.B., B.B., A.C. (Ana Cavatorta), A.C. (Agustina Cerri), A.C. (Andres Cordero), H.D., M.J.D.S., M.F.E., R.E., C.E., S.C., F.G., A.G., M.G., R.M.A., I.G., M.E.I., G.K., V.L., M.L.C., H.L., N.M., M.M. (Melina Mazzeo), A.S.M., L.M. (Luciana Montoto), M.M. (Marianne Muñoz), V.N., C.N., B.O., L.P., C.P., A.P., C.R., A.E.R., J.S., A.S., C.T. (Clara Theaux), G.T., E.T., R.T., V.V., G.W., C.Z., M.C.Z., S.Z. and P.P.; formal analysis, M.S.N.J.; funding acquisition, M.V.; investigation, M.S.N.J., C.T. (Carolina Torres) and M.V.; methodology, M.S.N.J., D.A., L.E.V., S.G., M.N., S.L., S.A., A.A. (Ariel Amadio), M.I., F.F. (Franco Fernandez), E.B., B.B., M.C., A.C. (Agustina Cerri), H.D., M.J.D.S., M.F.E., M.F., F.F. (Fabián Fay), A.F., F.F. (Florencia Ferrini), L.F., S.C., F.G., A.G., M.G., R.M.A., I.G., G.K., M.L.C., H.L., N.M., M.M. (Melina Mazzeo), M.M. (Marianne Muñoz), C.N., L.P., C.P., A.P., C.R., A.E.R., J.S., A.S., V.V., M.C.Z. and P.P.; project administration, M.V.; resources, M.S.N.J., L.M. (Laura Mojsiejczuk), D.A., L.E.V., S.G., M.N., S.L., S.A., A.A. (Ariel Amadio), M.I. (Matias Irazoqui), F.F. (Franco Fernandez), M.E.A., C.A.L., A.A. (Andres Angelletti), P.A., E.B., B.B., M.C., A.C. (Ana Cavatorta), A.C. (Agustina Cerri), A.C. (Andres Cordero), H.D., M.J.D.S., M.F.E., R.E., C.E., M.F., F.F. (Fabián Fay), A.F., F.F. (Florencia Ferrini), L.F., S.C., F.G., A.G., M.G., R.M.A., I.G., M.E.I., G.K., V.L., M.L.C., H.L., N.M., M.M. (Melina Mazzeo), A.S.M., L.M. (Luciana Montoto), M.M. (Marianne Muñoz), V.N., C.N., B.O., L.P., C.P., A.P., C.R., A.E.R., J.S., A.S., C.T. (Clara Theaux), G.T., E.T., R.T., V.V., G.W., C.Z., M.C.Z., S.Z., P.P. and M.V.; software, S.G., A.A. (Ariel Amadio), M.I., S.C., F.F. (Franco Fernandez) and M.C.; supervision, C.T. (Carolina Torres) and M.V.; visualization, M.S.N.J.; writing—original draft, M.S.N.J.; writing—review and editing, M.S.N.J., C.T. (Carolina Torres), S.G. and M.V. All authors have read and agreed to the published version of the manuscript.
Funding
The sequencing strategy was supported by Proyecto IP COVID-19 N°08 (Ministerio de Ciencia, Tecnología e Innovación, Argentina) and Focem COF 03/11 COVID-19 (Fondo para la Convergencia Estructural del MERCOSUR).
Institutional Review Board Statement
The study was conducted following the Declaration of Helsinki and approved by the Medical Ethics and Research Committees of ‘‘Ricardo Gutiérrez’’ Children’s Hospital, Buenos Aires, Argentina (DI-2020-165-GCABA-HGNRG).
Informed Consent Statement
Informed consent was waived due to patient information being anonymized and de-identified before analysis.
