Abstract
Mitochondria have been identified as the “powerhouse” of the cell, generating the cellular energy, ATP, for almost seven decades. Research over time has uncovered a multifaceted role of the mitochondrion in processes such as cellular stress signaling, generating precursor molecules, immune response, and apoptosis to name a few. Dysfunctional mitochondria resulting from a departure in homeostasis results in cellular degeneration. Viruses hijack host cell machinery to facilitate their own replication in the absence of a bonafide replication machinery. Replication being an energy intensive process necessitates regulation of the host cell oxidative phosphorylation occurring at the electron transport chain in the mitochondria to generate energy. Mitochondria, therefore, can be an attractive therapeutic target by limiting energy for viral replication. In this review we focus on the physiology of oxidative phosphorylation and on the limited studies highlighting the regulatory effects viruses induce on the electron transport chain.
1. Introduction
Mitochondria have traditionally been viewed as the energy hubs of the cell. The term “powerhouse” was coined almost seven decades ago [1]. Over the last few years this notion has expanded with mitochondria shown to play a moonlighting role in cellular pathophysiology since this organelle is not only vital in cellular metabolism but also in stress response, signaling, immune response, as well as apoptosis. The key ATP generating process in the mitochondria is oxidative phosphorylation (OxPhos), defined as the process wherein energy is generated from nutrients via reduction of oxygen (Figure 1). Viruses, being obligate intracellular pathogens, have to depend on host cells for energy required for replication. OxPhos is therefore one of the main cellular pathways regulated during viral infections. The effector site of OxPhos in the mitochondria is the electron transport chain (ETC), composed of a series of protein complexes embedded in the inner mitochondrial membrane (IMM) containing subunits encoded both on the nuclear and the mitochondrial genomes. Although mitochondria are known to play a vital role in the innate immune response to viral infections, in this review we will focus exclusively on the ETC and how viruses affect the functioning of the OxPhos system.
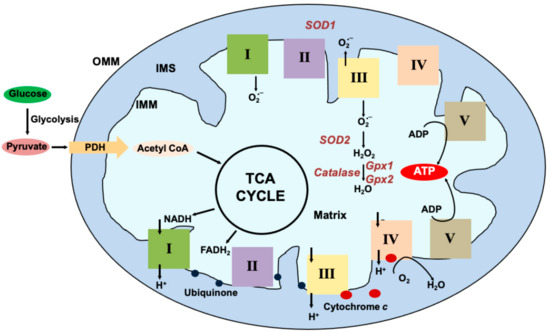
Figure 1.
Mitochondrial Oxidative Phosphorylation: Glucose, the primary energy substrate is metabolized via glycolysis into pyruvate. From the cytosol, pyruvate enters the mitochondria via the transporters and is decarboxylated by Pyruvate dehydrogenase (PDH) to form Acetyl Coenzyme A (Acetyl CoA) that is used in the Tricarboxylic Acid Cycle (TCA). Reducing equivalents generated in the form of NADH and FADH2 are funneled into Complex I (I) or Complex II (II) embedded in the inner mitochondrial membrane (IMM) respectively. Ubiquinone (blue circles) transfers the electrons from I and II to Complex III (III). Cytochrome c (red circle) transfers the electrons from III to Complex IV (IV) where it is used to reduce molecular oxygen to H2O. As the electrons pass through the complexes, protons (H+) are pumped into the intermembrane space (IMS) creating a gradient across the IMM. The energy from this gradient is used by complex V (V) also known as ATP synthase to generate ATP from ADP. Some electrons escape and react with molecular oxygen to form superoxide (O2•−) at I and III. Superoxide is generated on the matrix side at I and both on the matrix and IMS side at III. Superoxide dismutase (SOD) 1 and 2 localized to the IMS and the matrix respectively scavenge the superoxide to generate H2O2. Glutathione peroxidases further breakdown the peroxide to H2O.
2. Physiology of OxPhos
OxPhos is a key physiological program regulated in cells under viral infections. Four protein complexes make up the ETC, providing the potential energy that drives OxPhos through a fifth complex, ATP synthase. In addition, accessory proteins that interact with these complexes have the capacity to regulate OxPhos. Reducing equivalents (NADH and FADH2) generated during glycolysis and the Krebs cycle pass sequentially through the ETC. Coenzyme Q (CoQ) shuttles electrons between Complex I/II and III. Cytochrome c transfers the electrons between Complex III and IV. Complex IV, known as Cytochrome c oxidase (COX), is the terminal enzyme in the ETC responsible for reduction of molecular oxygen to water using the electrons provided by cytochrome c. As the electrons pass through the complexes, a proton gradient is generated across the IMM and, as noted, is used by Complex V (ATP synthase) to generate ATP.
The primary energy substrate utilized by most cells is glucose, which can be metabolized by two crucial pathways. The first of these pathways is glycolysis, which is a series of reactions that occur in the cytoplasm. The main end product of glycolysis is pyruvate, which is shuttled into the mitochondria by the enzyme complex pyruvate dehydrogenase. This complex sits in the inner mitochondria membrane and converts pyruvate to acetyl CoA. The acetyl CoA enters the other major glucose metabolic pathway—the tricarboxylic acid (TCA) cycle, also known as the Krebs cycle. The major role of this cycle is to generate the high energy compounds NADH and FADH2 used by the ETC to generate ATP; it also produces metabolic intermediates. The cycle consists of a series of reactions that are catalyzed by a number of dehydrogenase enzymes found in the mitochondria matrix (with the notable exception of succinate dehydrogenase found in the inner mitochondria membrane). The major dehydrogenase enzymes that generate NADH are glyceraldehyde-3-phospate dehydrogenase in glycolysis and isocitrate dehydrogenase, alpha ketoglutarate dehydrogenase, and malate dehydrogenase in the TCA cycle. The NADH and FADH2 reducing equivalents generated by the TCA cycle enter the ETC at Complex I and Complex II, respectively. Breakdown of one glucose molecule from the TCA cycle can generate more reducing equivalents as compared to glycolysis: the TCA cycle can generate 6 NADH molecules per glucose molecule whereas glycolysis can only generate 4 NADH. The net energy from glycolysis is reduced further because the first step of glycolysis consumes 2 molecules of ATP. The TCA cycle can provide further reducing equivalents in the form of 2 FADH2 molecules and one GTP molecule per glucose molecule.
Other sources that can be utilized by the mitochondria include glutamine and fatty acids. Glutamine, the most abundant amino acid in the blood, enters the cell via transporters on the plasma membrane (SLC38A1, SLC38A2, and SLC1A5). SLC1A5 imports glutamine into the mitochondria, where it is shuttled into the TCA cycle via conversion to alpha ketoglutarate by glutamate dehydrogenase (reviewed in [2]). Fatty acids, on the other hand, are converted into fatty-acyl CoA, which allows them to be broken down in consecutive steps that occur in the mitochondrial matrix to generate several molecules of NADH, FADH2, and acetyl CoA depending on the length of the fatty acid. This process is termed beta oxidation. Cardiac myocytes rely primarily on beta oxidation to generate energy. There are other pathways that can provide energy such as the one carbon metabolism (for breakdown of amino acids) and the pentose phosphate pathway that branches off from glycolysis that are beyond the scope of this review. However, all these pathways for breakdown of substrates including glucose, glutamine, or fatty acids and amino acids, ultimately converge upon OxPhos in order to generate ATP required for cellular functioning. Hence, many viruses directly or indirectly target this process to hijack the host cell’s energy metabolism for enhancing their own survival and propagation. Viruses are capable of modulating other steps including substate import, breakdown, and modification for entry into mitochondria (for example, conversion of fatty acids to fatty acyl CoA to enter beta oxidation). However, these are beyond the scope of this review but well summarized elsewhere [3].
One other key function of the ETC is the generation of reactive oxygen species (ROS). Some of the electrons passing through the ETC escape and react with molecular oxygen to form superoxide. Complex I of the ETC generates ROS in the mitochondrial matrix and Complex III generates it in both the matrix and IMS. In an intact physiological system, most of the superoxide generated is reduced by the action of ROS scavengers such as superoxide dismutases and glutathione peroxidases. However, when the ETC is dysfunctional, ROS production exceeds the scavenging capacity, resulting in increased ROS with attendant cellular damage. Total ROS levels, therefore, are the difference between production and scavenging capacity of a cell. Viral infections and ROS have also been discussed here.
3. Complex I of the ETC (NADH Dehydrogenase)
Complex I (CI) is made up of 45 subunits with 38 subunits encoded on the nuclear genome and seven on the mitochondrial genome [4]. This complex is identified by its L-shaped structure, with one arm embedded in the IMM and the other protruding into the mitochondrial matrix. Of the 45 subunits, 14 form the core structure (equivalent to the entire complex I in many bacteria) and are equally split between the two arms of the complex. The remaining 31 proteins are considered accessory subunits. While transferring the electrons from NADH to ubiquinone, CI can pump 4 protons across the inner mitochondrial membrane. Enhanced activity of this complex will result in enhanced mitochondrial respiration whereas inhibition would result in excessive ROS production.
4. Complex II (Succinate Dehydrogenase, SDH)
This enzyme is a part of both the TCA cycle and the ETC. The SDH complex is made up of four nuclear encoded subunits and is the only one that has no representation on the mitochondrial genome. Subunit A and B are the catalytic subunits, whereas C and D are the membrane anchors. This complex is responsible for the oxidation reaction converting succinate to fumarate. The electrons generated are fed into the ETC. Alternatively, they can be used to reduce the ubiquinone pool and contributes towards antioxidant function [5].
5. Complex III (Cytochrome bc1 Complex, CIII)
This complex is made up of 11 subunits with 10 encoded in the nucleus and one in the mitochondria [6]. CIII oxidizes ubiquinol with electrons transferred to cytochrome c. Mitochondrial complex III generates superoxide during the ubiquinone Q-cycle [7,8]. During this process, two electrons from CI and CII are transferred to ubiquinone, resulting in its reduction to ubiquinol (QH2). CIII then moves these two electrons to the single electron carrier cytochrome c. This results in the unstable radical ubisemiquinone (Q•−), which can donate its unpaired electron to oxygen to generate superoxide within the Q-cycle. Also, 2 protons are pumped across the inner mitochondrial membrane to contribute to the electrochemical gradient. In addition to electron transfer, CIII also helps reoxidation of CoQ, and also generates ROS [9].
6. Complex IV (Cytochrome c Oxidase, COX)
COX is the terminal enzyme in the ETC and is made up of 13 stoichiometric subunits with 10 encoded in the nucleus and three in the mitochondria. More than 90% of the oxygen consumed is reduced to water by COX. Being the rate-limiting enzyme makes COX a vital regulator of the OxPhos system [10]. This complex is unique in that the regulation can occur via multiple complex mechanisms such as allosteric regulation [11], organ specific isoforms [12], and post-translational modifications [13]. This enzyme also plays a vital role in cellular inflammatory pathways [14]. Specific knockdown of subunit 4 isoform 1 (COX4I1) in macrophages has been shown to induce ROS as well as activate pro-inflammatory cytokines [15].
