Abstract
Alternanthera yellow vein virus (AlYVV), a monopartite begomovirus, has been identified infecting a diverse range of crops and native plants in Pakistan, India, and China. However, distinctive yellow vein symptoms, characteristic of begomovirus infection, were observed on the Ageratum conyzoides weed in Oman, prompting a thorough genomic characterization in this study. The results unveiled a complete genome sequence of 2745 base pairs and an associated betasatellite spanning 1345 base pairs. In addition, Sequence Demarcation Tool analyses indicated the highest nucleotide identity of 92.8% with a previously reported AlYVV-[IN_abalpur_A_17:LC316182] strain, whereas the betasatellite exhibited a 99.8% nucleotide identity with isolates of tomato leaf curl betasatellite. Thus, our findings propose a novel AlYVV Oman virus (AlYVV-OM) variant, emphasizing the need for additional epidemiological surveillance to understand its prevalence and significance in Oman and the broader region. To effectively manage the spread of AlYVV-OM and minimize its potential harm to (agro)ecosystems, future research should focus on elucidating the genetic diversity of AlYVV-OM and its interactions with other begomoviruses.
1. Introduction
Circular Rep-encoding single-stranded (CRESS) DNA viruses, identified within the Cressdnaviricota phylum, encompass single-stranded DNA viruses encoding a replication-associated protein (Rep), believed to have a shared ancestral origin [1]. These viruses that infect plants fall into two families: Geminiviridae and Nanoviridae [2]. Among these, Begomoviruses stand as the largest genus among single-stranded DNA viruses, constituting 88% of the Geminiviridae family. They pose a significant threat to global crop yields, proficiently transmitted by the polyphagous whitefly vector, Bemisia tabaci, in a circulative manner, which comprises various biotypes [3]. This efficient vectorization extends across a wide range of hosts, spanning both cultivated and wild plant species [4,5]. Additionally, other species like Trialeurodes ricini and Trialeurodes vaporariorum have also been identified as potential transmitters of Begomoviruses [6]. Geminiviruses are divided into New-World (America) or Old-World (OW) (Australia, Asia, Mediterranean and Europe) viruses. According to the updated list of ICTV guidelines, the family Geminiviridae is divided into 14 genera (with 520 species) depending on (i) the host they infect, (ii) the insect vector which helps in transmission, and (iii) the genome organization. Among all genera, Begomovirus is the largest genus (with 445 known species), which infects a wide range of plant hosts, causing enormous economic losses to crops and ornamental plants globally [7]. Begomoviruses are further divided into two groups: (i) monopartite (having a single genome molecule, with the majority of them being associated with satellite molecules such as alphasatellites, betasatellites, and/or deltasatellites), and (ii) bipartite (having two genome molecules, known as DNA A and DNA B). The DNA A molecule of bipartite begomoviruses has a genome organization analogous to that of monopartite begomoviruses. Due to the small size of their genome (~2.7 kb), begomoviruses encode only a few proteins with diverse virulence functions. In general, the DNA-A component of both mono- and bipartite begomoviruses typically encode six proteins necessary for viral replication, movement, and the regulation of gene expression. Out of these two open reading frames (ORFs), V1 and V2 are located on the V-strand known as the virion strand, and four ORFs (C1–C4) are available on the C-strand known as the complimentary strand. The ORF V1 encodes for a coat protein (CP), V2 encodes for a pre-coat protein, C1 encodes for a replication-associated protein (Rep), C2 encodes for a transcriptional activation protein (TrAP), C3 encodes for a replication enhancer protein (REn), and C4 encodes for the C4 protein, which usually functions as a determinant of pathogenicity. The DNA-B molecule has two identified ORFs: one on the V-strand (BV1), which encodes for the nuclear shuttle protein (NSP), and the second on the C-strand (BC1), which encodes for the movement protein (MP). However, recent studies have identified an additional ORF named V3 in tomato yellow leaf curl virus (TYLCV). This ORF is suggested to play a role in full virus infection, potentially contributing to the virus’s ability to infect and propagate within host plants [8]. In addition, alphasatellites and betasatellites are crucial components for the majority of monopartite begomoviruses in the OW, each encoding a single recognized protein. Alphasatellites specifically code for the Rep protein, pivotal in the autonomous replication of satellites. However, betasatellites encode a solitaprotein on their C-strand, referred to as beta C1 (βC1), a well-established pathogenicity determinant, whose complex role has been thoroughly reviewed recently [9].