Data Availability Statement
All genome sequences and associated metadata generated for this study are published in GISAID’s EpiCoV database. To view the contributors of each individual sequence with details such as accession number, Virus name, Collection date, Originating Lab and Submitting Lab and the list of Authors, visit 10.55876/gis8.230613ux.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Thakur, S.; Sasi, S.; Pillai, S.G.; Nag, A.; Shukla, D.; Singhal, R.; Phalke, S.; Velu, G.S.K. SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines. Front. Med. 2022, 9, 815389. [Google Scholar] [CrossRef] [PubMed]
- Oude Munnink, B.B.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next Phase of SARS-CoV-2 Surveillance: Real-Time Molecular Epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Elbe, S.; Buckland-Merrett, G. Data, Disease and Diplomacy: GISAID’s Innovative Contribution to Global Health: Data, Disease and Diplomacy. Global Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Azman, A.S.; Chen, X.; Zou, J.; Tian, Y.; Sun, R.; Xu, X.; Wu, Y.; Lu, W.; Ge, S.; et al. Global Landscape of SARS-CoV-2 Genomic Surveillance and Data Sharing. Nat. Genet. 2022, 54, 499–507. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Tracking SARS-CoV-2 Variants. 2021. Available online: https://Www.Who.Int/En/Activities/Tracking-SARS-CoV-2-Variants/ (accessed on 25 February 2023).
- Sabino, E.C.; Buss, L.F.; Carvalho, M.P.S.; Prete, C.A.; Crispim, M.A.E.; Fraiji, N.A.; Pereira, R.H.M.; Parag, K.V.; Da Silva Peixoto, P.; Kraemer, M.U.G.; et al. Resurgence of COVID-19 in Manaus, Brazil, despite High Seroprevalence. Lancet 2021, 397, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Zahradník, J.; Nunvar, J.; Schreiber, G. Perspectives: SARS-CoV-2 Spike Convergent Evolution as a Guide to Explore Adaptive Advantage. Front. Cell. Infect. Microbiol. 2022, 12, 748948. [Google Scholar] [CrossRef] [PubMed]
- Consorcio Argentino de Genómica de SARS-CoV-2 Proyecto Argentino Interinstitucional de Genómica de SARS-CoV-2. Available online: http://Pais.Qb.Fcen.Uba.Ar/ (accessed on 25 February 2023).
- Ministerio de Salud de la Republica Argentina Monitor Público de Vacunación. Available online: https://Www.Argentina.Gob.Ar/Coronavirus/Vacuna/Aplicadas (accessed on 25 February 2023).
- Ministerio de Justicia y Derechos Humanos. Presidencia de la Republica Argentina Decreto 67/2021. Distanciamiento Social, Preventivo y Obligatorio y Aislamiento Social, Preventivo y Obligatorio. Available online: https://www.argentina.gob.ar/normativa/nacional/decreto-67-2021-346580 (accessed on 25 February 2023).
- Torres, C.; Mojsiejczuk, L.; Acuña, D.; Alexay, S.; Amadio, A.; Aulicino, P.; Debat, H.; Fay, F.; Fernández, F.; Giri, A.A.; et al. Cost-Effective Method to Perform SARS-CoV-2 Variant Surveillance: Detection of Alpha, Gamma, Lambda, Delta, Epsilon, and Zeta in Argentina. Front. Med. 2021, 8, 755463. [Google Scholar] [CrossRef]
- Zambrana Montaño, R.; Culasso, A.C.A.; Fernández, F.; Marquez, N.; Debat, H.; Salmerón, M.; Zamora, A.M.; Ruíz de Huidobro, G.; Costas, D.; Alabarse, G.; et al. Evolution of SARS-CoV-2 during the First Year of the COVID-19 Pandemic in Northwestern Argentina. Virus Res. 2023, 323, 198936. [Google Scholar] [CrossRef]
- World Health Organization (WHO). COVID-19 Weekly Epidemiological Update, 44th ed.; World Health Organization: Geneva, Switzerland, 2021. Available online: https://apps.who.int/iris/handle/10665/341904 (accessed on 25 February 2023).
- Josh Quick 2020. NCoV-2019 Sequencing Protocol. Protocols.Io. Available online: https://dx.doi.org/10.17504/protocols.io.bbmuik6w (accessed on 25 February 2023).
- Freed, N.E.; Vlková, M.; Faisal, M.B.; Silander, O.K. Rapid and Inexpensive Whole-Genome Sequencing of SARS-CoV-2 Using 1200 Bp Tiled Amplicons and Oxford Nanopore Rapid Barcoding. Biol. Methods Protoc. 2020, 5, bpaa014. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Fast Identification and Removal of Sequence Contamination from Genomic and Metagenomic Datasets. PLoS ONE 2011, 6, e17288. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap—Sourceforge.Net/Projects/Bbmap/. Available online: https://sourceforge.net/projects/bbmap/ (accessed on 25 February 2023).
