Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties
Abstract
1. Introduction
2. Methodology
2.1. Virus Culture and Inactivation
2.2. Atomic Force Microscopy (AFM)
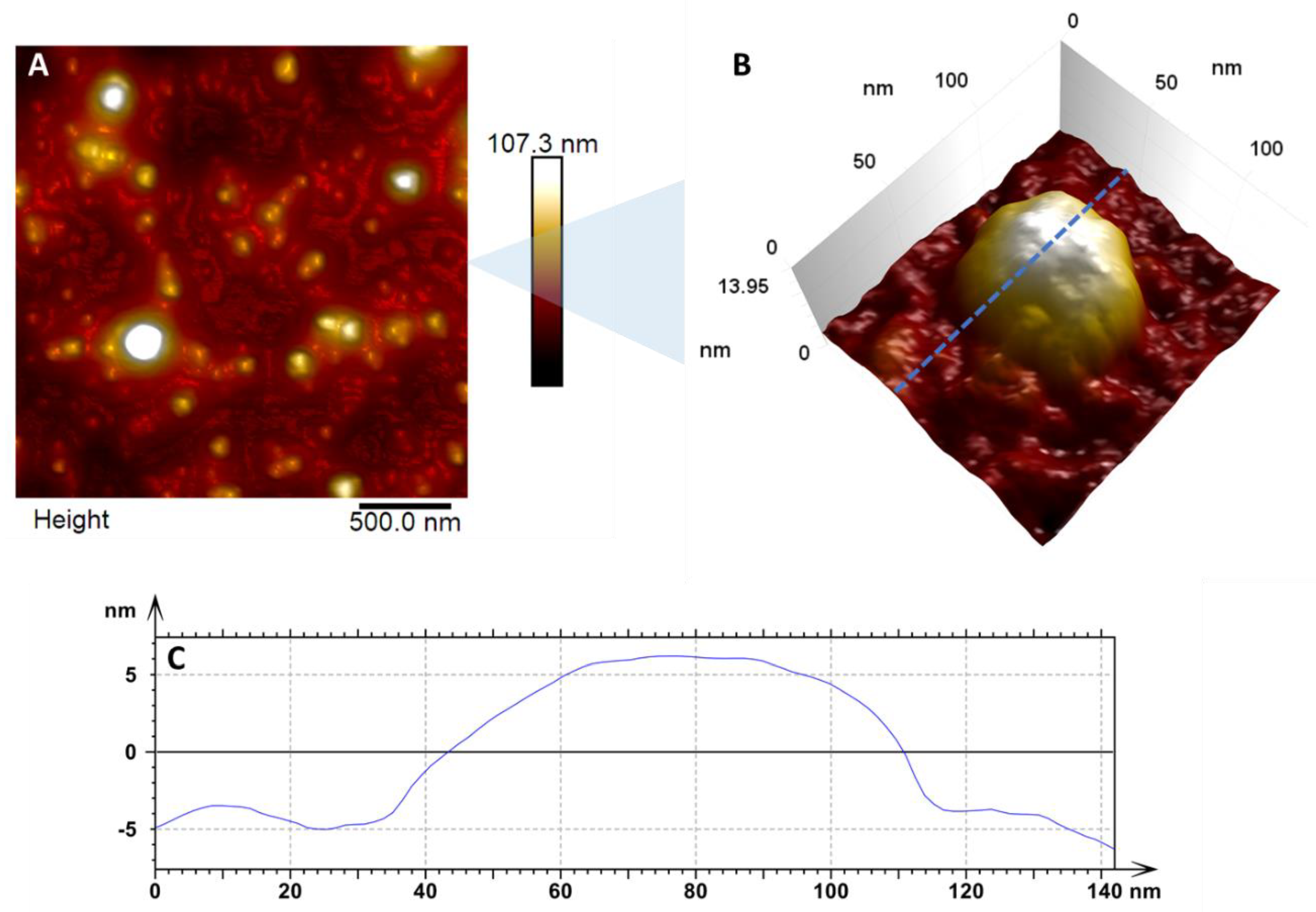
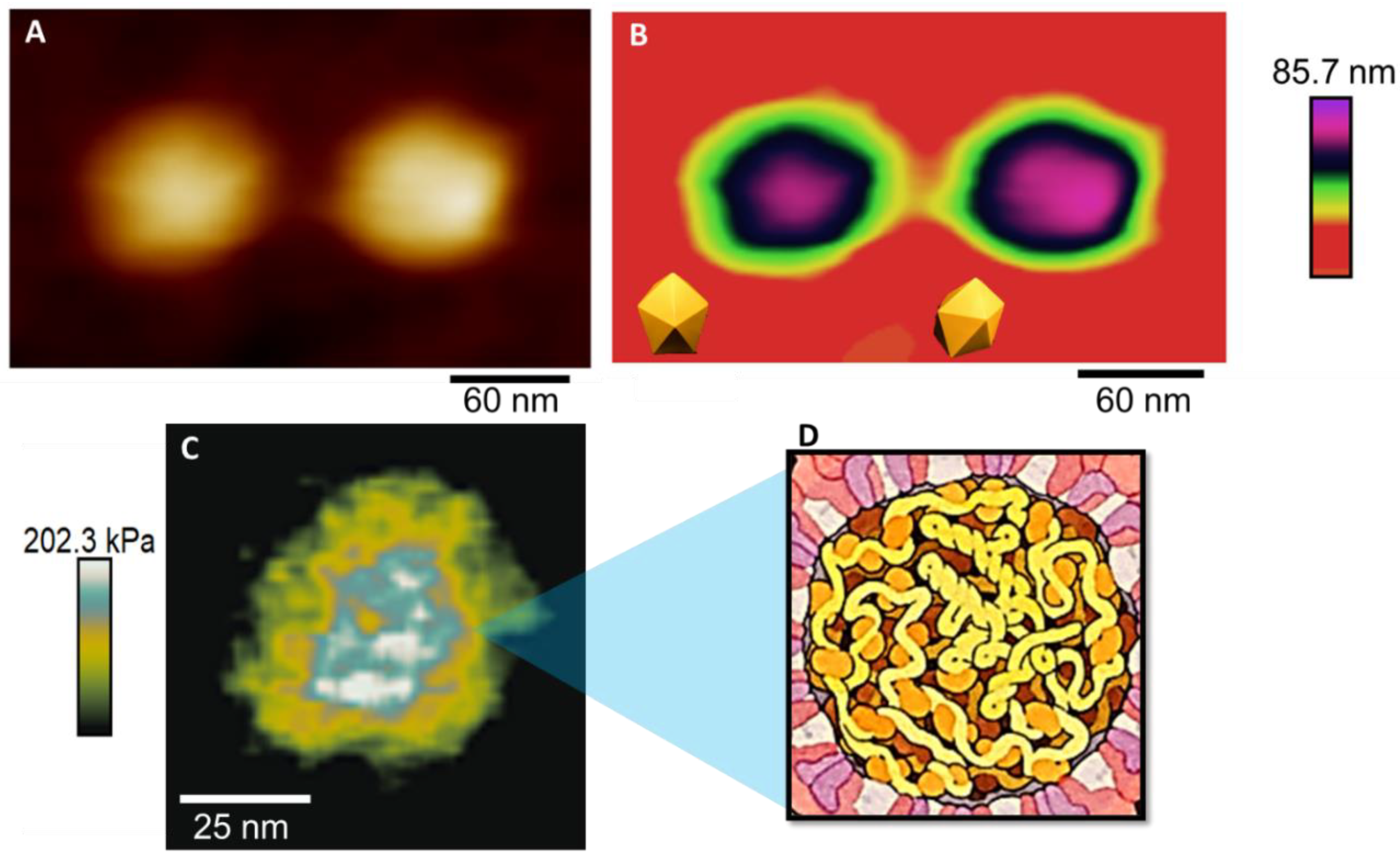
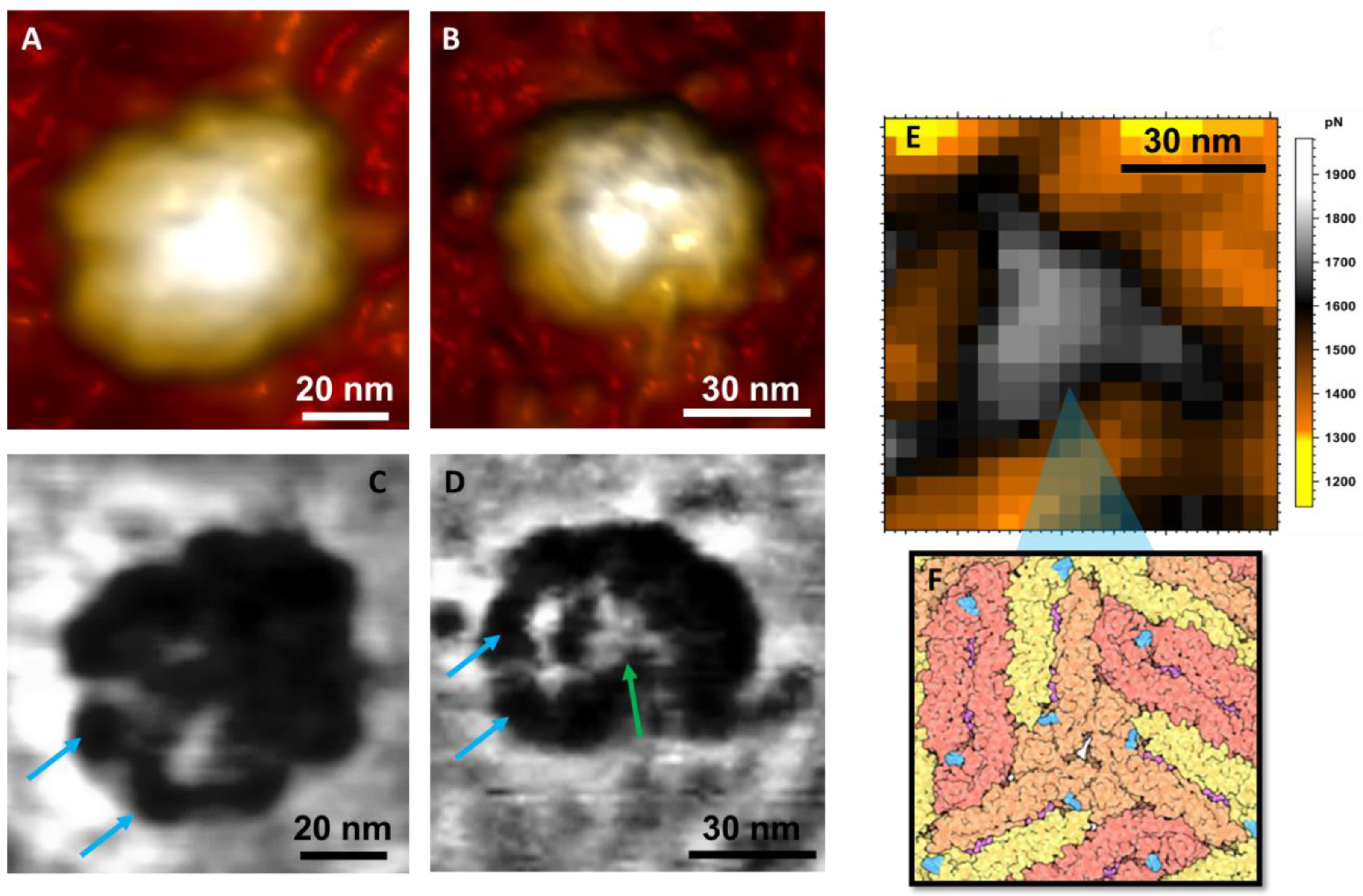
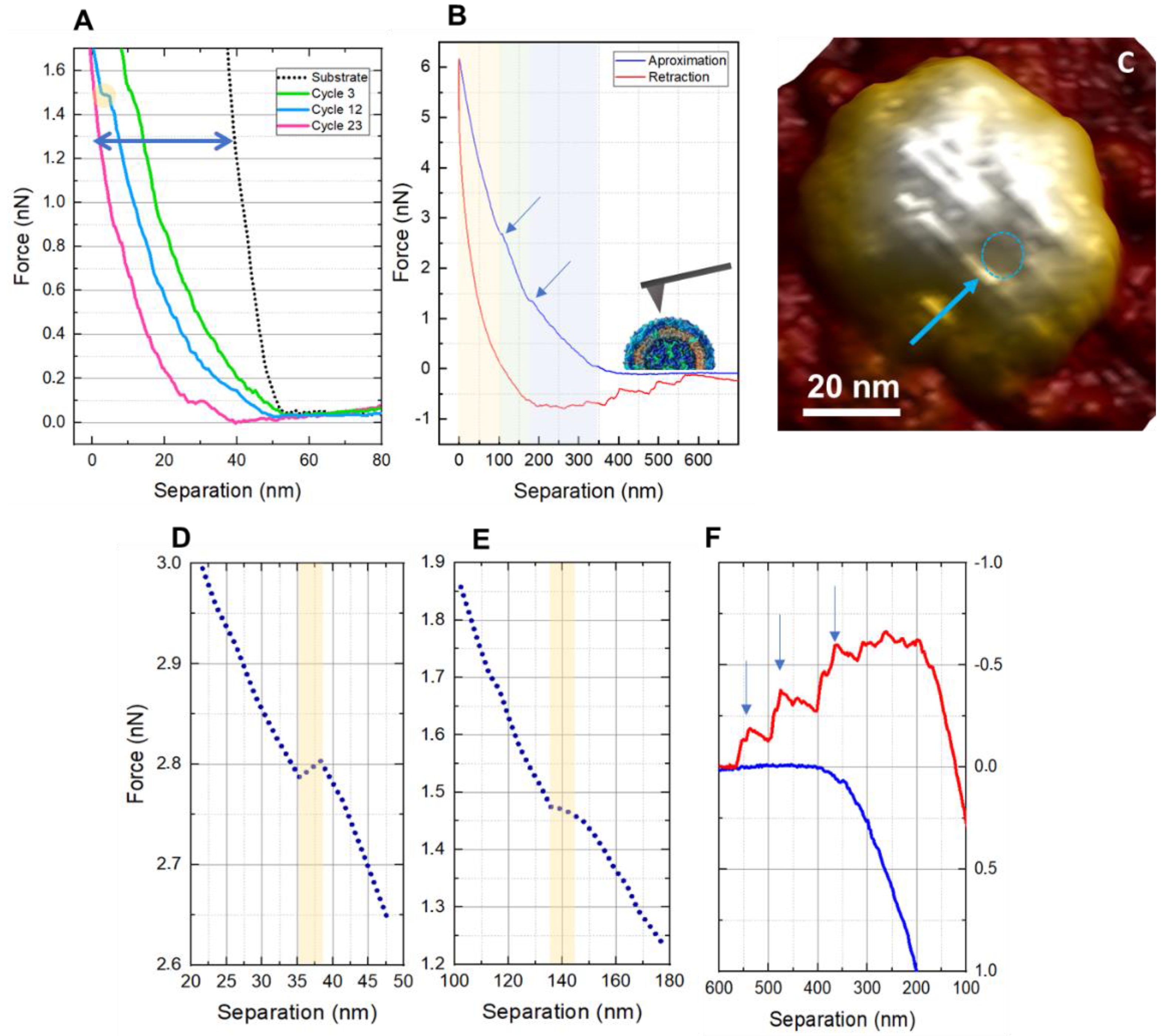
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pierson, T.C.; Diamond, M.S. The Continued Threat of Emerging Flaviviruses. Nat. Microbiol. 2020, 5, 796–812. [Google Scholar] [CrossRef] [PubMed]