Begomoviruses in Oman have been the subject of limited diversity, epidemiological exploration, and gene function studies, with only a handful of reports documenting exotic strains [10,11,12]. In the present study, we aimed to fill this gap by investigating the introduction of a new begomovirus, AlYVV, infecting Ageratum conyzoides in Oman.
2. Material and Methods
2.1. Specimen Collection and DNA Extraction
During 2020, distinctive yellow vein symptoms indicative of begomovirus infection were observed on six Ageratum conyzoides plants with an incidence of 20–35%, which were collected from open field crops from Musandam (coordinates 26.1644° N 56.2426° E), Oman (Figure 1A). Newly emerging leaves, six symptomatic and three asymptomatic (Figure 1), were harvested, placed in a cold box, and transported to the plant biotechnology lab for a comprehensive genomic study. Subsequently, total genomic DNA extraction was conducted using CTAB protocols, following previously suggested procedures [13] with minor adjustments.

Figure 1.
Ageratum conyzoides plant naturally infected by a bipartite AlYVV-OM, showing yellow vein symptoms (A), and asymptomatic plant (B).
2.2. Amplification, Cloning, and Sequencing
The isolated DNA was quantified and dilutions were prepared for use in PCR reaction using begomovirus-specific primers, amplifying an approximately 550 bp fragment of the CP gene in standardized PCR conditions [14] (Figure 2d). The full-length begomovirus genome was amplified in rolling circle amplification (RCA) using phi 29 DNA polymerase in the TempliPhi 100 Amplification Kit (GE Healthcare, Life Sciences, Piscataway, NJ, USA), following established protocols [15]. The concatemer of RCA products was then subjected to restriction fragment length polymorphism (RFLP) to release monomer molecules. Finally, utilizing the BamH1 restriction enzyme, a full-length linear molecule of the begomovirus was generated and subsequently cloned into pGEM5Zf+ (Promega, Madison, WI, USA) (Figure 2a–c). Simultaneously, the betasatellite was amplified with betasatellite-specific primers and cloned into the pTZ57R/T cloning vector (Thermo Fisher Scientific, Waltham, MA, USA) (Figure 2e,f).
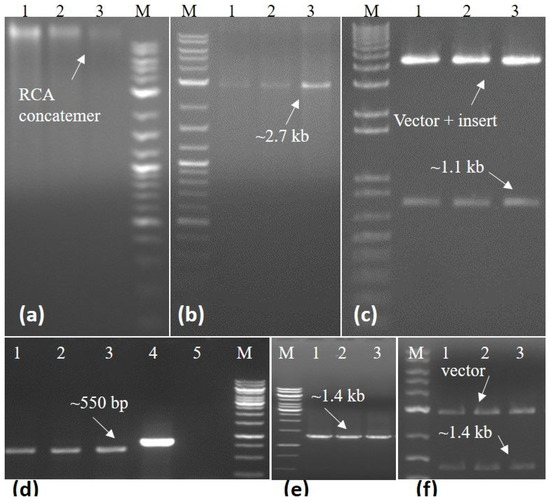
Figure 2.
Three Ageratum conyzoides plants confirming positive detection in PCR were selected for further full-length amplification of begomovirus through RCA and betasatellite by specific primers. Amplification of RCA concatemers using Phi29 DNA polymerase enzyme from plants 1–3 (a). The monomer molecule of begomovirus was produced through restricted digestion using the BamH1 enzyme for RCA from plants 1–3 (b). The production of recombinant begomovirus molecules from RCA restricted for plants 1–3 (c). Amplification of AlYVV-OM using PCR for plants 1–3 and lanes 4 and 5 served as the positive and negative controls, respectively (d). Amplification of tomato leaf curl betasatellite (ToLCB) through PCR (e). The insertion of ToLCB into a cloning vector amplified from plants 1–3 (f). “M” represents the 1 kb molecular marker employed to gauge the size of the DNA fragments in each case.
2.3. Bioinformatic Analysis
After the cloning and confirmation of full-length clones for begomovirus (n = 3) and betasatellite (n = 3), one begomovirus and betasatellite from each plant were subjected to complete Sanger sequencing at Macrogen Inc. (Seoul, Republic of Korea). Multiple sequence contigs were received and assembled using SeqMan, built in the DNAstar Lasergene package (Madison, WI, USA). Initial genome identification was performed in a BLASTn search (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 16 December 2021), and highly similar sequences were retrieved from GenBank for pairwise sequence analysis using the sequence demarcation tool [16]. Multiple sequence alignments were performed using MUSCLE, and phylogenetic tree was constructed in MEGAX using the neighbor-joining algorithm, selecting the best-fit Kimura-2 parameter with 1000 bootstrap values [17]. Potential recombination events were verified using the RDP4 program with default settings and a cut-off value of p ≤ 0.05 [18]. Predicted ORFs were identified using ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/, accessed on 5 January 2022).