7. Complex V (ATP Synthase, CV)
Complex V (ATP Synthase) transforms energy from the proton gradient created by the flow of electrons through the ETC to generate ATP. The nuclear mitochondrial distribution of the subunits that make up this complex is 14:2. The activity of this complex is driven by the proton gradient across the inner mitochondrial membrane to generate energy. The enzyme has two functional domains—one named F1, a soluble portion situated in the mitochondrial matrix, and the other Fo, in the inner mitochondrial membrane. There are 11 genes that form these two domains of which two are encoded by the mitochondrial genome. From these 11 genes, the F1 subunit is comprised of 5 genes and the remaining ones form the Fo subunit. An average of 30.63 ATP molecules are formed per glucose molecule via oxidative phosphorylation; by contrast, only 1.45 ATP/glucose molecule is formed by substrate level phosphorylation during glycolysis. An important quantity regarding CV function is its efficiency, the P/O ratio, which is defined as the molecules of ATP generated per molecule of oxygen consumed; the maximum P/O ration for 1 molecule of glucose is 2.79 [16].
8. Supercomplexes
Formation of higher order structures, called supercomplexes (SCs), are composed of complex I, III and IV of the ETC and have been identified from yeast to man [17,18]. They are thought to enhance efficiency of OxPhos [19] although contrary evidence has been presented [20]. SCs have defined stoichiometries, for example CI forms a supercomplex with CIII2 and CIV (SC I + III2 + IV, known as the respirasome), as well as with CIII2 alone (SC I + III2). CIII2 forms a supercomplex with CIV (SC III2 + IV), and CV forms dimers (CV2) [21]. Almost all of Complex I is exclusively detected as a part of various SC assemblies [22] whereas complex III can be found as homodimer and complex IV either as a homodimer or monomer. Besides the respirasome, other assemblies include CI + CIII2 and CIII2 + CIV. CI, which is present as a part of CI + CIII2, is much lower than the respirasome [22]. These configurations are important in lower organisms such as yeast, which lack a traditional complex I enzyme. Various subunits from each complex interact with each other to stabilize the supercomplexes. For example- in a CI + CIII2 assembly, there are 2 main interactions—one in the NDUFA11 and the UQCRB, UQCRQ, and UQCRH subunits of CIII, and a second one in the matrix between NDUFB4, NDUFB9, and the CIII subunit UQCRC [23]. On the other hand, the contacts formed between CI and CIII within the respirasome involve so-called supernumerary subunits. These supernumerary subunits are not found in bacteria and are considered to be eukaryotic origin [24].
There are several hypotheses that aim to explain the presence and role of supercomplexes. One of the most prevalent theories is that of they may be useful for substrate channeling. That is, the formation of complexes of enzymes that act sequentially in a pathway so that a specific substrate can be transferred from one enzymatic activity to the next without allowing free diffusion of the substrate into the bulk solution. In order for substrate channeling to occur, a dedicated pool of bound electron carriers (ubiquinone and cytochrome c) must be present. However, structural analyses reveled that the distance between the two cytochrome c binding sites on CIII and CIV in the supercomplex is too large (>6 nm), thereby precluding the substrate channeling hypothesis [25,26]. Other theories include the efficiency of electron transport rather than strict channeling. In this model, the supercomplex simply provides enhances electrostatic interactions where cytochrome c can “roll” between complex III and IV and also mix with the free pool. Other presumed functions include enhanced stability to help the assembly of complexes, in particular for the largest of the ETC complexes—complex I. This is called the cooperative assembly model [27]. The plasticity model [28] suggests that supercomplexes formation helps to adapt to changing metabolic requirements, and that supercomplexes prevent electron escape to reduce ROS [21]. Structurally, some supercomplexes are known to affect membrane curvature and shape. Complex V homodimers have been identified in yeast and appear important for IMM bending and cristae formation [29]. Though it was recently shown to participate in supercomplex formation [30] in a ciliate protist (Tetrahymena) and to affect membrane curvature, it is yet to be identified in mammalian supercomplexes.
There are known assembly factors that help to connect these complexes. These include cardiolipin, PHB1 (prohibitin), PHB2, and SCAF1 (supercomplex assembly factor 1) [30]. Of these, SCAF1 (also known as COX7A2L) is the only dedicated assembly factor for supercomplexes and is required for biogenesis and assembly of CIII2 + IV but does not affect the assembly of the respirasome [31]. A recent study also showed that, besides SCAF1 containing complexes (S-MRC, SCAF1 containing mitochondrial respiratory chain complex) a second type is also present, called C-MRC (COX7A2 containing mitochondrial respiratory chain complex) is also present. The SCAF1-dependent S-MRC includes the SCAF1-containing respirasome, which accounts for approximately half of total CIII and CIV levels. The remaining CIII and CIV are equally distributed between the CIII2 + CIV supercomplex and free complexes. The C-MRC organization displays a relatively low amount of the COX7A2-containing respirasome, no CIII2 + CIV supercomplex, and abundant free CIII (~60% of total CIII) and CIV (~80% of total CIV). The exclusive presence of one configuration or the other in knockout cells of the corresponding isoform led to some changes in mitochondrial bioenergetics. However, no differences in respiratory parameters were observed where the two MRC organizations co-exist [27]. There are several more details that have been identified regarding super complex components, assembly, and possible functions that are well reviewed elsewhere [32].
9. Viruses and Oxidative Phosphorylation
Viruses, being intracellular pathogens, depend on host cellular machinery and energy to facilitate their entry, replication, and exit. In the recent past, significant advances have been made towards understanding the role of cellular mitochondrial function and immune responses [33,34,35]. Studies have also been focused on the crosstalk between mitochondrial dynamics, including fusion-fission and mitophagy (reviewed in [36,37]). Although mitochondrial OxPhos regulates all these functional pathways, very few studies have evaluated the effect and the underlying mechanism of how viruses hijack the host mitochondrial OxPhos system. Here we will review the studies characterizing the effects of viruses on the ETC, specifically the mitochondrial complexes, ATP levels, and ROS. We will also discuss the details regarding the pathways that appear to regulate the ETC complexes in virally infected cells.
For the purpose of this review, we will use the Baltimore classification of viruses wherein the groups are classified on the basis of the viral genome [38]. Most of the work evaluating the role of mitochondrial OxPhos in viral infections has been done on viruses in group IV (+ sense single stranded RNA).
(+) ssRNA: This group of viruses harbors a single stranded RNA genome that produces functional mRNAs. An RNA-dependent RNA polymerase transcribes the genome to generate a polyprotein. Viral or host cellular proteases cleave the polyprotein into individual proteins. This group has eight families with either enveloped or non-enveloped capsids.
Flaviviruses: The viruses that have been studied in some detail for their role in regulating OxPhos are Hepatitis C virus (HCV), Zika virus (ZV), and West-Nile virus (WNV).
Hepatitis C virus: One of the earliest pieces of evidence of mitochondrial dysfunction in patients with HCV infection was the identification of antimitochondrial antibodies in serum [39]. Similarly, a defect in OxPhos along with increased oxidative stress markers were observed in liver biopsies from patients with chronic HCV infections [40]. In a transgenic mouse model for HCV genotype 1b strain N, defective activity of CI was observed along with an increase in ROS levels. The Core protein of HCV localizes to the mitochondrial outer membrane to cause enhanced Ca2+ flux into the mitochondria, resulting in CI dysfunction and increased ROS [41]. Similarly, using cell lines with inducible HCV replicons expressing the entire HCV polyprotein, enhanced calcium toxicity in the mitochondria was shown to cause an inhibition of CI activity and an increase in ROS [42], which were found to be reversible upon amantadine treatment [43]. It was also hypothesized on the basis of a case report that CIII dependent mitochondrial dysfunction underlies the myopathy phenotype in HCV [44]. HCV non-structural protein NS5A also localizes to the mitochondrial fraction and induces ROS via dysregulation of Ca2+ signaling [45,46]. Transcriptomic analysis of Huh-7.5 cells transfected with the full-length HCV genome displayed a reduction in expression of CI (ND1, ND3, ND4) and CIV (MT-CO2) subunits encoded on the mitochondrial genome [47]. Interactome analysis has identified HCV core, p7, and NS4B proteins to interact with the mitochondrial proteome in host cells [48]. MNRR1 (CHCHD2), a bi-organellar regulator of mitochondrial function that interacts with CIV and is required for its optimal function, was also identified as one of the top candidate host gene required for HCV replication [48]. MNRR1 was first identified as an HCV Non-structural protein 2 transregulated protein [49]. Although HCV inhibits mitochondria, the induction and requirement of MNRR1 could be hypothesized to be related to its anti-apoptotic or transcriptional regulatory function [50,51].
Zika virus: Zika virus rose to prominence in the recent past due to its association with microcephaly [52]. The presence of viral nucleic acids in fetal brains and placentas led to the causal association of microcephaly with viral infection [53]. Although there is a lack of evidence suggesting a direct effect of Zika viral proteins regulating the ETC, studies have shown Complexes II, IV, and V to be affected. Zika viral proteins such as NS4A and 4B do localize to the mitochondria to modulate mitochondrial dynamics and apoptosis [54,55].MNRR1 is also upregulated in ZIKV infected cells and may promote viral replication [56]. Zika viral infection of neurons generated the metabolite itaconate from the TCA cycle that inhibits CII activity, resulting in mitochondrial dysfunction [57]. The effect on oxygen consumption rate (OCR), a function of CIV, displayed a strain-specific effect. Using MRC-5 cells, only the MR766 strain was shown to inhibit OCR. Other strains, such as H/PF/2013, M-F37L, DN-1, and DN-2, were comparable to the uninfected cells for their effect on OCR [58]. Finally, Zika viral (and also other flaviviral) capsid proteins induce DAPIT [59], an assembly subunit of CV [60].
West Nile virus: This virus infects keratinocytes and dendritic cells in skin as well as cells in the central nervous system [61,62]. Using neuroblastoma cells A172, significant downregulation was observed for nuclear encoded genes for CII (SDHB), CIV (COX5B and 6B), and CV (ATP5G1, 5C1, 5J, 5B, 5A1, 5O, 5F1), suggestive of an inhibitory effect on ETC and mitochondrial function [63]. In virally infected Vero cells, oxidative phosphorylation was inhibited with a shift towards glycolysis [64]. The modulation of other mitochondrial pathways by West Nile virus has been reviewed previously [65,66].
Coronaviruses:
SARS-CoV-2: The recent COVID pandemic overburdened the economic and health care sectors across the globe. Research was focused towards identifying therapeutic targets and a vaccine. The initial studies performed in multiple cell and tissue types identified an inhibitory effect of viral infection on nuclear encoded CI subunits including NDUFS2, NDUFS6, NDUFB7 [67]. CoV-2 was also shown to inhibit both nuclear as well as mitochondrially encoded mitochondrial genes. The gene profile was evaluated across disease progression. At the initial stage, minimal effects on gene expression were observed in lungs. Downregulation of mitochondrial genes was observed when viral titers peaked. The downregulated genes involved those encoding the structural and assembly subunits of the OxPhos complexes. Upon clearing of the virus, the inhibitory effect on mitochondrial genes was reversed in the lung, but not other organs such as the heart, liver, and kidneys [68]. Downregulation of CI was proposed to be responsible for the hypoxemic phenotype associated with the disease [69]. Cytokine storm underlies the pathogenicity of COVID. Monocytes infected with CoV-2 displayed downregulation of subunits from complexes I, II, III, and V, resulting in dysfunctional mitochondria and enhanced ROS that contributed to the cytokine production [70]. OCR was significantly reduced in peripheral blood mononuclear cells from COVID patients [71]. Additionally, multiple viral proteins such as ORF-3C localize to the mitochondria and induce organellar dysfunction [72]. Moreover, NSP10 interacts with ND4L and COXII to modulate complex activity [73]. Enzyme remodeling by subunit switch has also been observed specifically in SARS-CoV-2 infected cells. The C15orf48 subunit is induced upon infection and replaces its paralog, NDUFA4, in CIV [14]. Finally, levels of OxPhos regulators such as MNRR1 were also shown to be lower in SARS-CoV-2 patient hearts and may potentially contribute towards the cardiac complications of the disease [74].