- Paixão, E.S.; Teixeira, M.G.; Rodrigues, L.C. Zika, Chikungunya and Dengue: The Causes and Threats of New and Re-Emerging Arboviral Diseases. BMJ Glob. Health 2018, 3, e000530. [Google Scholar] [CrossRef] [PubMed]
- Saiz, J.-C.; Vázquez-Calvo, Á.; Blázquez, A.B.; Merino-Ramos, T.; Escribano-Romero, E.; Martín-Acebes, M.A. Zika Virus: The Latest Newcomer. Front. Microbiol. 2016, 7, 496. [Google Scholar] [CrossRef]
- Dai, L.; Wang, Q.; Song, H.; Gao, G.F. Zika Virus Envelope Protein and Antibody Complexes. Subcell. Biochem. 2018, 88, 147–168. [Google Scholar] [CrossRef] [PubMed]
- Barba-Spaeth, G.; Dejnirattisai, W.; Rouvinski, A.; Vaney, M.-C.; Medits, I.; Sharma, A.; Simon-Lorière, E.; Sakuntabhai, A.; Cao-Lormeau, V.-M.; Haouz, A.; et al. Structural Basis of Potent Zika-Dengue Virus Antibody Cross-Neutralization. Nature 2016, 536, 48–53. [Google Scholar] [CrossRef]
- Hasan, S.S.; Sevvana, M.; Kuhn, R.J.; Rossmann, M.G. Structural Biology of Zika Virus and Other Flaviviruses. Nat. Struct. Mol. Biol. 2018, 25, 13–20. [Google Scholar] [CrossRef]