2.4. Population Study
To identify the genetic diversity of the virus population, different parameters of the DnaSP (v. 6.12) program were used [19,20]. The multiple aligned sequences were studied for the entire segregating sites (s), the average number of nucleotide differences between sequences (kt), the total number of mutations (Eta), the sequence nucleotide diversity (π), and Watterson’s estimate of the population mutation rate based on the total number of segregating sites (θ − w), and the total number of mutations (θ–η) were also analyzed along with the number of haplotypes (h) and the haplotype diversity (Hd) [21].
3. Results
3.1. Alternanthera Yellow Vein Oman Virus Is Associated with Ageratum conyzoides
The presence of the monopartite begomovirus in symptomatic A. conyzoides was confirmed through PCR analysis. Notably, six symptomatic samples exhibited bands of approximately 700 base pairs when subjected to detection primers. None of the healthy plants exhibited any amplification of the PCR product. Utilizing Phi29 DNA polymerase and hexamer primers in RCA amplification from three samples resulted in the generation of concatemers composed of circular DNA molecules. Subsequent RFLP analysis, employing the BamH1 restriction enzyme, revealed that the monomer molecule of the expected begomovirus might be the causative agent in the infected A. conyzoides plants.
Three full-length begomoviruses, each derived from a different plant, were sequenced using the Sanger method, and all sequencing contigs were assembled to produce a predictable begomovirus genome. The complete genome sequence of this begomovirus spanned 2791 bp and exhibited the typical genome orientation observed in OW monopartite begomoviruses with six ORFs (V2 and CP in the virion-sense and Rep, TrAP, Ren, and C4 protein in the complementary sense; Table 1). To ensure accuracy, any ambiguities in the complete genome sequences were resolved before submission to the NCBI GenBank, currently accessible under numbers ON186760-ON186762. The pairwise sequence analysis using the STD tool revealed the highest nucleotide identity at 92.8% with an isolate of alternanthera yellow vein virus (AlYVV) AlYVV-[IN: abalpur:A:17] (LC316182), which infects Alternanthera sessilis in India [22]. Subsequently, nucleotide identities ranged from 92.5% to 90.7% with other AlYVV isolates, while a distinct range of 72.2% to 74% of nucleotide identity was observed with various other begomovirus species (Figure 3a and Table 2). The identified isolate undergoes scrutiny as a novel variant, AlYVV-OM, which specifically infects the A. conyzoides weed in Oman [23]. The phylogenetic tree further reinforced this conclusion by clustering Omani isolates (AlYVV-OM-wed39-4, AlYVV-OM-wed39-5, and AlYVV-OM-wed39-6) with AlYVV isolates described earlier in the same group supported by 1000 bootstrap values (Figure 3b).

Table 1.
Features of alternanthera yellow vein Oman virus and tomato leaf curl betasatellite isolated from Ageratum conyzoides plants.
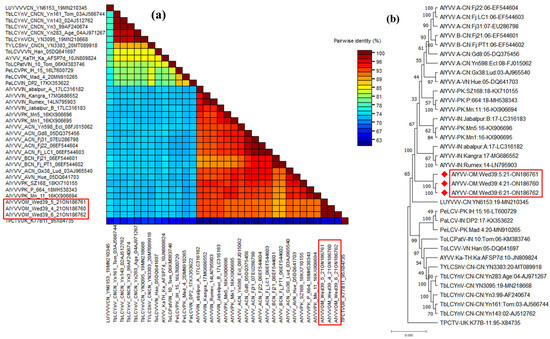
Figure 3.
Pairwise sequence analysis using sequence demarcation tool (SDT V1.2) of AlYVV-OM isolates highlighted in red boxes identified from this study and used for analysis (a), and phylogenetic dendrograms based on complete nucleotide sequences of DNA A of AlYVV-OM genome components (b). To calculate mutation distances, vertical and horizontal branches are arbitrary and proportional, respectively. The virus tree was arbitrarily rooted on the sequence of tomato curly top virus. The begomovirus acronyms used were ludwigia yellow vein Vietnam virus (LUYVVV), tobacco leaf curl Yunnan virus (TbLCYNV), tomato leaf curl Vietnam virus (TOLCVV), ageratum yellow vein virus (AYVV), tomato leaf curl Patna virus isolate (ToLCPV), pedilanthus leaf curl virus (PeLCV), papaya leaf curl virus (PaLCV), and alternanthera yellow vein virus (AYVV). A diverse DNA sequence from the tomato pseudo-curly top virus (TPCTV) was employed for comparative purposes.