Others: The three other viruses in the + ssRNA group include Rubella virus, Coxsackie B3, and Hepatitis E virus. Rubella virus causing German measles, in contrast to the others in the group, actually induced mitochondrial OxPhos by enhancing activities of CI, II, III, and IV in A549 cells 24 h post infection using isolated mitochondria [75]. Subunits SDHA, SDHB (CI), UQCRC2 (CIII), and COX4I1 (CIV) were also induced upon acute infection. The induction of OxPhos was found to be strain specific with Wb-12 strain showing maximal induction and 07-00426 showing minimal increase [76]. The induction of OxPhos in rubella virus infected cells has been attributed to the energy requirement of viral replication owing to the observation that the mitochondria in the infected cells are in close proximity to the viral replication complex [77]. Host cellular p32 protein facilitates the interaction of viral capsid with the mitochondria [78].
Coxsackie virus B3 (CVB3) mediated effects on OxPhos depend on the immune responsiveness of the host. Studies using C57/BL6 mice (that efficiently eliminates the virus) and A.SW/SnJ (unable to eliminate the virus) show a completely variable response. Hearts from C57/BL6 show an increase in CI and CIII activities whereas A.SW/SnJ hearts show a significant reduction [79] suggesting that mitochondrial function has a potential role to play in the viral replication cycle as well as in the host cellular response to infection.
Hepatitis E virus (HEV) is the causal agent of acute viral hepatitis. Recently, cell culture models have identified CIII function to be required for the replication of HEV [80] making it an attractive drug target. OxPhos dysfunction was also evident in primary human brain microvascular endothelial cells wherein the infected cells displayed a significant reduction in the protein levels of ATP5A1, a catalytic subunit of CV, resulting in bioenergetic deficit and apoptosis [81].
(-) ssRNA: The three viruses in this group on which studies have been performed characterizing OxPhos are Influenza, Rabies, and Respiratory syncytial virus (RSV).
Influenza: This virus is responsible for causing seasonal epidemics as well as pandemics (reviewed in [82]. One of the early studies documenting the effect of influenzas virus on mitochondrial function identified an ~50% reduction in MDCK cellular oxygen consumption rate in the infected cells compared to the mock control [83]. In contrast, mass spectrometric analysis of A549 cells infected with swine influenza virus identified NDUFS8 and ATP5B and 5D subunits to be upregulated [84] whereas H1N1 infection did not affect protein levels of ETC subunits [84]. Recently, H5N1 influenza viral infected cells were shown to have significantly higher levels of COX subunit 4 isoform 1 (COX4I1). Further, a CRISPR/Cas9 knockout of COX4I1 resulted in a ~200-fold reduction in viral titers. Lycorine, a compound inhibiting viral replication, was shown to function by inhibition of this isoform of COX [85]. Influenza virus may also indirectly affect the expression of certain subunits such as COX6C via regulation of microRNAs [86]. The M1 protein from influenza virus interacts with and inhibits the functioning of CIV [87]. These effects on mitochondrial function suggest that the virus probably hijacks mitochondrial metabolism depending on the stage of its replication cycle such that activation is induced via multiple pathways when energy is required [88].
Rabies: This virus, responsible for causing fatal encephalitis, induces mitochondrial dysfunction underlying the pathogenic phenotype. Mitochondrial function was evaluated in baby hamster kidney cells using the challenge virus standard-11 strain. A significant reduction in intracellular ATP levels was observed in these cells along with increased ROS levels. Both of these were attributed to high mitochondrial membrane potential resulting from increased activities of CI and CIV generating ROS and hydrolysis of ATP [89]. The same group later identified rabies viral phosphoprotein to interact with CI and regulate its function [90]. Extensive analysis was also performed on postmortem brain tissues from rabies encephalitis. Increased activities of CI, IV, and V were observed along with an increase in multiple subunit proteins that constitute individual complexes of the ETC [91].
Respiratory syncytial virus: This virus causes acute lower respiratory tract infections especially in the young and immunocompromised. RSV infected cells display a perinuclear clustering of the mitochondria suggestive of cellular stress. A time dependent reduction in basal oxygen consumption was observed in A549 cells with an increase in glycolysis and ROS levels [92]. These changes were shown to be CI dependent. Reduced activity of CI along with increased ROS levels were conducive for RSV replication in these cells and these effects were induced by the matrix protein of the virus [93]. A downregulation of mitochondrial biogenesis was also a feature of RSV infected cells [94].
ssRNA-RT: This group includes retroviruses with the most common being Human Immunodeficiency Virus (HIV). The viruses in this category have a reverse transcriptase enzyme that generates a cDNA intermediate from the RNA genome. One of the earliest pieces of evidence of HIV virus affecting mitochondrial function was described almost four decades ago. HIV positive ACH-2 cells were shown to have mitochondrially localized viral RNA and proportionally defective mitochondrial morphology [95]. Shortly thereafter, using Saccharomyces cerevisiae as a model system, it was shown that the HIV protein Vpr induced mitochondrial dysfunction by reducing activities of the entire ETC [96]. In strong contrast, an increase in expression of individual subunits and activity of CIV was observed [97]. HIV-1 infection also inhibits CI activity by a specific downregulation of the NDUFA6 subunit [98]. PBMCs from non-treated HIV-infected patients were found to have reduced CII, III, and IV activities [99]. We have recently shown in glial cells that the inhibitory effect of antiretrovirals on SDH is abrogated in the presence of latent or active HIV infection [100]. Effects of viral proteins on the ETC as a result of direct interaction have also been described. A direct interaction between the p2 peptide of the Gag and Gag-Pol precursors of HIV and COXI during acute phase of infection results in increased ATP levels [101]. Tat protein of HIV, however, inhibits COX and induces mitochondrial membrane permeabilization [102]. This property has allowed the use of Tat as a COX inhibitor in experimental settings. The ATP synthase β-subunit is required for optimal HIV viral transfer from the antigen presenting cell to the CD4+ T-cells. Although the mechanism of the localization of an inner mitochondrial protein to the cell surface is unclear, these findings made ATP synthase an attractive therapeutic target for HIV [103]. Defects in mitochondrial function (CIV) measured as OCR also depends on the stage of infection. Viral infection proportionally inhibited OCR rates with minimal effects on glycolysis [104]. These contrasting results on ETC in HIV infected cells could potentially point towards cell and strain specific effects. Comprehensive studies towards this avenue are required for a better understanding of how HIV subverts mitochondrial OxPhos towards its replicative benefit. HIV viral proteins also regulate multiple physiological processes of the mitochondria (reviewed in [105]).
dsDNA-RT: These viruses have a DNA genome with an RNA intermediate. Hepatitis B (HBV) is an important virus in this group, responsible for liver disease that can lead to cirrhosis and hepatocellular carcinoma. A protein encoded by the HBV genome, ORF X (HBx), interacts with the OMM and induces apoptosis [106,107]. Using a two-hybrid assay system it was also shown that HBx interacted with subunit 3 of CIV (COXIII) [108]. This results in an increase in mitochondrial function and cell growth [109]. A significant downregulation of the ETC complex levels along with activity was observed in hepatoma cells expressing HBx [110] with a resultant increase in ROS levels. A ~50% reduction in CII activity was also associated with chronic HBV infection as evaluated using liver biopsy specimens [111]. HBV, in contrast, induces OxPhos in macrophages and this increase is required to downregulate the immune response [112]. HBV DNA also can integrate into the mitochondrial genome coding for the subunits of ETC and may contribute towards organellar dysfunction in infected cells [113]. Correcting mitochondrial dysfunction is a potential therapeutic target in chronic HBV [114]. These results indicate that the virus differentially regulates mitochondrial function in cell types conducive towards its own replication. In some cells it increases mitochondrial function, whereas in others it decreases them with enhanced ROS.
dsDNA: Three DNA viruses have been studied for their effects on mitochondrial OxPhos and are described here.
Human Cytomegalovirus (HCMV): This herpesvirus is highly seroprevalent in the population. A majority of HCMV infections are congenital and result in neurodevelopmental anomalies [115]. HCMV depends on host cell energy for its replication. HCMV infected cells induce both OxPhos and glycolysis [116]. Metabolomic analysis also show an increase in the TCA cycle as well as glycolytic intermediates, supporting the induction of OxPhos and glycolysis [117]. A viral protein, pUL13, is responsible for the effect on OxPhos since virus with a deletion of pUL13 fails to induce OxPhos. pUL13 has been shown to interact with the MICOS complex responsible for maintenance of cristae that harbor the individual OxPhos complexes [118]. Another viral protein, pUL37x1, induces mitochondrial biogenesis and contributes towards OxPhos induction [119]. Viral infection also induces factors critical towards maintenance of the mitochondrial genome as well as those responsible for the assembly of the individual OxPhos complexes and for mitoribosome biogenesis [120]. Finally, GRIM-19 (Gene associated with retinoic acid and interferon-β-induced mortality-19) is another assembly factor of CI [121]. This protein relocalizes to other cellular niches such as the nucleus in response to mitochondrial stress to induce apoptosis. In HCMV infected cells, the β2.7 RNA transcript was shown to interact with GRIM19 to prevent its nuclear localization and thereby inhibit apoptosis of the infected cells [122].
Epstein-Barr virus (EBV): EBV is also seroprevalent with latent infection. Conditions of immunosuppression result in infection [123]. During early stages of infection, an induction of glycolysis takes place [124]. As the infection proceeds, OxPhos induction also occurs indirectly via activation of one-carbon metabolism [125]. One carbon metabolism is a series of reactions providing methyl groups for a multitude of cellular pathways including OxPhos [126]. Additionally, like CI, CII also has SDUFA1-4 that are responsible for the assembly of the complex [127,128,129]. However, recent studies have identified SENP2 to regulate sumoylation and assembly of CII under nutrient stressed condition [130]. This study identified desumoylation of SDHA subunit of CII under conditions of glutamine deprivation to result in an inhibitory effect on CII assembly and function. Epstein-Barr Viral (EBV) protein LMP1 reduces functioning of SENP2 [131]. However, this study did not evaluate the effect on mitochondrial function.
Human Papilloma virus (HPV): HPV, the causal agent of cervical cancers, also regulates host cellular OxPhos. The E2 protein plays a key role in viral genome replication [132]. E2 from high-risk HPV-16 and 18 interacts with UQCRC2 and UQCRFS1 (CIII) and COXII (CIV) to induce ROS generation by the mitochondria [133]. Recently, cells stably expressing the oncoprotein E7 of HPV-16 was shown to interact with the ATP5B subunit of CV, causing an increase in mitochondrial function. A mild increase was also observed with E7 of HPV-8 [134]. The E2 protein also regulates mitochondrial function indirectly via induction of p32 [135], an RNA-binding protein associated with TFAM [136]. TFAM is required for mitochondrial transcription and translation (reviewed in [137]).