- Zika Virus Biology, Transmission, and Pathways—1st Edition. Available online: https://www.elsevier.com/books/zika-virus-biology-transmission-and-pathways/martin/978-0-12-820268-5 (accessed on 22 July 2022).
- Zika Virus. Available online: https://www.who.int/news-room/fact-sheets/detail/zika-virus (accessed on 22 July 2022).
- Shen, S.; Xiao, W.; Zhang, L.; Lu, J.; Funk, A.; He, J.; Tu, S.; Yu, J.; Yang, L.; Fontanet, A.; et al. Prevalence of Congenital Microcephaly and Its Risk Factors in an Area at Risk of Zika Outbreaks. BMC Pregnancy Childbirth 2021, 21, 214. [Google Scholar] [CrossRef]
- Vazeille, M.; Zouache, K.; Vega-Rúa, A.; Thiberge, J.-M.; Caro, V.; Yébakima, A.; Mousson, L.; Piorkowski, G.; Dauga, C.; Vaney, M.-C.; et al. Importance of Mosquito ′′Quasispecies′′ in Selecting an Epidemic Arthropod-Borne Virus. Sci. Rep. 2016, 6, 29564. [Google Scholar] [CrossRef]
- The Flaviviruses: Structure, Replication and Evolution, Volume 59-1st Edition. Available online: https://www.elsevier.com/books/the-flaviviruses-structure-replication-and-evolution/chambers/978-0-12-039859-1 (accessed on 22 July 2022).