Table 2.
Percent nucleotide sequence identities of alternanthera yellow vein Oman virus (AlYVV-OM) with other related begomoviruses.
Population structure analyses, performed to evaluate the intensity of genetic variability among the begomovirus sequences under study, confirmed the total number of polymorphic segregating sites (S = 1309), with 681 InDel sites, the haplotype diversity (hd = 0.998), the total mutation (Eta = 2285), the average number of nt. differences (Kt = 460), and the nucleotide diversity (π = 0.17) for AlYVV-OM datasets (Table 3). Similarly, the analysis performed for ORF datasets revealed the maximum number of polymorphic sites (s) and number of mutations (η) in AC1 (Rep) and AV1 (CP) genes with nucleotide diversity π = 0.1. Moreover, for betasatellites, the total number of polymorphic sites (s) was 834, with a total number of mutations (η) of 1482 and a nucleotide diversity (π) of 0.30509 (π = 0.3), which were also very high. However, the obtained maximum π value explains the non-random distribution of nucleotides throughout viral and sub-viral genome regions, which significantly contributes to a high degree of genetic variability. Therefore, this estimation suggests the presence of highly diverse populations, within and among populations.

Table 3.
Nucleotide diversity of alternanthera yellow vein Oman virus (AlYVV-OM) and ToLCB.
Furthermore, strong evidence of population genetic diversity is evident through both the total number of haplotypes (H = 33) and the high haplotype diversity (Hd = 0.99) (Table 3). Simultaneously, within the ORF datasets, the values for H and Hd span ranges of 31–42 and 0.96 to 0.99, respectively. This result underscores the varying contribution of genes to DNA polymorphism. Similarly, for betasatellites, the values for H and Hd were 17 and 1.000, respectively, indicating a low level of sequence divergence, but a notable frequency of unique mutations (Table 3).
3.2. Tomato Leaf Curl Betasatellite Associated with the Disease
Three full-length clones (wed39-7, wed39-8, and wed39-9), each spanning 1375 base pairs, were identified and subsequently submitted to NCBI GenBank under the accession numbers ON186763-ON186765.
These isolates exhibit a high degree of similarity, presenting betasatellite genomes characterized by a single predicted beta C1 ORF on the virion-sense strand. Additionally, a notable feature includes a region rich in adenine sequences (A-rich) and a conserved satellite region (SCR), a commonality observed across all betasatellites. In pairwise sequence analysis using SDT, these isolates displayed a maximum nt. identity at 99.8–99.9% with tomato leaf curl betasatellite ToLCB- [OM: Sen10:20] (MT188562), previously reported in Oman [14]. Subsequently, they exhibited identities of 88.2%, 83.5%, and 84.7% with ToLCKB, CLCMuB, and ChiLCB isolates, respectively (Figure 4a). In the phylogenetic analysis, these isolates formed a distinct cluster with other ToLCB isolates, supported by 1000 bootstrap values, confirming their association with ToLCB (Figure 4b). According to the ICTV guidelines set for betasatellite classification (>79% identity), the identified clones are recognized as a novel isolate of ToLCB, linked with AlYVV-OM, and infecting a new host—A. conyzoides in Oman.
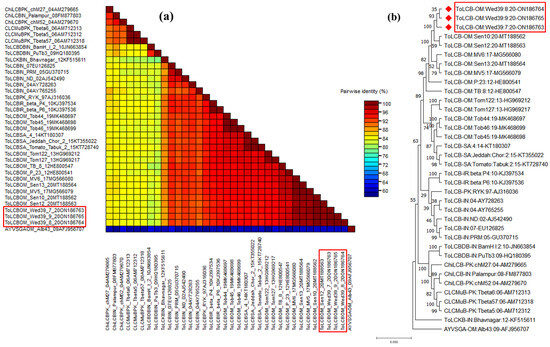
Figure 4.