10. Viruses and Mitochondrial Reactive Oxygen Species
Multiple studies have reported the generation of ROS upon direct or indirect (for example via gene regulation) interaction of viral proteins with the host cell mitochondria. Examples are HCV mediated inhibition of CI activity [42], downregulation of assembly factors for CIII in SARS-CoV-2 infected cells [138], and Rabies viral phosphoprotein interaction with CI induce ROS production [90]. Others have reported an increase in mitochondrial ROS via (a) regulation of proteins involved in cristae structure such as prohibitins [139], (b) dysregulated calcium homeostasis resulting in a mitochondrial overload and ROS generation as seen with HBsAg, the surface antigen of HBV [140], (c) regulation of the mitochondrial membrane channels resulting in membrane depolarization and ROS by Tat protein of HIV [102], and (d) downregulation of the ROS scavenging enzymes such as SOD2 as seen in SARS-CoV-2 infections [138].
Excessive ROS is deleterious to the host cell and therefore would not be conducive for viral replication. Therefore, the ROS generated must be within levels that can facilitate viral replication and prevent host cell death. So why do viruses induce ROS unless it’s beneficial? The role of ROS as a signaling molecule [141] in the host cells could underlie the induction observed in virally infected cells.
The two major reactive species generated by the mitochondria are the superoxide anion (O2•−)and hydrogen peroxide (H2O2) [142]. Superoxide anion, for example, has been shown to activate the Raf/MEK/ERK pathway [143]. This pathway is required for replication of SARS-CoV-2 [144]. H2O2 activates the p38-MAPK pathway to facilitate replication of HCMV [145]. HCV induced ROS also facilitates viral replication via NFκB-dependent induction of DR6, which interacts with the viral protein NS5A to induce viral replication [146]. Similarly, studies have shown increased ROS to stabilize HIF-1 [147,148]. HIF-1 causes enhanced infectivity and replication of HIV in host cells [149,150].
ROS, in addition to regulating cellular signaling pathways, can also modify viral proteins to enhance its functionality. Oxidation induces dimerization and guanylation of the NS5A protein of Dengue virus, enhancing RNA-capping and replication [151]. Methionine oxidation of Kaposi Sarcoma Herpes Viral helicase also enhances its stability and function [152]. Although ROS are beneficial for some viral infections, high levels of ROS would be deleterious to the host cell and therefore result in abortive replication of the virus. Thus, viruses also induce antioxidant genes when the ROS levels in cells reach levels to activate apoptotic cascades. HPV E7 protein induces the enzyme catalase to degrade H2O2 [153]. Similarly, HBV induces NRF2 to activate antioxidant genes [154]. NS5A of HCV induces Glutathione peroxidase 1 (GPX1) and GPX4. Induction of GPX4 counteracts lipid peroxidation, resulting in enhanced infectivity of the progeny virus [155]. Viruses such as HCMV [156,157] and Influenza (reviewed in [158]) induce ROS acutely to facilitate induction of viral promoters and then induces ROS scavengers to reduce ROS. As ROS also induces apoptosis, viruses counteract the apoptotic pathway by multiple mechanisms such as transcriptional inhibition of proapoptotic proteins like Bim by EBNA3A and EBNA3C of EBV [159], or induction of proteins that inhibit multiple targets in the apoptotic cascade (reviewed in [160]).
11. Summary
In summary, although an exact mechanism is lacking, there appears to be a fine regulatory system in play to ensure optimal viral replication and evasion of immune response in the host cell. Viruses either induce or inhibit OxPhos, depending on its life cycle, either by direct interaction with the OxPhos complexes and their assembly factors or indirectly by regulating transcription of specific subunits and assembly factors. As ROS is a product of ETC function, viruses also regulate ROS generated via the ETC to support their own replication and modulate host signaling pathways. Table 1 summarizes the effects observed on OxPhos in virally infected cells. Finally, detailed studies characterizing a common mechanism used by multiple viruses are required. Mechanistic studies on mitochondrial supercomplexes would help uncover novel molecular mechanisms hijacked by viruses. This would allow the characterization of potential therapeutic targets for viral infection that would be of immense benefit during viral pandemics.

Table 1.
Effects of different viruses on OxPhos complexes.
Author Contributions
Conceptualization, S.A.; data curation, S.A., N.P. and E.G.; writing—original draft preparation, S.A., N.P. and E.G.; writing—review and editing, S.A., N.P. and L.I.G.; visualization S.A. and N.P., supervision, S.A.; project administration, S.A.; funding acquisition, S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research and APC were funded by startup funds from the Wayne State University School of Medicine (S.A.) and Henry L. Brasza endowment (L.I.G).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Siekevitz, P. Powerhouse of the Cell. Sci. Am. 1957, 197, 131–144. [Google Scholar] [CrossRef]
- Yoo, H.C.; Yu, Y.C.; Sung, Y.; Han, J.M. Glutamine reliance in cell metabolism. Exp. Mol. Med. 2020, 52, 1496–1516. [Google Scholar] [CrossRef] [PubMed]
- Thaker, S.K.; Ch’Ng, J.; Christofk, H.R. Viral hijacking of cellular metabolism. BMC Biol. 2019, 17, 59. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; King, M.S.; Yu, M.; Klipcan, L.; Leslie, A.G.; Hirst, J. Structure of subcomplex Ibeta of mammalian respiratory complex I leads to new supernumerary subunit assignments. Proc. Natl. Acad. Sci. USA 2015, 112, 12087–12092. [Google Scholar] [CrossRef] [PubMed]
- Ernster, L.; Dallner, G. Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 1995, 1271, 195–204. [Google Scholar] [CrossRef]
- Ndi, M.; Marin-Buera, L.; Salvatori, R.; Singh, A.P.; Ott, M. Biogenesis of the bc1 Complex of the Mitochondrial Respiratory Chain. J. Mol. Biol. 2018, 430, 3892–3905. [Google Scholar] [CrossRef]
- Turrens, J.F.; Alexandre, A.; Lehninger, A.L. Ubisemiquinone is the electron donor for superoxide formation by complex III of heart mitochondria. Arch. Biochem. Biophys. 1985, 237, 408–414. [Google Scholar] [CrossRef]
- Muller, F.L.; Liu, Y.; Van Remmen, H. Complex III Releases Superoxide to Both Sides of the Inner Mitochondrial Membrane. J. Biol. Chem. 2004, 279, 49064–49073. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, R.; Purhonen, J.; Kallijärvi, J. The mitochondrial coenzyme Q junction and complex III: Biochemistry and pathophysiology. FEBS J. 2021, 289, 6936–6958. [Google Scholar] [CrossRef] [PubMed]
- Arnold, S. The power of life—Cytochrome c oxidase takes center stage in metabolic control, cell signalling and survival. Mitochondrion 2012, 12, 46–56. [Google Scholar] [CrossRef]
- Napiwotzki, J.; Kadenbach, B. Extramitochondrial ATP/ADP-Ratios Regulate Cytochrome c Oxidase Activity via Binding to the Cytosolic Domain of Subunit IV. Biol. Chem. 1998, 379, 335–340. [Google Scholar] [CrossRef]
- Hüttemann, M.; Kadenbach, B.; Grossman, L.I. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 2001, 267, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Steenaart, N.A.; Shore, G.C. Mitochondrial cytochrome c oxidase subunit IV is phosphorylated by an endogenous kinase. FEBS Lett. 1997, 415, 294–298. [Google Scholar] [CrossRef]
- Clayton, S.A.; Daley, K.K.; MacDonald, L.; Fernandez-Vizarra, E.; Bottegoni, G.; O’Neil, J.D.; Major, T.; Griffin, D.; Zhuang, Q.; Adewoye, A.B.; et al. Inflammation causes remodeling of mitochondrial cytochrome c oxidase mediated by the bifunctional gene C15orf48. Sci. Adv. 2021, 7, eabl5182. [Google Scholar] [CrossRef] [PubMed]
- Angireddy, R.; Kazmi, H.R.; Srinivasan, S.; Sun, L.; Iqbal, J.; Fuchs, S.Y.; Guha, M.; Kijima, T.; Yuen, T.; Zaidi, M.; et al. Cytochrome c oxidase dysfunction enhances phagocytic function and osteoclast formation in macrophages. FASEB J. 2019, 33, 9167–9181. [Google Scholar] [CrossRef] [PubMed]
- Mookerjee, S.A.; Gerencser, A.A.; Nicholls, D.G.; Brand, M.D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 2017, 292, 7189–7207. [Google Scholar] [CrossRef]
- Caruana, N.J.; Stroud, D.A. The road to the structure of the mitochondrial respiratory chain supercomplex. Biochem. Soc. Trans. 2020, 48, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Blaza, J.N.; Larsson, N.-G.; Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metab. 2017, 25, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Lapuente-Brun, E.; Moreno-Loshuertos, R.; Acín-Pérez, R.; Latorre-Pellicer, A.; Colás, C.; Balsa, E.; Perales-Clemente, E.; Quirós, P.M.; Calvo, E.; Rodríguez-Hernández, M.A.; et al. Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain. Science 2013, 340, 1567–1570. [Google Scholar] [CrossRef] [PubMed]
- Milenkovic, D.; Misic, J.; Hevler, J.F.; Molinié, T.; Chung, I.; Atanassov, I.; Li, X.; Filograna, R.; Mesaros, A.; Mourier, A.; et al. Preserved respiratory chain capacity and physiology in mice with profoundly reduced levels of mitochondrial respirasomes. Cell Metab. 2023, 35, 1799–1813.e7. [Google Scholar] [CrossRef]
- Letts, J.A.; Sazanov, L.A. Clarifying the supercomplex: The higher-order organization of the mitochondrial electron transport chain. Nat. Struct. Mol. Biol. 2017, 24, 800–808. [Google Scholar] [CrossRef] [PubMed]
- Schagger, H.; Pfeiffer, K. The Ratio of Oxidative Phosphorylation Complexes I–V in Bovine Heart Mitochondria and the Composition of Respiratory Chain Supercomplexes. J. Biol. Chem. 2001, 276, 37861–37867. [Google Scholar] [CrossRef] [PubMed]
- Letts, J.A.; Fiedorczuk, K.; Sazanov, L.A. The architecture of respiratory supercomplexes. Nature 2016, 537, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012, 4, a011403. [Google Scholar] [CrossRef] [PubMed]