- Gavino-Leopoldino, D.; Figueiredo, C.M.; da Silva, M.O.L.; Barcellos, L.G.; Neris, R.L.S.; Pinto, L.D.M.; Araújo, S.M.B.; Ladislau, L.; Benjamim, C.F.; Da Poian, A.T.; et al. Skeletal Muscle Is an Early Site of Zika Virus Replication and Injury, Which Impairs Myogenesis. J. Virol. 2021, 95, e00904-21. [Google Scholar] [CrossRef]
- Figueiredo, C.P.; Barros-Aragão, F.G.Q.; Neris, R.L.S.; Frost, P.S.; Soares, C.; Souza, I.N.O.; Zeidler, J.D.; Zamberlan, D.C.; de Sousa, V.L.; Souza, A.S.; et al. Zika Virus Replicates in Adult Human Brain Tissue and Impairs Synapses and Memory in Mice. Nat. Commun. 2019, 10, 3890. [Google Scholar] [CrossRef]
- Allard, A.; Althouse, B.M.; Hébert-Dufresne, L.; Scarpino, S.V. The Risk of Sustained Sexual Transmission of Zika Is Underestimated. PLoS Pathog. 2017, 13, e1006633. [Google Scholar] [CrossRef] [PubMed]
- Leier, H.C.; Weinstein, J.B.; Kyle, J.E.; Lee, J.-Y.; Bramer, L.M.; Stratton, K.G.; Kempthorne, D.; Navratil, A.R.; Tafesse, E.G.; Hornemann, T.; et al. A Global Lipid Map Defines a Network Essential for Zika Virus Replication. Nat. Commun. 2020, 11, 3652. [Google Scholar] [CrossRef] [PubMed]
- Shahrizaila, N.; Lehmann, H.C.; Kuwabara, S. Guillain-Barré Syndrome. Lancet 2021, 397, 1214–1228. [Google Scholar] [CrossRef]
- Chiu, C.-F.; Chu, L.-W.; Liao, I.-C.; Simanjuntak, Y.; Lin, Y.-L.; Juan, C.; Ping, Y. The Mechanism of the Zika Virus Crossing the Placental Barrier and the Blood-Brain Barrier. Front. Microbiol. 2020, 11, 214. [Google Scholar] [CrossRef]
- Ventura, C.V.; Ventura, L.O. Ophthalmologic Manifestations Associated With Zika Virus Infection. Pediatrics 2018, 141, S161–S166. [Google Scholar] [CrossRef]
- Azevedo, R.S.; de Sousa, J.R.; Araujo, M.T.F.; Filho, A.J.M.; de Alcantara, B.N.; Araújo, F.; Queiroz, M.G.D.; Cruz, A.C.; Vasconcelos, B.H.B.; Chiang, J.O.; et al. In Situ Immune Response and Mechanisms of Cell Damage in Central Nervous System of Fatal Cases Microcephaly by Zika Virus. Sci. Rep. 2017, 8, 5. [Google Scholar] [CrossRef]
- Zanluca, C.; de Noronha, L.; Duarte dos Santos, C.N. Maternal-Fetal Transmission of the Zika Virus: An Intriguing Interplay. Tissue Barriers 2018, 6, e1402143. [Google Scholar] [CrossRef]
- Regla-Nava, J.A.; Wang, Y.-T.; Fontes-Garfias, C.R.; Liu, Y.; Syed, T.; Susantono, M.; Gonzalez, A.; Viramontes, K.M.; Verma, S.K.; Kim, K.; et al. A Zika Virus Mutation Enhances Transmission Potential and Confers Escape from Protective Dengue Virus Immunity. Cell Rep. 2022, 39, 110655. [Google Scholar] [CrossRef]
- Cardoso-Lima, R.; Souza, P.F.N.; Guedes, M.I.F.; Santos-Oliveira, R.; Alencar, L.M.R. SARS-CoV-2 Unrevealed: Ultrastructural and Nanomechanical Analysis. Langmuir 2021, 37, 10762–10769. [Google Scholar] [CrossRef]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika Virus Strain Causes Birth Defects in Experimental Models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef]
- Moser, L.A.; Boylan, B.T.; Moreira, F.R.; Myers, L.J.; Svenson, E.L.; Fedorova, N.B.; Pickett, B.E.; Bernard, K.A. Growth and Adaptation of Zika Virus in Mammalian and Mosquito Cells. PLoS Negl. Trop. Dis. 2018, 12, e0006880. [Google Scholar] [CrossRef]