Pairwise sequence analysis using sequence demarcation tool (SDT V1.2) of ToLCB isolates highlighted in red boxes identified from this study and used for analysis (a), and phylogenetic dendrograms based on complete nucleotide sequences of ToLCB genome components (b). To calculate mutation distances, vertical and horizontal branches are arbitrary and proportional, respectively. The betasatellite tree was arbitrarily rooted on the sequence of ageratum yellow vein Singapore alphasatellite. The betasatellite acronyms used are chilli leaf curl betasatellite (ChiLCB), cotton leaf curl Multan betasatellite (CLCMuB), and tomato leaf curl betasatellite (ToLCB). A varied DNA sequence from the ageratum yellow vein Singapore alphasatellite (AYVSGA) was utilized for comparative analysis.
3.3. Recombination Analysis
To explore the potential role of recombination in AlYVV-OM, a comprehensive recombination analysis was conducted using complete genome sequences of AlYVV-OM and 100 isolates of other begomoviruses retrieved from GenBank under default settings. Although the predicted ORFs of AlYVV-OM did not reveal any variable sequence identity with the corresponding ORFs of other begomoviruses, and therefore indicating that AlYVV-OM is not a recombinant virus, it is noteworthy that AlYVV-OM has been involved as a major/minor parent, potentially contributing to recombination events that might influence the evolution of other begomoviruses. Therefore, in the Recombination Detection Program (RDP) analysis, AlYVV-OM was identified as a significant contributor in the recombination of AlYVV (LC316182), serving as a major parent according to at least four algorithms: Maxchi (1.38 × 10−6), Chimaera (3.54 × 10−5), SiSscan (1.26 × 10−24), and 3Seq (2.91 × 10−6). In addition, for ageratum yellow vein China virus (KU954382), AlYVV-OM was recognized as a minor parent, with GENECONV (4.34 × 10−4) and SiScan (1.99 × 10−73) algorithms considered as reliable indicators of this involvement.
3.4. Population Study
Virus population analysis was applied to calculate the degree of genetic variability (>0.08) within and among populations (Table 3). However, we observed a total of 1309 polymorphic sites (s), encompassing 2285 mutations (η), and a nucleotide diversity of 0.17 (π = 0.1) for AlYVV-OM. Likewise, in the case of betasatellites, we identified a total of 900 polymorphic sites (s), with a noteworthy 1200 mutations (η), resulting in a substantial nucleotide diversity (π) of 0.12 (π = 0.3). However, the obtained maximum π value explains the non-random distribution of nucleotides throughout viral and sub-viral genome regions, which significantly contributes to a high degree of genetic variability. Therefore, the estimation suggests diverse populations within and among populations. In addition, the genetic diversity within and among populations was also determined by the number of haplotypes (H) and haplotype diversity (Hd). Therefore, using DnaSP software (v. 6.0) [20], analysis was performed for sequence datasets that contained a haplotype distribution among reference DNA sequences of the begomovirus population, and we found a total number of haplotypes (H) of 33, where its haplotype diversity was identified as close to 1, i.e., Hd = 0.99. Therefore, the overall result explains the low level of sequence divergence but the high frequency of unique mutations (Table 4).

Table 4.
Geographical distribution of alternanthera yellow vein virus and its host range.
4. Discussion
Over the past two decades, various factors such as the transport of infected materials, vector population dynamics, and biological and environmental changes have played a vital role in the emergence of new begomovirus species and/or strains in different geographical areas, infecting diverse host plant species [34]. Consequently, previously unrecognized hosts, including crops, ornamental plants, weeds, and trees, have become susceptible to different begomoviruses. Despite limited studies reporting the incidence of AlYVV, we present the first-ever documentation of a novel variant, AlYVV-OM, in Oman. The occurrence of this virus has been observed in limited geographical areas and the complete genome sequence of AlYVV-OM was identified from the Ageratum conyzoides host.
Comparison with other publicly available AlYVV isolates (12 sequences) revealed that AlYVVOM exhibited the highest nucleotide identity (92.5%) with an Indian isolate, highlighting its close relation to an Indian isolate among the publicly available AlYVV sequences. AlYVV was initially discovered in China in 2005, primarily infecting Alternanthera philoxeroides, sharing a notable genomic similarity and exhibiting the highest nucleotide identity with ageratum yellow vein virus AYVV [33]. Its subsequent appearances in Vietnam and Pakistan during 2008 and 2010, respectively, associated the virus with diverse DNA satellites, encompassing ageratum yellow leaf curl betasatellite, cotton leaf curl Multan betasatellite (CLCuMuB), potato leaf curl alphasatellite (PotLCuA), and hi-biscus leaf curl alphasatellite (HLCuA) [35]. Furthermore, a report in India highlighted its presence alongside CLCuMuB, infecting Picrorhiza kurroa plants [24] (Table 2). This intercontinental prevalence underscores the adaptability and broad host range of AlYVV.