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data. 2010. Available online: http://www.Bioinformatics.Babraham.Ac.Uk/Projects/Fastqc/ (accessed on 25 February 2023).
- Li, H.; Durbin, R. Fast and Accurate Long-Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve Years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Benton, M. 2021 Nanopore Guppy GPU Basecalling on Windows Using WSL2. 2022. Available online: https://medium.com/analytics-vidhya/explained-output-of-nvidia-smi-utility-fc4fbee3b124 (accessed on 3 August 2022).
- ARTIC SARS-CoV-2 Workflow. EPI2ME. Oxford Nanopore Technologies Ltd. Available online: https://github.com/epi2me-labs/wf-artic (accessed on 25 February 2023).
- Medaka. Oxford Nanopore Technologies Ltd. Available online: https://github.com/nanoporetech/medaka (accessed on 25 February 2023).
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; Von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Pango Designation. Available online: https://github.com/cov-lineages/pango-designation (accessed on 10 June 2022).
- Rambaut, A.; Lam, T.T.; Max Carvalho, L.; Pybus, O.G. Exploring the Temporal Structure of Heterochronous Sequences Using TempEst (Formerly Path-O-Gen). Virus Evol. 2016, 2, vew007. [Google Scholar] [CrossRef]
- Drummond, A.J.; Ho, S.Y.W.; Phillips, M.J.; Rambaut, A. Relaxed Phylogenetics and Dating with Confidence. PLoS Biol. 2006, 4, e88. [Google Scholar] [CrossRef]
- Drummond, A.J. Bayesian Coalescent Inference of Past Population Dynamics from Molecular Sequences. Mol. Biol. Evol. 2005, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian Phylogenetic and Phylodynamic Data Integration Using BEAST 1.10. Virus Evol. 2018, 4. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- FigTree v1.4.4. Available online: https://github.com/rambaut/figtree/releases (accessed on 1 March 2019).
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol. Biol. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef] [PubMed]
- Tzou, P.L.; Tao, K.; Pond, S.L.K.; Shafer, R.W. Coronavirus Resistance Database (CoV-RDB): SARS-CoV-2 Susceptibility to Monoclonal Antibodies, Convalescent Plasma, and Plasma from Vaccinated Persons. PLoS ONE 2022, 17, e0261045. [Google Scholar] [CrossRef]
- Ministerio de Hacienda Resolución 426-E/2017. Available online: https://www.argentina.gob.ar/normativa/nacional/resoluci%C3%B3n-426-2017-279545 (accessed on 10 June 2022).
- Organización Panamericana de la Salud Situación de COVID-19 En Argentina. Distribución Espacio-Temporal de Casos y Muertes. Available online: https://paho-covid19-response-who.hub.arcgis.com/pages/paho-argentina-covid-19-response (accessed on 15 November 2022).
- Torres, C.; Nabaes Jodar, M.; Acuña, D.; Montaño, R.M.Z.; Culasso, A.C.A.; Amadio, A.F.; Aulicino, P.; Ceballos, S.; Cacciabue, M.; Debat, H.; et al. Omicron Waves in Argentina: Dynamics of SARS-CoV-2 Lineages BA.1, BA.2 and the Emerging BA.2.12.1 and BA.4/BA.5. Viruses 2023, 15, 312. [Google Scholar] [CrossRef]
- C.37 Lineage Defining Mutations. Available online: https://cov-lineages.org/constellations.html (accessed on 25 January 2022).
- Romero, P.E.; Dávila-Barclay, A.; Gonzáles, L.; Salvatierra, G.; Cuicapuza, D.; Solis, L.; Marcos, P.; Huancachoque, J.; Carhuaricra, D.; Rosadio, R.; et al. Novel Sublineage within B.1.1.1 Currently Expanding in Peru and Chile, with a Convergent Deletion in the ORF1a Gene (Δ3675-3677) and a Novel Deletion in the Spike Gene (Δ246-252, G75V, T76I, L452Q, F490S, T859N). Available online: https://virological.org/t/novel-sublineage-within-b-1-1-1-currently-expanding-in-peru-and-chile-with-a-convergent-deletion-in-the-orf1a-gene-3675-3677-and-a-novel-deletion-in-the-spike-gene-246-252-g75v-t76i-l452q-f490s-t859n/685/1 (accessed on 1 June 2021).