- Kuralay, F.; Dükar, N.; Bayramlı, Y. Poly-L-lysine Coated Surfaces for Ultrasensitive Nucleic Acid Detection. Electroanalysis 2018, 30, 1556–1565. [Google Scholar] [CrossRef] [PubMed]
- Mazia, D.; Schatten, G.; Sale, W. Adhesion of Cells to Surfaces Coated with Polylysine. Applications to Electron Microscopy. J. Cell Biol. 1975, 66, 198–200. [Google Scholar] [CrossRef] [PubMed]
- Oropesa, R.; Ramos, J.R.; Falcón, V.; Felipe, A. Characterization of Virus-like Particles by Atomic Force Microscopy in Ambient Conditions. Adv. Nat. Sci. Nanosci. Nanotechnol. 2013, 4, 025007. [Google Scholar] [CrossRef][Green Version]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef] [PubMed]
- Cortese, M.; Goellner, S.; Acosta, E.G.; Neufeldt, C.J.; Oleksiuk, O.; Lampe, M.; Haselmann, U.; Funaya, C.; Schieber, N.; Ronchi, P.; et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017, 18, 2113–2123. [Google Scholar] [CrossRef]
- Barreto-Vieira, D.F.; Barth, O.M.; da Silva, M.A.N.; Santos, C.C.; da Santos, A.S.; Batista, F.J.B.; de Filippis, A.M.B. Ultrastructure of Zika Virus Particles in Cell Cultures. Mem. Inst. Oswaldo Cruz 2016, 111, 532–534. [Google Scholar] [CrossRef][Green Version]
- Genome Number and Size Polymorphism in Zika Virus Infectious Units. J. Virol. 2021, 95, e00787-20. [CrossRef]
- Cui, Y.; Grant, D.G.; Lin, J.; Yu, X.; Franz, A.W.E. Zika Virus Dissemination from the Midgut of Aedes Aegypti Is Facilitated by Bloodmeal-Mediated Structural Modification of the Midgut Basal Lamina. Viruses 2019, 11, 1056. [Google Scholar] [CrossRef]
- Louten, J. (Ed.) Chapter 2-Virus Structure and Classification; Essential Human Virology; Academic Press: Cambridge, MA, USA, 2016; pp. 19–29. ISBN 9780128009475. [Google Scholar] [CrossRef]
- Zandi, R.; Reguera, D.; Bruinsma, R.F.; Gelbart, W.M.; Rudnick, J. Origin of Icosahedral Symmetry in Viruses. Proc. Natl. Acad. Sci. USA 2004, 101, 15556–15560. [Google Scholar] [CrossRef]
- Pierson, T.C.; Diamond, M.S. Degrees of maturity: The complex structure and biology of flaviviruses. Curr. Opin. Virol. 2012, 2, 168–175. [Google Scholar] [CrossRef] [PubMed]
- Mateu, M.G. Mechanical Properties of Viruses Analyzed by Atomic Force Microscopy: A Virological Perspective. Virus Res. 2012, 168, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Sotcheff, S.; Routh, A. Understanding Flavivirus Capsid Protein Functions: The Tip of the Iceberg. Pathogens 2020, 9, 42. [Google Scholar] [CrossRef] [PubMed]
- Roos, W.H. AFM Nanoindentation of Protein Shells, Expanding the Approach beyond Viruses. Semin. Cell Dev. Biol. 2018, 73, 145–152. [Google Scholar] [CrossRef]
- Bhella, D. The Role of Cellular Adhesion Molecules in Virus Attachment and Entry. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140035. [Google Scholar] [CrossRef]
- Valente, A.P.; Moraes, A.H. Zika Virus Proteins at an Atomic Scale: How Does Structural Biology Help Us to Understand and Develop Vaccines and Drugs against Zika Virus Infection? J. Venom. Anim. Toxins Incl. Trop. Dis. 2019, 25, e20190013. [Google Scholar] [CrossRef]