The majority of begomoviruses prevalent in South Asia (China, India, and Pakistan) differ from those in the Arabian Peninsula, owing to the natural sea barrier. However, the very close genetic relationship between AlYVV-OM and Indian isolates suggests a likely origin from India, with the possibility that trade via sea or air routes facilitated the distribution of this virus into Oman. Additionally, it is noteworthy that AlYVV identified in China was not reported to be accompanied by betasatellites. In contrast, Ha et al. [36] reported an AlYVV isolate from Vietnam to be associated with alternanthera yellow vein betasatellite. This suggests that AlYVV has the capability to infect plants in the field, regardless of the presence or absence of a betasatellite [37]. Notably, the association of betasatellites may enhance begomoviruses’ ability to overcome host defense responses, broaden host ranges, and indirectly impact in planta virus accumulation [38]. Although betasatellite molecules have no precise nucleotide sequence identity with the helper (mono-or-bipartite) begomoviruses, they exhibit conserved nonanucleotide (TAATATT/AC) sequences [39]. Betasatellites are very adaptable in their trans-replication with diverse begomoviruses, including other members of the Geminiviridae family. The Rep protein in the majority of begomoviruses supports the replication of different betasatellites, even in cases where they lack cognate-related virus iteron sequences [40]. Thus, it is assumed that virus–betasatellite interactions are non-specific and related to trans-replication, long-distance movement, and transmission [41]. The particular sequences in betasatellites mimic the iterons for the rep binding, which illustrates its indiscriminate replicative nature [42]. Due to a similar purpose, it is very likely that AlYVV can effectively trans-replicate ToLCB. The vector (whitefly) population and its polyphagous nature also contributed to the evolution of these begomoviruses.
The identification of various begomoviruses and betasatellites in Ageratum conyzoides underscores the potential importance of weeds as alternate hosts, acting as reservoirs for the emergence of diverse begomovirus species worldwide. Thus, we hypothesize that AlYVV-OM may be an ancient virus that has evolved over time within Ageratum conyzoides, possessing the capability to interact with ToLCB. Furthermore, our findings indicate that ToLCB can form associations with AlYVV-OM, potentially facilitating its interaction with various begomoviruses, such as TYLCV, a globally distributed begomovirus frequently observed in Oman. The current study also forms a foundation for further studies on the epidemiology and genetic diversity of AlYVV-OM in Oman and the wider region.
5. Conclusions
The complete genomes of novel AlYVV-OM and ToLCB were meticulously examined within the weed host Ageratum conyzoides, offering valuable insights into their epidemiology and emphasizing the importance of comprehending their population structure. Considering the potential evolution of diverse begomoviral strains within specific geographical areas and their possible spread to other regions through various transmission routes, it becomes imperative to implement protective measures. To curtail the transmission of these pathogens, it is highly recommended to regulate trade products through enhanced phytosanitary measures and establish facilities for the early detection of viruses. Implementing such measures is crucial for preventing and controlling the spread of viruses across regions, thereby protecting against potential harm and ensuring the long-term sustainability of (agro)ecosystems.
Author Contributions
M.S.S. conceived the study and oversaw its execution. M.S. conducted the experiments. M.S. and M.S.S. analyzed and interpreted the data and drafted the initial manuscript. The manuscript was further edited by G.O. and A.M.A.-S. All authors have read and agreed to the published version of the manuscript.
Funding
Sultan Qaboos University provided support for this study under SR/AGR/CROP/16/01, IG/AGR/CROP/23/02, and CL/SQU-ZJU/AGR/23/01 projects.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data generated during this research have been included in this published article.
Acknowledgments
We are thankful to the University of Sultan Qaboos for performing this study.
Conflicts of Interest
The authors have no conflict of interest to disclose.