- Rathore, S.; Berndtsson, J.; Marin-Buera, L.; Conrad, J.; Carroni, M.; Brzezinski, P.; Ott, M. Cryo-EM structure of the yeast respiratory supercomplex. Nat. Struct. Mol. Biol. 2018, 26, 50–57. [Google Scholar] [CrossRef]
- Vercellino, I.; Sazanov, L.A. Structure and assembly of the mammalian mitochondrial supercomplex CIII2CIV. Nature 2021, 598, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vizarra, E.; Ugalde, C. Cooperative assembly of the mitochondrial respiratory chain. Trends Biochem. Sci. 2022, 47, 999–1008. [Google Scholar] [CrossRef]
- Acín-Pérez, R.; Fernández-Silva, P.; Peleato, M.L.; Pérez-Martos, A.; Enriquez, J.A. Respiratory Active Mitochondrial Supercomplexes. Mol. Cell 2008, 32, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Muhleip, A.; Flygaard, R.K.; Baradaran, R.; Haapanen, O.; Gruhl, T.; Tobiasson, V.; Maréchal, A.; Sharma, V.; Amunts, A. Structural basis of mitochondrial membrane bending by the I-II-III(2)-IV(2) supercomplex. Nature 2023, 615, 934–938. [Google Scholar] [CrossRef]
- Azuma, K.; Ikeda, K.; Inoue, S. Functional Mechanisms of Mitochondrial Respiratory Chain Supercomplex Assembly Factors and Their Involvement in Muscle Quality. Int. J. Mol. Sci. 2020, 21, 3182. [Google Scholar] [CrossRef]
- Lobo-Jarne, T.; Nývltová, E.; Pérez-Pérez, R.; Timón-Gómez, A.; Molinié, T.; Choi, A.; Mourier, A.; Fontanesi, F.; Ugalde, C.; Barrientos, A. Human COX7A2L Regulates Complex III Biogenesis and Promotes Supercomplex Organization Remodeling without Affecting Mitochondrial Bioenergetics. Cell Rep. 2018, 25, 1786–1799.e4. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Barrientos, A.; Fontanesi, F.; Ott, M. The functional significance of mitochondrial respiratory chain supercomplexes. Embo Rep. 2023, 24, e57092. [Google Scholar] [CrossRef]
- Garaude, J.; Acín-Pérez, R.; Martínez-Cano, S.; Enamorado, M.; Ugolini, M.; Nistal-Villán, E.; Hervás-Stubbs, S.; Pelegrín, P.; Sander, L.E.; Enríquez, J.A.; et al. Mitochondrial respiratory-chain adaptations in macrophages contribute to antibacterial host defense. Nat. Immunol. 2016, 17, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Hanada, Y.; Ishihara, N.; Wang, L.; Otera, H.; Ishihara, T.; Koshiba, T.; Mihara, K.; Ogawa, Y.; Nomura, M. MAVS is energized by Mff which senses mitochondrial metabolism via AMPK for acute antiviral immunity. Nat. Commun. 2020, 11, 5711. [Google Scholar] [CrossRef] [PubMed]
- Koshiba, T.; Yasukawa, K.; Yanagi, Y.; Kawabata, S.-I. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci. Signal. 2011, 4, ra7. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-J.; Ahn, D.-G.; Syed, G.H.; Siddiqui, A. The essential role of mitochondrial dynamics in antiviral immunity. Mitochondrion 2017, 41, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, K.; Zeng, S.; Zou, L.; Li, X.; Xu, C.; Li, B.; Liu, X.; Li, Z.; Zhu, W.; et al. The Role of Mitophagy in Viral Infection. Cells 2022, 11, 711. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Krupovic, M.; Agol, V.I. The Baltimore Classification of Viruses 50 Years Later: How Does It Stand in the Light of Virus Evolution? Microbiol. Mol. Biol. Rev. 2021, 85, e0005321. [Google Scholar] [CrossRef]
- Grimbert, S.; Johanet, C.; Bendjaballah, F.; Homberg, J.; Poupon, R.; Beaugrand, M. Antimitochondrial antibodies in patients with chronic hepatitis C. Liver Int. 1996, 16, 161–165. [Google Scholar] [CrossRef]
- Barbaro, G.; Di Lorenzo, G.; Asti, A.; Ribersani, M.; Belloni, G.; Grisorio, B.; Filice, G.; Barbarini, G. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: Ultrastructural and biochemical findings. Am. J. Gastroenterol. 1999, 94, 2198–2205. [Google Scholar] [CrossRef]
- Korenaga, M.; Wang, T.; Li, Y.; Showalter, L.A.; Chan, T.; Sun, J.; Weinman, S.A. Hepatitis C Virus Core Protein Inhibits Mitochondrial Electron Transport and Increases Reactive Oxygen Species (ROS) Production. J. Biol. Chem. 2005, 280, 37481–37488. [Google Scholar] [CrossRef] [PubMed]
- Piccoli, C.; Scrima, R.; Quarato, G.; D’Aprile, A.; Ripoli, M.; Lecce, L.; Boffoli, D.; Moradpour, D.; Capitanio, N. Hepatitis C virus protein expression causes calcium-mediated mitochondrial bioenergetic dysfunction and nitro-oxidative stress. Hepatology 2007, 46, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Quarato, G.; Scrima, R.; Ripoli, M.; Agriesti, F.; Moradpour, D.; Capitanio, N.; Piccoli, C. Protective role of amantadine in mitochondrial dysfunction and oxidative stress mediated by hepatitis C virus protein expression. Biochem. Pharmacol. 2014, 89, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Cortelli, P.; Mandrioli, J.; Zeviani, M.; Lodi, R.; Prata, C.; Pecorari, M.; Orlando, G.; Guaraldi, G. Mitochondrial complex III deficiency in a case of HCV related noninflammatory myopathy. J. Neurol. 2007, 254, 1450–1452. [Google Scholar] [CrossRef] [PubMed]
- Jassey, A.; Liu, C.-H.; Changou, C.A.; Richardson, C.D.; Hsu, H.-Y.; Lin, L.-T. Hepatitis C Virus Non-Structural Protein 5A (NS5A) Disrupts Mitochondrial Dynamics and Induces Mitophagy. Cells 2019, 8, 290. [Google Scholar] [CrossRef]
- Gong, G.; Waris, G.; Tanveer, R.; Siddiqui, A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc. Natl. Acad. Sci. USA 2001, 98, 9599–9604. [Google Scholar] [CrossRef] [PubMed]
- Gerresheim, G.K.; Bathke, J.; Michel, A.M.; Andreev, D.E.; Shalamova, L.A.; Rossbach, O.; Hu, P.; Glebe, D.; Fricke, M.; Marz, M.; et al. Cellular Gene Expression during Hepatitis C Virus Replication as Revealed by Ribosome Profiling. Int. J. Mol. Sci. 2019, 20, 1321. [Google Scholar] [CrossRef] [PubMed]
- Ramage, H.R.; Kumar, G.R.; Verschueren, E.; Johnson, J.R.; Von Dollen, J.; Johnson, T.; Newton, B.; Shah, P.; Horner, J.; Krogan, N.J.; et al. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol. Cell 2015, 57, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Xiu, W.; Xu, J.; Chen, X.; Wang, G.; Duan, J.; Sun, L.; Liu, B.; Xie, W.; Pu, G.; et al. Increased CHCHD2 expression promotes liver fibrosis in nonalcoholic steatohepatitis via Notch/osteopontin signaling. J. Clin. Investig. 2022, 7, e162402. [Google Scholar] [CrossRef]
- Aras, S.; Bai, M.; Lee, I.; Springett, R.; Hüttemann, M.; Grossman, L.I. MNRR1 (formerly CHCHD2) is a bi-organellar regulator of mitochondrial metabolism. Mitochondrion 2015, 20, 43–51. [Google Scholar] [CrossRef]
- Aras, S.; Purandare, N.; Gladyck, S.; Somayajulu-Nitu, M.; Zhang, K.; Wallace, D.C.; Grossman, L.I. Mitochondrial Nuclear Retrograde Regulator 1 (MNRR1) rescues the cellular phenotype of MELAS by inducing homeostatic mechanisms. Proc. Natl. Acad. Sci. USA 2020, 117, 32056–32065. [Google Scholar] [CrossRef] [PubMed]
- Beckham, J.D.; Pastula, D.M.; Massey, A.; Tyler, K.L. Zika Virus as an Emerging Global Pathogen: Neurological Complications of Zika Virus. JAMA Neurol. 2016, 73, 875–879. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, J.; Rabeneck, D.B.; Martines, R.B.; Reagan-Steiner, S.; Ermias, Y.; Estetter, L.B.; Suzuki, T.; Ritter, J.; Keating, M.K.; Al, J.B.E.; et al. Zika Virus RNA Replication and Persistence in Brain and Placental Tissue. Emerg. Infect. Dis. 2017, 23, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Shin, O.S. Zika virus modulates mitochondrial dynamics, mitophagy, and mitochondria-derived vesicles to facilitate viral replication in trophoblast cells. Front. Immunol. 2023, 14, 1203645. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, J.; Yang, Y.; Qu, S.; Wan, F.; Zhang, Z.; Wang, R.; Li, G.; Cong, H. Zika virus infection induced apoptosis by modulating the recruitment and activation of pro-apoptotic protein Bax. J. Virol. 2021, 95, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Xin, Q.L.; Deng, C.L.; Chen, X.; Wang, J.; Wang, S.B.; Wang, W.; Deng, F.; Zhang, B.; Xiao, G.; Zhang, L.K. Quantitative Proteomic Analysis of Mosquito C6/36 Cells Reveals Host Proteins Involved in Zika Virus Infection. J. Virol. 2017, 91, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Daniels, B.P.; Kofman, S.B.; Smith, J.R.; Norris, G.T.; Snyder, A.G.; Kolb, J.P.; Gao, X.; Locasale, J.W.; Martinez, J.; Gale, M.; et al. The Nucleotide Sensor ZBP1 and Kinase RIPK3 Induce the Enzyme IRG1 to Promote an Antiviral Metabolic State in Neurons. Immunity 2019, 50, 64–76.e4. [Google Scholar] [CrossRef] [PubMed]
- Yau, C.; Low, J.Z.; Gan, E.S.; Kwek, S.S.; Cui, L.; Tan, H.C.; Mok, D.Z.; Chan, C.Y.; Sessions, O.M.; Watanabe, S.; et al. Dysregulated metabolism underpins Zika-virus-infection-associated impairment in fetal development. Cell Rep. 2021, 37, 110118. [Google Scholar] [CrossRef] [PubMed]
- Airo, A.M.; Felix-Lopez, A.; Mancinelli, V.; Evseev, D.; Lopez-Orozco, J.; Shire, K.; Paszkowski, P.; Frappier, L.; Magor, K.E.; Hobman, T.C. Flavivirus Capsid Proteins Inhibit the Interferon Response. Viruses 2022, 14, 968. [Google Scholar] [CrossRef]
- He, J.; Ford, H.C.; Carroll, J.; Douglas, C.; Gonzales, E.; Ding, S.; Fearnley, I.M.; Walker, J.E. Assembly of the membrane domain of ATP synthase in human mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 2988–2993. [Google Scholar] [CrossRef]
- Lim, P.-Y.; Behr, M.J.; Chadwick, C.M.; Shi, P.-Y.; Bernard, K.A. Keratinocytes Are Cell Targets of West Nile Virus In Vivo. J. Virol. 2011, 85, 5197–5201. [Google Scholar] [CrossRef]
- Sampson, B.A.; Ambrosi, C.; Charlot, A.; Reiber, K.; Veress, J.F.; Armbrustmacher, V. The pathology of human West Nile virus infection. Hum. Pathol. 2000, 31, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Koh, W.-L.; Ng, M.-L. Molecular Mechanisms of West Nile Virus Pathogenesis in Brain Cells. Emerg. Infect. Dis. 2005, 11, 629–632. [Google Scholar] [CrossRef] [PubMed]