- Padilla-Rojas, C.; Jimenez-Vasquez, V.; Hurtado, V.; Mestanza, O.; Molina, I.S.; Barcena, L.; Morales Ruiz, S.; Acedo, S.; Lizarraga, W.; Bailon, H.; et al. Genomic Analysis Reveals a Rapid Spread and Predominance of Lambda (C.37) SARS-CoV-2 Lineage in Peru despite Circulation of Variants of Concern. J. Med. Virol. 2021, 93, 6845–6849. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Herrera, N.; Araujo-Castillo, R.V.; Mestanza, O.; Galarza, M.; Rojas-Serrano, N.; Solari-Zerpa, L. SARS-CoV-2 Lambda and Gamma Variants Competition in Peru, a Country with High Seroprevalence. Lancet Reg. Health—Am. 2022, 6, 100112. [Google Scholar] [CrossRef] [PubMed]
- Quispe-Ricalde, M.A.; Castelán-Sánchez, H.G.; Meza-Rodríguez, P.M.; Dávila-Ramos, S.; Sierra, J.L.; Batista-Garcia, R.; Concha-Velasco, F.; Lucana, S.F.; De Santa Cruz, J.; Zea, V.; et al. Evidence of Natural Selection and Dominance of SARS-CoV-2 Variant Lambda (C.37) over Variants of Concern in Cusco, Peru. Arch. Virol. 2023, 168, 88. [Google Scholar] [CrossRef]
- Kashima, S.; Slavov, S.N.; Giovanetti, M.; Rodrigues, E.S.; Patané, J.S.L.; Viala, V.L.; Santos, E.V.; Evaristo, M.; Lima, L.P.O.; Martins, A.J.; et al. Introduction of SARS-CoV-2 C.37 (WHO VOI Lambda) in the Sao Paulo State, Southeast Brazil. J. Med. Virol. 2022, 94, 1206–1211. [Google Scholar] [CrossRef]
- Molina-Mora, J.A.; Reales-González, J.; Camacho, E.; Duarte-Martínez, F.; Tsukayama, P.; Soto-Garita, C.; Brenes, H.; Cordero-Laurent, E.; Ribeiro Dos Santos, A.; Guedes Salgado, C.; et al. Overview of the SARS-CoV-2 Genotypes Circulating in Latin America during 2021. Front. Public Health 2023, 11, 1095202. [Google Scholar] [CrossRef] [PubMed]
- Stadtmüller, M.; Laubner, A.; Rost, F.; Winkler, S.; Patrasová, E.; Šimůnková, L.; Reinhardt, S.; Beil, J.; Dalpke, A.H.; Yi, B. Emergence and Spread of a Sub-Lineage of SARS-CoV-2 Alpha Variant, B.1.1.7 in Europe, and with Further Evolution of Spike Mutation Accumulations Shared with the Beta and Gamma Variants. Virus Evol. 2022, 8, veac010. [Google Scholar] [CrossRef] [PubMed]
- Presidencia de la Nacion Argentina CIERRE DE FRONTERAS. Decisión Administrativa 268/2021. Available online: https://www.argentina.gob.ar/normativa/nacional/decisi%c3%b3n_administrativa-268-2021-348256 (accessed on 15 February 2023).
- Torjesen, I. Covid-19: Delta Variant Is Now UK’s Most Dominant Strain and Spreading through Schools. BMJ 2021, n1445. [Google Scholar] [CrossRef] [PubMed]
- Bolze, A.; Luo, S.; White, S.; Cirulli, E.T.; Wyman, D.; Dei Rossi, A.; Machado, H.; Cassens, T.; Jacobs, S.; Schiabor Barrett, K.M.; et al. SARS-CoV-2 Variant Delta Rapidly Displaced Variant Alpha in the United States and Led to Higher Viral Loads. Cell Rep. Med. 2022, 3, 100564. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Althaus, C.L.; Giovanetti, M.; San, J.E.; Giandhari, J.; Pillay, S.; Naidoo, Y.; Ramphal, U.; Msomi, N.; et al. Rapid Replacement of the Beta Variant by the Delta Variant in South. Africa. Epidemiology. 2021. Available online: https://www.medrxiv.org/content/10.1101/2021.09.23.21264018v1 (accessed on 15 February 2023).