- Dai, L.; Song, J.; Lu, X.; Deng, Y.-Q.; Musyoki, A.M.; Cheng, H.; Zhang, Y.; Yuan, Y.; Song, H.; Haywood, J.; et al. Structures of the Zika Virus Envelope Protein and Its Complex with a Flavivirus Broadly Protective Antibody. Cell Host Microbe 2016, 19, 696–704. [Google Scholar] [CrossRef]
- Sirohi, D.; Chen, Z.; Sun, L.; Klose, T.; Pierson, T.C.; Rossmann, M.G.; Kuhn, R.J. The 3.8 Å Resolution Cryo-EM Structure of Zika Virus. Science 2016, 352, 467–470. [Google Scholar] [CrossRef]
- Roos, W.H.; Bruinsma, R.; Wuite, G.J.L. Physical Virology. Nat. Phys. 2010, 6, 733–743. [Google Scholar] [CrossRef]
- Hellwig, J.; Karlsson, R.-M.P.; Wågberg, L.; Pettersson, T. Measuring Elasticity of Wet Cellulose Beads with an AFM Colloidal Probe Using a Linearized DMT Model. Anal. Methods 2017, 9, 4019–4022. [Google Scholar] [CrossRef]
- Aguayo, S.; Bozec, L. Mechanics of Bacterial Cells and Initial Surface Colonisation. In Biophysics of Infection; Leake, M.C., Ed.; Advances in Experimental Medicine and Biology; Springer International Publishing: Cham, Switzerland, 2016; pp. 245–260. ISBN 978-3-319-32189-9. [Google Scholar]
- Grzeszczuk, Z.; Rosillo, A.; Owens, Ó.; Bhattacharjee, S. Atomic Force Microscopy (AFM) As a Surface Mapping Tool in Microorganisms Resistant Toward Antimicrobials: A Mini-Review. Front. Pharmacol. 2020, 11, 517165. [Google Scholar] [CrossRef] [PubMed]
- Usselman, R.J.; Qazi, S.; Aggarwal, P.; Eaton, S.S.; Eaton, G.R.; Russek, S.; Douglas, T. Gadolinium-Loaded Viral Capsids as Magnetic Resonance Imaging Contrast Agents. Appl. Magn. Reson. 2015, 46, 349–355. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Oltrogge, L.M.; Broomell, C.C.; Liepold, L.O.; Prevelige, P.E.; Young, M.; Douglas, T. Controlled Assembly of Bifunctional Chimeric Protein Cages and Composition Analysis Using Noncovalent Mass Spectrometry. J. Am. Chem. Soc. 2008, 130, 16527–16529. [Google Scholar] [CrossRef] [PubMed]
- Schlicksup, C.J.; Zlotnick, A. Viral Structural Proteins as Targets for Antivirals. Curr. Opin. Virol. 2020, 45, 43–50. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Araujo Dorneles, M.L.; Cardoso-Lima, R.; Souza, P.F.N.; Santoro Rosa, D.; Magne, T.M.; Santos-Oliveira, R.; Alencar, L.M.R. Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties. Viruses 2022, 14, 1727. https://doi.org/10.3390/v14081727
de Araujo Dorneles ML, Cardoso-Lima R, Souza PFN, Santoro Rosa D, Magne TM, Santos-Oliveira R, Alencar LMR. Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties. Viruses. 2022; 14(8):1727. https://doi.org/10.3390/v14081727
Chicago/Turabian Stylede Araujo Dorneles, Maria Luiza, Ruana Cardoso-Lima, Pedro Filho Noronha Souza, Daniela Santoro Rosa, Tais Monteiro Magne, Ralph Santos-Oliveira, and Luciana Magalhães Rebelo Alencar. 2022. "Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties" Viruses 14, no. 8: 1727. https://doi.org/10.3390/v14081727
APA Stylede Araujo Dorneles, M. L., Cardoso-Lima, R., Souza, P. F. N., Santoro Rosa, D., Magne, T. M., Santos-Oliveira, R., & Alencar, L. M. R. (2022). Zika Virus (ZIKV): A New Perspective on the Nanomechanical and Structural Properties. Viruses, 14(8), 1727. https://doi.org/10.3390/v14081727