- Mingo-Casas, P.; Blázquez, A.-B.; de Cedrón, M.G.; San-Félix, A.; Molina, S.; Escribano-Romero, E.; Calvo-Pinilla, E.; de Oya, N.J.; de Molina, A.R.; Saiz, J.-C.; et al. Glycolytic shift during West Nile virus infection provides new therapeutic opportunities. J. Neuroinflamm. 2023, 20, 217. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.-H.; Wang, T. West Nile Virus Induced Cell Death in the Central Nervous System. Pathogens 2019, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Petersen, L.R.; Brault, A.C.; Nasci, R.S. West Nile Virus: Review of the Literature. JAMA 2013, 310, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.; Silverstein, A.; Flores, M.; Cao, K.; Kumagai, H.; Mehta, H.H.; Yen, K.; Kim, S.J.; Cohen, P. Host mitochondrial transcriptome response to SARS-CoV-2 in multiple cell models and clinical samples. Sci. Rep. 2021, 11, 3. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Soto Albrecht, Y.; Murdock, D.G.; Angelin, A.; Singh, L.N.; et al. Core mitochondrial genes are down-regulated during SARS-CoV-2 infection of rodent and human hosts. Sci. Transl. Med. 2023, 15, eabq1533. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.L.; Dasgupta, A.; Chen, K.H.; Wu, D.; Baid, K.; Mamatis, J.E.; Gonzalez, V.; Read, A.; Bentley, R.E.; Martin, A.Y.; et al. SARS-CoV-2 mitochondriopathy in COVID-19 pneumonia exacerbates hypoxemia. Redox Biol. 2022, 58, 102508. [Google Scholar] [CrossRef] [PubMed]
- Codo, A.C.; Davanzo, G.G.; de Brito Monteiro, L.; de Souza, G.F.; Muraro, S.P.; Virgilio-da-Silva, J.V.; Prodonoff, J.S.; Carregari, V.C.; de Biagi Junior, C.A.O.; Crunfli, F.; et al. Elevated Glucose Levels Favor SARS-CoV-2 Infection and Monocyte Response through a HIF-1alpha/Glycolysis-Dependent Axis. Cell Metab. 2020, 32, 437–446.e5. [Google Scholar] [CrossRef]
- Ajaz, S.; McPhail, M.J.; Singh, K.K.; Mujib, S.; Trovato, F.M.; Napoli, S.; Agarwal, K. Mitochondrial metabolic manipulation by SARS-CoV-2 in peripheral blood mononuclear cells of patients with COVID-19. Am. J. Physiol. Cell Physiol. 2021, 320, C57–C65. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, A.; Oldani, M.; Forcella, M.E.; Vantaggiato, C.; Cappelletti, G.; Pontremoli, C.; Valenti, F.; Forni, D.; Saresella, M.; Biasin, M.; et al. SARS-CoV-2 ORF3c impairs mitochondrial respiratory metabolism, oxidative stress, and autophagic flux. iScience 2023, 26, 107118. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, L.; Dong, C.; Che, Y.; Jiang, L.; Liu, L.; Zhao, H.; Liao, Y.; Sheng, Y.; Dong, S.; et al. The interaction of the SARS coronavirus non-structural protein 10 with the cellular oxido-reductase system causes an extensive cytopathic effect. J. Clin. Virol. 2005, 34, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Kakavandi, S.; Zare, I.; VaezJalali, M.; Dadashi, M.; Azarian, M.; Akbari, A.; Ramezani Farani, M.; Zalpoor, H.; Hajikhani, B. Structural and non-structural proteins in SARS-CoV-2: Potential aspects to COVID-19 treatment or prevention of progression of related diseases. Cell Commun. Signal. 2023, 21, 110. [Google Scholar] [CrossRef] [PubMed]
- Claus, C.; Schönefeld, K.; Hübner, D.; Chey, S.; Reibetanz, U.; Liebert, U.G. Activity Increase in Respiratory Chain Complexes by Rubella Virus with Marginal Induction of Oxidative Stress. J. Virol. 2013, 87, 8481–8492. [Google Scholar] [CrossRef] [PubMed]
- Bilz, N.C.; Jahn, K.; Lorenz, M.; Lüdtke, A.; Hübschen, J.M.; Geyer, H.; Mankertz, A.; Hübner, D.; Liebert, U.G.; Claus, C. Rubella Viruses Shift Cellular Bioenergetics to a More Oxidative and Glycolytic Phenotype with a Strain-Specific Requirement for Glutamine. J. Virol. 2018, 92, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Bowden, D.S.; Marshall, J.A. Membrane junctions associated with rubella virus infected cells. J. Submicrosc. Cytol. Pathol. 1996, 28, 101–108. [Google Scholar] [PubMed]
- Beatch, M.D.; Hobman, T.C. Rubella Virus Capsid Associates with Host Cell Protein p32 and Localizes to Mitochondria. J. Virol. 2000, 74, 5569–5576. [Google Scholar] [CrossRef] [PubMed]
- Ebermann, L.; Wika, S.; Klumpe, I.; Hammer, E.; Klingel, K.; Lassner, D.; Völker, U.; Erben, U.; Zeichhardt, H.; Schultheiss, H.-P.; et al. The mitochondrial respiratory chain has a critical role in the antiviral process in Coxsackievirus B3-induced myocarditis. Lab. Investig. 2012, 92, 125–134. [Google Scholar] [CrossRef]
- Qu, C.; Zhang, S.; Wang, W.; Li, M.; Wang, Y.; Heijde-Mulder, M.; Shokrollahi, E.; Hakim, M.S.; Raat, N.J.H.; Peppelenbosch, M.P.; et al. Mitochondrial electron transport chain complex III sustains hepatitis E virus replication and represents an antiviral target. FASEB J. 2018, 33, 1008–1019. [Google Scholar] [CrossRef]
- Tian, J.; Shi, R.; Xiao, P.; Liu, T.; She, R.; Wu, Q.; An, J.; Hao, W.; Soomro, M. Hepatitis E Virus Induces Brain Injury Probably Associated With Mitochondrial Apoptosis. Front. Cell. Infect. Microbiol. 2019, 9, 433. [Google Scholar] [CrossRef] [PubMed]
- Kuiken, T.; Taubenberger, J.K. Pathology of human influenza revisited. Vaccine 2008, 26, D59–D66. [Google Scholar] [CrossRef] [PubMed]
- Derakhshan, M.; Willcocks, M.M.; Salako, M.A.; Kass, G.E.N.; Carter, M.J. Human herpesvirus 1 protein US3 induces an inhibition of mitochondrial electron transport. J. Gen. Virol. 2006, 87, 2155–2159. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wang, H.; Bai, L.; Yu, Y.; Sun, Z.; Yan, Y.; Zhou, J. Mitochondrial proteomic analysis of human host cells infected with H3N2 swine influenza virus. J. Proteom. 2013, 91, 136–150. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Huang, H.; Li, B.; Li, H.; Zhao, Y.; Li, Y.; Ye, W.; Qi, W.; Tang, W.; Wang, L. Identification of cytochrome c oxidase subunit 4 isoform 1 as a positive regulator of influenza virus replication. Front. Microbiol. 2022, 13, 862205. [Google Scholar] [CrossRef] [PubMed]
- Othumpangat, S.; Noti, J.D.; Beezhold, D.H. Lung epithelial cells resist influenza A infection by inducing the expression of cytochrome c oxidase VIc which is modulated by miRNA 4276. Virology 2014, 468–470, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Oleynikov, I.P.; Sudakov, R.V.; Radyukhin, V.A.; Arutyunyan, A.M.; Azarkina, N.V.; Vygodina, T.V. Interaction of Amphipathic Peptide from Influenza Virus M1 Protein with Mitochondrial Cytochrome Oxidase. Int. J. Mol. Sci. 2023, 24, 4119. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Solaymani-Mohammadi, F.; Namdari, H.; Arjeini, Y.; Mousavi, M.J.; Rezaei, F. Metabolic host response and therapeutic approaches to influenza infection. Cell. Mol. Biol. Lett. 2020, 25, 15. [Google Scholar] [CrossRef] [PubMed]
- Alandijany, T.; Kammouni, W.; Chowdhury, S.K.R.; Fernyhough, P.; Jackson, A.C. Mitochondrial dysfunction in rabies virus infection of neurons. J. Neuro Virol. 2013, 19, 537–549. [Google Scholar] [CrossRef]
- Kammouni, W.; Wood, H.; Saleh, A.; Appolinario, C.M.; Fernyhough, P.; Jackson, A.C. Rabies virus phosphoprotein interacts with mitochondrial Complex I and induces mitochondrial dysfunction and oxidative stress. J. Neuro Virol. 2015, 21, 370–382. [Google Scholar] [CrossRef]
- Harsha, P.K.; Ranganayaki, S.; Yale, G.; Dey, G.; Mangalaparthi, K.K.; Yarlagadda, A.; Sagar, B.K.C.; Mahadevan, A.; Bharath, M.M.S.; Mani, R.S. Mitochondrial Dysfunction in Rabies Virus-Infected Human and Canine Brains. Neurochem. Res. 2022, 47, 1610–1636. [Google Scholar] [CrossRef]
- Hu, M.; Schulze, K.E.; Ghildyal, R.; Henstridge, D.C.; Kolanowski, J.L.; New, E.J.; Hong, Y.; Hsu, A.C.; Hansbro, P.M.; Wark, P.A.; et al. Respiratory syncytial virus co-opts host mitochondrial function to favour infectious virus production. Elife 2019, 8, e42448. [Google Scholar] [CrossRef]
- Hu, M.; Bogoyevitch, M.A.; Jans, D.A. Respiratory Syncytial Virus Matrix Protein Is Sufficient and Necessary to Remodel Host Mitochondria in Infection. Cells 2023, 12, 1311. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Hoshi, M.; Ikeda, F.; Fujiyuki, T.; Yoneda, M.; Kai, C. Downregulation of mitochondrial biogenesis by virus infection triggers antiviral responses by cyclic GMP-AMP synthase. PLoS Pathog. 2021, 17, e1009841. [Google Scholar] [CrossRef] [PubMed]
- Somasundaran, M.; Zapp, M.L.; Beattie, L.K.; Pang, L.; Byron, K.S.; Bassell, G.J.; Sullivan, J.L.; Singer, R.H. Localization of HIV RNA in mitochondria of infected cells: Potential role in cytopathogenicity. J. Cell Biol. 1994, 126, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Macreadie, I.G.; Thorburn, D.R.; Kirby, D.M.; Castelli, L.A.; de Rozario, N.L.; Azad, A.A. HIV-1 protein Vpr causes gross mitochondrial dysfunction in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997, 410, 145–149. [Google Scholar] [CrossRef]
- Tripathy, M.K.; Mitra, D. Differential modulation of mitochondrial OXPHOS system during HIV-1 induced T-cell apoptosis: Up regulation of Complex-IV subunit COX-II and its possible implications. Apoptosis 2010, 15, 28–40. [Google Scholar] [CrossRef] [PubMed]
- Ladha, J.S.; Tripathy, M.K.; Mitra, D. Mitochondrial complex I activity is impaired during HIV-1-induced T-cell apoptosis. Cell Death Differ. 2005, 12, 1417–1428. [Google Scholar] [CrossRef]
- Miró, O.; López, S.; Martínez, E.; Pedrol, E.; Milinkovic, A.; Deig, E.; Garrabou, G.; Casademont, J.; Gatell, J.M.; Cardellach, F. Mitochondrial Effects of HIV Infection on the Peripheral Blood Mononuclear Cells of HIV-Infected Patients Who Were Never Treated with Antiretrovirals. Clin. Infect. Dis. 2004, 39, 710–716. [Google Scholar] [CrossRef]
- Kaur, H.; Minchella, P.; Alvarez-Carbonell, D.; Purandare, N.; Nagampalli, V.K.; Blankenberg, D.; Hulgan, T.; Gerschenson, M.; Karn, J.; Aras, S.; et al. Contemporary Antiretroviral Therapy Dysregulates Iron Transport and Augments Mitochondrial Dysfunction in HIV-Infected Human Microglia and Neural-Lineage Cells. Int. J. Mol. Sci. 2023, 24, 12242. [Google Scholar] [CrossRef]