- Giovanetti, M.; Fonseca, V.; Wilkinson, E.; Tegally, H.; San, E.J.; Althaus, C.L.; Xavier, J.; Nanev Slavov, S.; Viala, V.L.; Ranieri Jerônimo Lima, A.; et al. Replacement of the Gamma by the Delta Variant in Brazil: Impact of Lineage Displacement on the Ongoing Pandemic. Virus Evol. 2022, 8, veac024. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary Analysis of the Delta and Delta Plus Variants of the SARS-CoV-2 Viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef]
- Focosi, D.; Quiroga, R.; McConnell, S.; Johnson, M.C.; Casadevall, A. Convergent Evolution in SARS-CoV-2 Spike Creates a Variant Soup from Which New COVID-19 Waves Emerge. IJMS 2023, 24, 2264. [Google Scholar] [CrossRef]
- Kimura, I.; Kosugi, Y.; Wu, J.; Zahradnik, J.; Yamasoba, D.; Butlertanaka, E.P.; Tanaka, Y.L.; Uriu, K.; Liu, Y.; Morizako, N.; et al. The SARS-CoV-2 Lambda Variant Exhibits Enhanced Infectivity and Immune Resistance. Cell Rep. 2022, 38, 110218. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.-M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 Spike Mutations That Attenuate Monoclonal and Serum Antibody Neutralization. Cell Host Microbe 2021, 29, 477–488.e4. [Google Scholar] [CrossRef]
- Venkatakrishnan, A.J.; Anand, P.; Lenehan, P.; Ghosh, P.; Suratekar, R.; Siroha, A.; Chowdhury, D.R.; O’Horo, J.C.; Yao, J.D.; Pritt, B.S.; et al. Antigenic Minimalism of SARS-CoV-2 Is Linked to Surges in COVID-19 Community Transmission and Vaccine Breakthrough Infections. Infectious Diseases (except HIV/AIDS). 2021. Available online: https://www.medrxiv.org/content/10.1101/2021.05.23.21257668v3.full (accessed on 15 February 2023).
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F.; et al. N-Terminal Domain Antigenic Mapping Reveals a Site of Vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, G.; Guo, Y.; Zhou, T.; Gorman, J.; Lee, M.; Rapp, M.; Reddem, E.R.; Yu, J.; Bahna, F.; Bimela, J.; et al. Potent SARS-CoV-2 Neutralizing Antibodies Directed against Spike N-Terminal Domain Target a Single Supersite. Cell Host Microbe 2021, 29, 819–833.e7. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent Deletions in the SARS-CoV-2 Spike Glycoprotein Drive Antibody Escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef] [PubMed]
- Vogels, C.B.F.; Breban, M.I.; Ott, I.M.; Alpert, T.; Petrone, M.E.; Watkins, A.E.; Kalinich, C.C.; Earnest, R.; Rothman, J.E.; Goes De Jesus, J.; et al. Multiplex QPCR Discriminates Variants of Concern to Enhance Global Surveillance of SARS-CoV-2. PLoS Biol. 2021, 19, e3001236. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Chan, J.F.-W.; Liu, H.; Liu, Y.; Chai, Y.; Shi, J.; Shuai, H.; Hou, Y.; Huang, X.; Yuen, T.T.-T.; et al. Spike Mutations Contributing to the Altered Entry Preference of SARS-CoV-2 Omicron BA.1 and BA.2. Emerg. Microbes Infect. 2022, 11, 2275–2287. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Kemp, S.A.; Papa, G.; Datir, R.; Ferreira, I.A.T.M.; Marelli, S.; Harvey, W.T.; Lytras, S.; Mohamed, A.; Gallo, G.; et al. Recurrent Emergence of SARS-CoV-2 Spike Deletion H69/V70 and Its Role in the Alpha Variant, B.1.1.7. Cell Rep. 2021, 35, 109292. [Google Scholar] [CrossRef]
- Shapira, G.; Patalon, T.; Gazit, S.; Shomron, N. Immunosuppression as a Hub for SARS-CoV-2 Mutational Drift. Viruses 2023, 15, 855. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).