References
- Krupovic, M.; Varsani, A.; Kazlauskas, D.; Breitbart, M.; Delwart, E.; Rosario, K.; Zerbini, F.M. Cressdnaviricota: A virus phylum unifying seven families of rep-encoding viruses with single-stranded, circular DNA genomes. J. Virol. 2020, 94, e00582-20. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Krupovic, M.; Koonin, E.V. Deep roots and splendid boughs of the global plant virome. Annu. Rev. Phytopathol. 2020, 58, 23–53. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.L.; Batuman, O.; Webster, C.G.; Adkins, S. Role of the Insect Supervectors Bemisia tabaci and Frankliniella occidentalis in the Emergence and Global Spread of Plant Viruses. Annu. Rev. Virol. 2015, 2, 67–93. [Google Scholar] [CrossRef] [PubMed]
- Zerbini, F.M.; Briddon, R.W.; Idris, A.; Martin, D.P.; Moriones, E.; Navas-Castillo, J.; Ictv Report, C. ICTV Virus Taxonomy Profile: Geminiviridae. J. Gen. Virol. 2017, 98, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olive, E.; Navas-Castillo, J. Molecular and Biological Characterization of a New World Mono-/Bipartite Begomovirus/Deltasatellite Complex Infecting Corchorus siliquosus. Front. Microbiol. 2020, 11, 1755. [Google Scholar] [CrossRef] [PubMed]
- Idriss, M.; Abdallah, N.; Aref, N.; Haridy, G.; Madkour, M. Biotypes of the castor bean whitefly Trialeurodes ricini (Misra) (Hom., Aleyrodidae) in Egypt: Biochemical characterization and efficiency of geminivirus transmission. J. Appl. Entomol. 1997, 121, 501–509. [Google Scholar] [CrossRef]
- Walker, J.; Siddell, S.G.; Lefkowitz, E.J.; Mushegian, A.R.; Adriaenssens, E.M.; Alfenas-Zerbini, P.; Zerbini, F.M. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch. Virol. 2021, 166, 2633–2648. [Google Scholar] [CrossRef]
- Gong, P.; Tan, H.; Zhao, S.; Li, H.; Liu, H.; Ma, Y.; Zhou, X. Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 2021, 12, 4278. [Google Scholar] [CrossRef]
- Gnanasekaran, P.; Kishorekumar, R.; Bhattacharyya, D.; Vinoth Kumar, R.; Chakraborty, S. Multifaceted role of geminivirus associated betasatellite in pathogenesis. Mol. Plant Pathol. 2019, 20, 1019–1033. [Google Scholar] [CrossRef]
- Shahid, M.S.; Shafiq, M.; Ilyas, M.; Raza, A.; Al-Sadrani, M.N.; Al-Sadi, A.M.; Briddon, R.W. Frequent occurrence of Mungbean yellow mosaic India virus in tomato leaf curl disease affected tomato in Oman. Sci. Rep. 2019, 9, 16634. [Google Scholar] [CrossRef]
- Al Shihi, A.A.; Al Sadi, A.M.; Deadman, M.; Briddon, R.W.; Shahid, M.S. Identification of a distinct strain of Cotton leaf curl Gezira virus infecting tomato in Oman. J. Phytopathol. 2018, 166, 199–205. [Google Scholar] [CrossRef]
- Shahid, M.S.; Al-Sulaimani, H.; Al-Sadi, A.M. Squash Leaf Curl Virus: A New World Bipartite Begomovirus Threatening Squash Production in Oman. Plant Dis. 2020, 104, 2533. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Al-Mabsli, S.S.; Al-Wahaibi, A.K.; Al-Sadi, A.M.; Shahid, M.S. Association of a monopartite begomovirus and associated betasatellite with yellow vein disease of a weed host, Senna italica Mill. in Oman. Virusdisease 2021, 32, 378–380. [Google Scholar] [CrossRef] [PubMed]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Lima, A.T.M.; Silva, J.C.F.; Silva, F.N.; Castillo-Urquiza, G.P.; Silva, F.F.; Seah, Y.M.; Zerbini, F.M. The diversification of begomovirus populations is predominantly driven by mutational dynamics. Virus Evol. 2017, 3, vex005. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-Delbarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Shafiq, M.; Sattar, M.N.; Shahid, M.S.; Al-Sadi, A.M.; Briddon, R.W. Interaction of watermelon chlorotic stunt virus with satellites. Australas. PlantPathol. 2021, 50, 117–128. [Google Scholar] [CrossRef]
- Marabi, R.S.; Das, S.B.; Tripathi, N.; Wada, T.; Noda, H. Identification of begomoviruses from legume crop and weed plants and viruliferous status of the whitefly Bemisia tabaci in Central India. Curr. Sci. 2021, 120, 1240–1246. [Google Scholar] [CrossRef]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Varsani, A. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Kulshreshtha, A.; Kumar, R.; Hallan, V. First report of natural infection of alternanthera yellow vein virus and cotton leaf curl Multan betasatellite on a new host Picrorhiza kurroa, an important endangered medicinal herb. J. Plant Pathol. 2019, 101, 149–153. [Google Scholar] [CrossRef]