- Ogawa, M.; Takemoto, Y.; Sumi, S.; Inoue, D.; Kishimoto, N.; Takamune, N.; Shoji, S.; Suzu, S.; Misumi, S. ATP generation in a host cell in early-phase infection is increased by upregulation of cytochrome c oxidase activity via the p2 peptide from human immunodeficiency virus type 1 Gag. Retrovirology 2015, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Lecoeur, H.; Borgne-Sanchez, A.; Chaloin, O.; El-Khoury, R.; Brabant, M.; Langonné, A.; Porceddu, M.; Brière, J.-J.; Buron, N.; Rebouillat, D.; et al. HIV-1 Tat protein directly induces mitochondrial membrane permeabilization and inactivates cytochrome c oxidase. Cell Death Dis. 2012, 3, e282. [Google Scholar] [CrossRef] [PubMed]
- Yavlovich, A.; Viard, M.; Zhou, M.; Veenstra, T.D.; Wang, J.M.; Gong, W.; Heldman, E.; Blumenthal, R.; Raviv, Y. Ectopic ATP synthase facilitates transfer of HIV-1 from antigen-presenting cells to CD4+ target cells. Blood 2012, 120, 1246–1253. [Google Scholar] [CrossRef] [PubMed]
- Castellano, P.; Prevedel, L.; Valdebenito, S.; Eugenin, E.A. HIV infection and latency induce a unique metabolic signature in human macrophages. Sci. Rep. 2019, 9, 3941. [Google Scholar] [CrossRef] [PubMed]
- Schank, M.; Zhao, J.; Moorman, J.P.; Yao, Z.Q. The Impact of HIV- and ART-Induced Mitochondrial Dysfunction in Cellular Senescence and Aging. Cells 2021, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Shirakata, Y.; Kaneniwa, N.; Koike, K. Association of hepatitis B virus X protein with mitochondria causes mitochondrial aggregation at the nuclear periphery, leading to cell death. Oncogene 1999, 18, 6965–6973. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, Z.; Huh, K.-W.; Lasher, R.; Siddiqui, A. Hepatitis B Virus X Protein Colocalizes to Mitochondria with a Human Voltage-Dependent Anion Channel, HVDAC3, and Alters Its Transmembrane Potential. J. Virol. 2000, 74, 2840–2846. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.Z.; Yu, J.P.; Chen, Z.X.; Huang, Y.H.; Tao, Q.M. Cytochrome C oxidase III interacts with hepatitis B virus X protein in vivo by yeast two-hybrid system. World J. Gastroenterol. 2004, 10, 2805–2808. [Google Scholar] [CrossRef]
- Zou, L.-Y.; Zheng, B.-Y.; Fang, X.-F.; Li, D.; Huang, Y.-H.; Chen, Z.-X.; Zhou, L.-Y.; Wang, X.-Z. HBx co-localizes with COXIII in HL-7702 cells to upregulate mitochondrial function and ROS generation. Oncol. Rep. 2015, 33, 2461–2467. [Google Scholar] [CrossRef]
- Lee, Y.I.; Hwang, J.M.; Im, J.H.; Lee, Y.I.; Kim, N.S.; Kim, D.G.; Yu, D.Y.; Moon, H.B.; Park, S.K. Human Hepatitis B Virus-X Protein Alters Mitochondrial Function and Physiology in Human Liver Cells. J. Biol. Chem. 2004, 279, 15460–15471. [Google Scholar] [CrossRef]
- Honkoop, P.; De Man, R.A.; Scholte, H.R.; Zondervan, P.E.; Van Den Berg, J.W.; Rademakers, L.H.; Schalm, S.W. Effect of lamivudine on morphology and function of mitochondria in patients with chronic hepatitis B. Hepatology 1997, 26, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, Y.; Feng, S.; Ishida, Y.; Chiu, T.P.; Saito, T.; Wang, S.; Ann, D.K.; Ou, J.H.J. Macrophages activated by hepatitis B virus have distinct metabolic profiles and suppress the virus via IL-1beta to downregulate PPARalpha and FOXO3. Cell Rep. 2022, 38, 110284. [Google Scholar] [CrossRef] [PubMed]
- Giosa, D.; Lombardo, D.; Musolino, C.; Chines, V.; Raffa, G.; di Tocco, F.C.; D’aliberti, D.; Caminiti, G.; Saitta, C.; Alibrandi, A.; et al. Mitochondrial DNA is a target of HBV integration. Commun. Biol. 2023, 6, 684. [Google Scholar] [CrossRef] [PubMed]
- Fisicaro, P.; Barili, V.; Montanini, B.; Acerbi, G.; Ferracin, M.; Guerrieri, F.; Salerno, D.; Boni, C.; Massari, M.; Cavallo, M.C.; et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 2017, 23, 327–336. [Google Scholar] [CrossRef]
- Britt, W.J. Maternal Immunity and the Natural History of Congenital Human Cytomegalovirus Infection. Viruses 2018, 10, 405. [Google Scholar] [CrossRef] [PubMed]
- Combs, J.A.; Norton, E.B.; Saifudeen, Z.R.; Bentrup, K.H.Z.; Katakam, P.V.; Morris, C.A.; Myers, L.; Kaur, A.; Sullivan, D.E.; Zwezdaryk, K.J. Human Cytomegalovirus Alters Host Cell Mitochondrial Function during Acute Infection. J. Virol. 2020, 94, 10-1128. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.; Bajad, S.U.; Coller, H.A.; Shenk, T.; Rabinowitz, J.D. Dynamics of the Cellular Metabolome during Human Cytomegalovirus Infection. PLoS Pathog. 2006, 2, e132. [Google Scholar] [CrossRef]
- Betsinger, C.N.; Jankowski, C.S.; Hofstadter, W.A.; Federspiel, J.D.; Otter, C.J.; Beltran, P.M.J.; Cristea, I.M. The human cytomegalovirus protein pUL13 targets mitochondrial cristae architecture to increase cellular respiration during infection. Proc. Natl. Acad. Sci. USA 2021, 118, e2101675118. [Google Scholar] [CrossRef]
- Kaarbø, M.; Ager-Wick, E.; Osenbroch, P.; Kilander, A.; Skinnes, R.; Müller, F.; Eide, L. Human cytomegalovirus infection increases mitochondrial biogenesis. Mitochondrion 2011, 11, 935–945. [Google Scholar] [CrossRef]
- Karniely, S.; Weekes, M.P.; Antrobus, R.; Rorbach, J.; van Haute, L.; Umrania, Y.; Smith, D.L.; Stanton, R.J.; Minczuk, M.; Lehner, P.J.; et al. Human Cytomegalovirus Infection Upregulates the Mitochondrial Transcription and Translation Machineries. mBio 2016, 7, e00029. [Google Scholar] [CrossRef]
- Huang, G.; Lu, H.; Hao, A.; Ng, D.C.H.; Ponniah, S.; Guo, K.; Lufei, C.; Zeng, Q.; Cao, X. GRIM-19, a Cell Death Regulatory Protein, Is Essential for Assembly and Function of Mitochondrial Complex, I. Mol. Cell. Biol. 2004, 24, 8447–8456. [Google Scholar] [CrossRef] [PubMed]
- Reeves, M.B.; Davies, A.A.; McSharry, B.P.; Wilkinson, G.W.; Sinclair, J.H. Complex I Binding by a Virally Encoded RNA Regulates Mitochondria-Induced Cell Death. Science 2007, 316, 1345–1348. [Google Scholar] [CrossRef]
- Damania, B.; Kenney, S.C.; Raab-Traub, N. Epstein-Barr virus: Biology and clinical disease. Cell 2022, 185, 3652–3670. [Google Scholar] [CrossRef] [PubMed]
- Mrozek-Gorska, P.; Buschle, A.; Pich, D.; Schwarzmayr, T.; Fechtner, R.; Scialdone, A.; Hammerschmidt, W. Epstein–Barr virus reprograms human B lymphocytes immediately in the prelatent phase of infection. Proc. Natl. Acad. Sci. USA 2019, 116, 16046–16055. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.W.; Shen, H.; Nobre, L.; Ersing, I.; Paulo, J.A.; Trudeau, S.; Wang, Z.; Smith, N.A.; Ma, Y.; Reinstadler, B.; et al. Epstein-Barr-Virus-Induced One-Carbon Metabolism Drives B Cell Transformation. Cell Metab. 2019, 30, 539–555.e11. [Google Scholar] [CrossRef] [PubMed]
- Schober, F.A.; Moore, D.; Atanassov, I.; Moedas, M.F.; Clemente, P.; Végvári, A.; El Fissi, N.; Filograna, R.; Bucher, A.-L.; Hinze, Y.; et al. The one-carbon pool controls mitochondrial energy metabolism via complex I and iron-sulfur clusters. Sci. Adv. 2021, 7, eabf0717. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Maklashina, E.; Cecchini, G.; Iverson, T.M. The roles of SDHAF2 and dicarboxylate in covalent flavinylation of SDHA, the human complex II flavoprotein. Proc. Natl. Acad. Sci. USA 2020, 117, 23548–23556. [Google Scholar] [CrossRef]
- Van Vranken, J.G.; Bricker, D.K.; Dephoure, N.; Gygi, S.P.; Cox, J.E.; Thummel, C.S.; Rutter, J. SDHAF4 Promotes Mitochondrial Succinate Dehydrogenase Activity and Prevents Neurodegeneration. Cell Metab. 2014, 20, 241–252. [Google Scholar] [CrossRef]
- Na, U.; Yu, W.; Cox, J.; Bricker, D.K.; Brockmann, K.; Rutter, J.; Thummel, C.S.; Winge, D.R. The LYR Factors SDHAF1 and SDHAF3 Mediate Maturation of the Iron-Sulfur Subunit of Succinate Dehydrogenase. Cell Metab. 2014, 20, 253–266. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, K.; Thorne, R.F.; Shi, R.; Zhang, Q.; Wu, M.; Liu, L. Mitochondrial SENP2 regulates the assembly of SDH complex under metabolic stress. Cell Rep. 2023, 42, 112041. [Google Scholar] [CrossRef]
- Selby, T.L.; Biel, N.; Varn, M.; Patel, S.; Patel, A.; Hilding, L.; Ray, A.; Ross, T.; Cramblet, W.T.; Moss, C.R.; et al. The Epstein-Barr Virus Oncoprotein, LMP1, Regulates the Function of SENP2, a SUMO-protease. Sci. Rep. 2019, 9, 9523. [Google Scholar] [CrossRef]
- McBride, A.A. The papillomavirus E2 proteins. Virology 2013, 445, 57–79. [Google Scholar] [CrossRef] [PubMed]
- Lai, D.; Tan, C.L.; Gunaratne, J.; Quek, L.S.; Nei, W.; Thierry, F.; Bellanger, S. Localization of HPV-18 E2 at Mitochondrial Membranes Induces ROS Release and Modulates Host Cell Metabolism. PLoS ONE 2013, 8, e75625. [Google Scholar] [CrossRef] [PubMed]
- Kirschberg, M.; Heuser, S.; Marcuzzi, G.P.; Hufbauer, M.; Seeger, J.M.; Đukić, A.; Tomaić, V.; Majewski, S.; Wagner, S.; Wittekindt, C.; et al. ATP synthase modulation leads to an increase of spare respiratory capacity in HPV associated cancers. Sci. Rep. 2020, 10, 17339. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.-J.; Gu, P.-Q.; Zhao, W.; Ding, W.-Y.; Zhao, X.-Q.; Guo, S.-Y.; Zhong, T.-Y. The role of globular heads of the C1q receptor in HPV 16 E2-induced human cervical squamous carcinoma cell apoptosis is associated with p38 MAPK/JNK activation. J. Transl. Med. 2013, 11, 118. [Google Scholar] [CrossRef]
- Kanki, T.; Ohgaki, K.; Gaspari, M.; Gustafsson, C.M.; Fukuoh, A.; Sasaki, N.; Hamasaki, N.; Kang, D. Architectural Role of Mitochondrial Transcription Factor A in Maintenance of Human Mitochondrial DNA. Mol. Cell. Biol. 2004, 24, 9823–9834. [Google Scholar] [CrossRef] [PubMed]
- Boczonadi, V.; Ricci, G.; Horvath, R. Mitochondrial DNA transcription and translation: Clinical syndromes. Essays Biochem. 2018, 62, 321–340. [Google Scholar] [CrossRef] [PubMed]
- Guarnieri, J.W.; Dybas, J.M.; Fazelinia, H.; Kim, M.S.; Frere, J.; Zhang, Y.; Albrecht, Y.S.; Murdock, D.G.; Angelin, A.; Singh, L.N.; et al. Targeted down Regulation of Core Mitochondrial Genes during SARS-CoV-2 Infection. bioRxiv 2022. Available online: https://www.biorxiv.org/content/10.1101/2022.02.19.481089v1 (accessed on 1 October 2023).
- Too, I.H.K.; Bonne, I.; Tan, E.L.; Chu, J.J.H.; Alonso, S. Prohibitin plays a critical role in Enterovirus 71 neuropathogenesis. PLoS Pathog. 2018, 14, e1006778. [Google Scholar] [CrossRef]
- Lee, I.-K.; Lee, S.-A.; Kim, H.; Won, Y.-S.; Kim, B.-J. Induction of endoplasmic reticulum-derived oxidative stress by an occult infection related S surface antigen variant. World J. Gastroenterol. 2015, 21, 6872–6883. [Google Scholar] [CrossRef]
- Krylatov, A.V.; Maslov, L.N.; Voronkov, N.S.; Boshchenko, A.A.; Popov, S.V.; Gomez, L.; Wang, H.; Jaggi, A.S.; Downey, J.M. Reactive Oxygen Species as Intracellular Signaling Molecules in the Cardiovascular System. Curr. Cardiol. Rev. 2018, 14, 290–300. [Google Scholar] [CrossRef] [PubMed]
- Thannickal, V.J.; Fanburg, B.L.; Yasuoka, H.; Garrett, S.M.; Nguyen, X.-X.; Artlett, C.M.; Feghali-Bostwick, C.A.; Zimmer, A.; Bagchi, A.K.; Vinayak, K.; et al. Reactive oxygen species in cell signaling. Am. J. Physiol. Cell. Mol. Physiol. 2000, 279, L1005–L1028. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Chio, C.C.; Chi, K.H.; Wu, H.M.; Lin, W.W. Superoxide anion-dependent Raf/MEK/ERK activation by peroxisome proliferator activated receptor gamma agonists 15-deoxy-delta(12,14)-prostaglandin J(2), ciglitazone, and GW1929. Exp. Cell Res. 2002, 277, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Ghasemnejad-Berenji, M.; Pashapour, S. SARS-CoV-2 and the Possible Role of Raf/MEK/ERK Pathway in Viral Survival: Is This a Potential Therapeutic Strategy for COVID-19? Pharmacology 2021, 106, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Deng, J.; Lv, L.; Kang, Q.; Ma, P.; Yan, F.; Song, X.; Gao, B.; Zhang, Y.; Xu, J. Hydrogen Peroxide Induce Human Cytomegalovirus Replication through the Activation of p38-MAPK Signaling Pathway. Viruses 2015, 7, 2816–2833. [Google Scholar] [CrossRef] [PubMed]
- Luong, T.T.D.; Tran, G.V.Q.; Shin, D.-J.; Lim, Y.-S.; Hwang, S.B. Hepatitis C Virus Exploits Death Receptor 6-mediated Signaling Pathway to Facilitate Viral Propagation. Sci. Rep. 2017, 7, 6445. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.-N.; Yang, W.K.; Kim, J.; Kim, H.S.; Kim, E.J.; Yun, H.; Park, H.; Kim, S.S.; Choe, W.; Kang, I.; et al. Reactive oxygen species stabilize hypoxia-inducible factor-1 alpha protein and stimulate transcriptional activity via AMP-activated protein kinase in DU145 human prostate cancer cells. Carcinogenesis 2008, 29, 713–721. [Google Scholar] [CrossRef]
- Willson, J.A.; Arienti, S.; Sadiku, P.; Reyes, L.; Coelho, P.; Morrison, T.; Rinaldi, G.; Dockrell, D.H.; Whyte, M.K.B.; Walmsley, S.R. Neutrophil HIF-1α stabilization is augmented by mitochondrial ROS produced via the glycerol 3-phosphate shuttle. Blood 2022, 139, 281–286. [Google Scholar] [CrossRef]
- Duette, G.; Gerber, P.P.; Rubione, J.; Perez, P.S.; Landay, A.L.; Crowe, S.M.; Liao, Z.; Witwer, K.W.; Holgado, M.P.; Salido, J.; et al. Induction of HIF-1α by HIV-1 Infection in CD4 + T Cells Promotes Viral Replication and Drives Extracellular Vesicle-Mediated Inflammation. mBio 2018, 9, e00757-18. [Google Scholar] [CrossRef]
- Lan, X.; Cheng, K.; Chandel, N.; Lederman, R.; Jhaveri, A.; Husain, M.; Malhotra, A.; Singhal, P.C. High glucose enhances HIV entry into T cells through upregulation of CXCR4. J. Leukoc. Biol. 2013, 94, 769–777. [Google Scholar] [CrossRef]
- Gullberg, R.C.; Steel, J.J.; Moon, S.; Soltani, E.; Geiss, B.J. Oxidative stress influences positive strand RNA virus genome synthesis and capping. Virology 2015, 475, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Dong, J.; Liu, Z.; Rao, Y.; Feng, P.; Lan, K. Viperin catalyzes methionine oxidation to promote protein expression and function of helicases. Sci. Adv. 2019, 5, eaax1031. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.-H.; Kim, K.-H.; Cho, Y.-S.; Choi, H.-S.; Song, E.Y.; Myung, P.-K.; Kang, J.S.; Suh, S.-K.; Park, S.N.; Yoon, D.-Y. Protective effect of oxidative stress in HaCaT keratinocytes expressing E7 oncogene. Amino Acids 2007, 34, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, S.; Krause, J.; Himmelsbach, K.; Carvajal-Yepes, M.; Lieder, F.; Klingel, K.; Nassal, M.; Weiss, T.S.; Werner, S.; Hildt, E. Hepatitis B Virus Induces Expression of Antioxidant Response Element-regulated Genes by Activation of Nrf2. J. Biol. Chem. 2010, 285, 41074–41086. [Google Scholar] [CrossRef] [PubMed]
- Brault, C.; Lévy, P.; Duponchel, S.; Michelet, M.; Sallé, A.; Pécheur, E.-I.; Plissonnier, M.-L.; Parent, R.; Véricel, E.; Ivanov, A.V.; et al. Glutathione peroxidase 4 is reversibly induced by HCV to control lipid peroxidation and to increase virion infectivity. Gut 2014, 65, 144–154. [Google Scholar] [CrossRef]
- Speir, E.; Shibutani, T.; Yu, Z.-X.; Ferrans, V.; Epstein, S.E. Role of Reactive Oxygen Intermediates in Cytomegalovirus Gene Expression and in the Response of Human Smooth Muscle Cells to Viral Infection. Circ. Res. 1996, 79, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Tilton, C.; Clippinger, A.J.; Maguire, T.; Alwine, J.C. Human Cytomegalovirus Induces Multiple Means To Combat Reactive Oxygen Species. J. Virol. 2011, 85, 12585–12593. [Google Scholar] [CrossRef]
- Chen, K.-K.; Minakuchi, M.; Wuputra, K.; Ku, C.-C.; Pan, J.-B.; Kuo, K.-K.; Lin, Y.-C.; Saito, S.; Lin, C.-S.; Yokoyama, K.K. Redox control in the pathophysiology of influenza virus infection. BMC Microbiol. 2020, 20, 214. [Google Scholar] [CrossRef]
- Anderton, E.; Yee, J.; Smith, P.; Crook, T.; White, R.E.; Allday, M.J. Two Epstein-Barr virus (EBV) oncoproteins cooperate to repress expression of the proapoptotic tumour-suppressor Bim: Clues to the pathogenesis of Burkitt’s lymphoma. Oncogene 2008, 27, 421–433. [Google Scholar] [CrossRef]
- You, Y.; Cheng, A.-C.; Wang, M.-S.; Jia, R.-Y.; Sun, K.-F.; Yang, Q.; Wu, Y.; Zhu, D.; Chen, S.; Liu, M.-F.; et al. The suppression of apoptosis by α-herpesvirus. Cell Death Dis. 2017, 8, e2749. [Google Scholar] [CrossRef]
- Peischard, S.; Ho, H.T.; Theiss, C.; Strutz-Seebohm, N.; Seebohm, G. A Kidnapping Story: How Coxsackievirus B3 and Its Host Cell Interact. Cell. Physiol. Biochem. 2019, 53, 121–140. [Google Scholar] [CrossRef]
- Fislová, T.; Thomas, B.; Graef, K.M.; Fodor, E. Association of the Influenza Virus RNA Polymerase Subunit PB2 with the Host Chaperonin CCT. J. Virol. 2010, 84, 8691–8699. [Google Scholar] [CrossRef]
- Arakawa, C.; Endo, A.; Kohira, R.; Fujita, Y.; Fuchigami, T.; Mugishima, H.; Ohtake, A.; Murayama, K.; Mori, M.; Miyata, R.; et al. Liver-specific mitochondrial respiratory chain complex I deficiency in fatal influenza encephalopathy. Brain Dev. 2012, 34, 115–117. [Google Scholar] [CrossRef]
- Gorai, T.; Goto, H.; Noda, T.; Watanabe, T.; Kozuka-Hata, H.; Oyama, M.; Takano, R.; Neumann, G.; Watanabe, S.; Kawaoka, Y. F1Fo-ATPase, F-type proton-translocating ATPase, at the plasma membrane is critical for efficient influenza virus budding. Proc. Natl. Acad. Sci. USA 2012, 109, 4615–4620. [Google Scholar] [CrossRef]
- Hu, M.; Bogoyevitch, M.A.; Jans, D.A. Subversion of Host Cell Mitochondria by RSV to Favor Virus Production is Dependent on Inhibition of Mitochondrial Complex I and ROS Generation. Cells 2019, 8, 1417. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).