- Murtaza, G.; Mubin, M.; Nawaz-Ul-Rehman, M.S.; Amrao, L. Genetic analysis of alternanthera yellow vein virus (Ayvv) infecting Eclipta prostrata plant in Pakistan. Pak. J. Agric. Sci. 2018, 55, 505–512. [Google Scholar]
- Nawaz-Ul-Rehman, M.; Liaqat, I.; Nahid, N.; Saleem, F.; Alkahtani, S.; Al Qahtani, A.; Mubin, M. Alternanthera yellow vein virus (AYVV); A betasatellite independent begomovirus infecting Sonchus palustris in Pakistan. Braz. J.Biol. 2022, 82, e262248. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.S.; Shakir, S.; Farooq, M.; Amin, I.; Mansoor, S. First report of Alternanthera yellow vein virus from Eclipta prostrata in Pakistan. Plant Dis. 2017, 101, 266. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, S.P.; Yang, C.X.; Liu, Z.; Wu, Z.J. Detection and molecular characterization of three begomoviruses associated with yellow vein disease of Eclipta prostrata in fujian, China. J. Plant Pathol. 2015, 97, 161–165. [Google Scholar]
- Mubin, M.; Shahid, M.S.; Tahir, M.N.; Briddon, R.W.; Mansoor, S. Characterization of begomovirus components from a weed suggests that begomoviruses may associate with multiple distinct DNA satellites. Virus Genes 2010, 40, 452–457. [Google Scholar] [CrossRef]
- Ding, M.; Yang, L.; Zhao, Z.W.; Zhang, Z.K. First report of alternanthera yellow vein virus in Eclipta prostrata in China. J. Plant Pathol. 2009, 91, 232. [Google Scholar]
- He, Z.F.; Mao, M.J.; Yu, H.; Wang, X.M.; Li, H.P. First report of a strain of Alternanthera yellow vein virus infecting Eclipta prostrate (L.) L. (compositae) in China. J. Phytopathol. 2008, 156, 496–498. [Google Scholar] [CrossRef]
- Huang, J.F.; Jiang, T.; Zhou, X.P. Molecular characterization of begomoviruses infecting Ludwigia hyssopifolia. J. Plant Pathol. 2006, 88, 83–88. [Google Scholar]
- Guo, X.; Zhou, X. Molecular characterization of Alternanthera yellow vein virus: A new Begomovirus species infecting Alternanthera philoxeroides. J. Phytopathol. 2005, 153, 694–696. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.M.; Shahid, M.S.; Briddon, R.W.; Khan, A.; Zhu, J.-K.; Brown, J.K. An unusual alphasatellite associated with monopartite begomoviruses attenuates symptoms and reduces betasatellite accumulation. J. Gen.Virol. 2011, 92, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Ha, C.; Coombs, S.; Revill, P.; Harding, R.; Vu, M.; Dale, J. Molecular characterization of begomoviruses and DNA satellites from Vietnam: Additional evidence that the New World geminiviruses were present in the Old World prior to continental separation. J. Gen. Virol. 2008, 89, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Brown, J.K.; Moriones, E.; Stanley, J.; Zerbini, M.; Zhou, X.; Fauquet, C.M. Recommendations for the classification and nomenclature of the DNA-beta satellites of begomoviruses. Arch. Virol. 2008, 153, 763–781. [Google Scholar] [CrossRef]
- Briddon, R.W.; Markham, G. Complementation of bipartite begomovirus movement functions by topocuviruses and curtoviruses. Arch. Virol. 2001, 146, 1811–1819. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999, 18, 71–106. [Google Scholar] [CrossRef]
- Kharazmi, S.; Behjatnia, S.A.; Hamzehzarghani, H.; Niazi, A. Cotton leaf curl Multan betasatellite as a plant gene delivery vector trans-activated by taxonomically diverse geminiviruses. Arch. Virol. 2012, 157, 1269–1279. [Google Scholar] [CrossRef]
- Saunders, K. Analysis of geminivirus DNA replication by 2-D gel. Methods Mol. Biol. 2008, 451, 135–143. [Google Scholar]
- Nawaz-Ul-Rehman, M.S.; Nahid, N.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Post-transcriptional gene silencing suppressor activity of two non-pathogenic alphasatellites associated with a begomovirus. Virology 2010, 405, 300–308. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).