The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
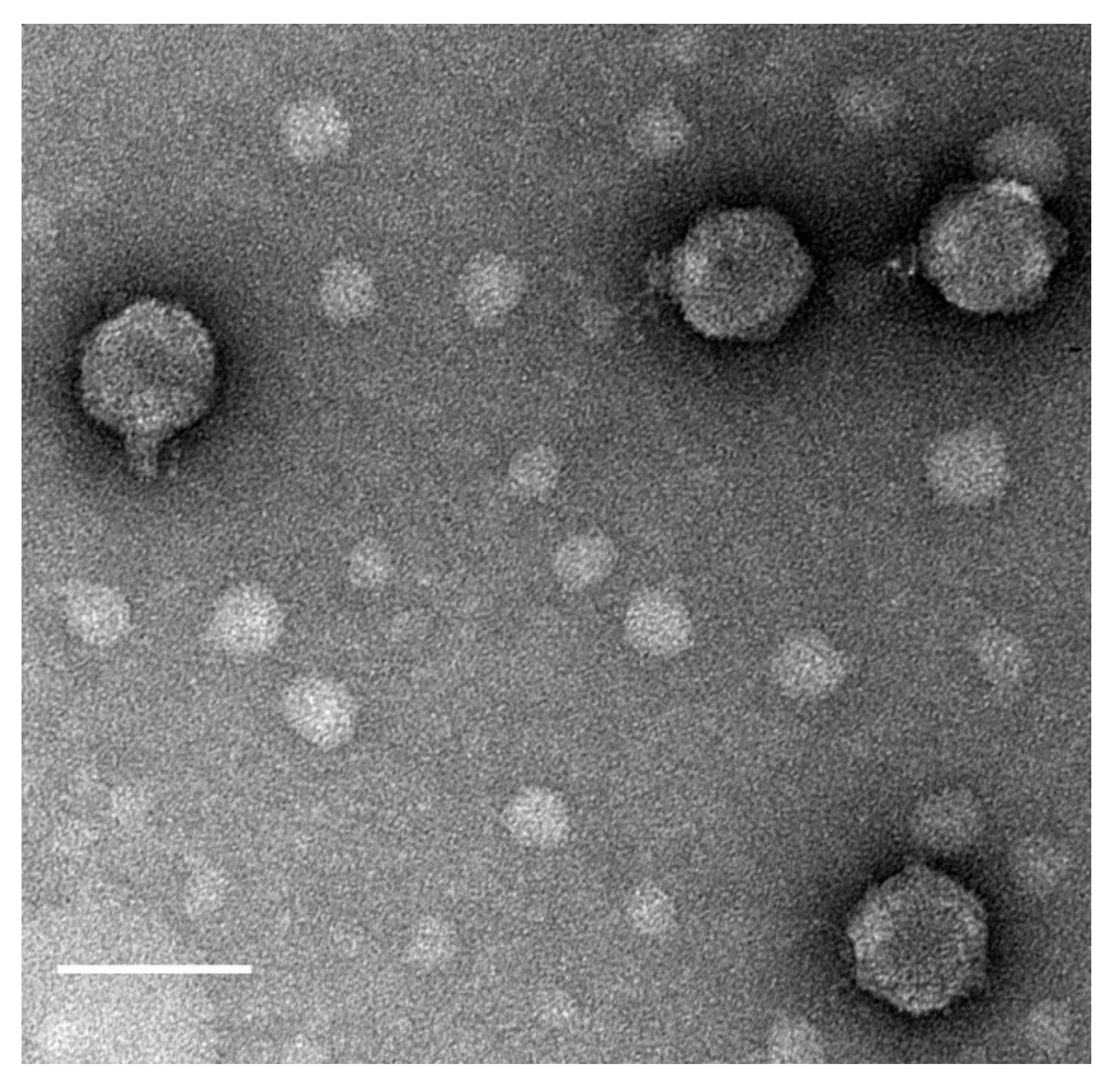
2.2. Phage Isolation, Propagation, Host Range Analysis, and Electron Microscopy
2.3. Phage DNA Isolation and Sequencing
2.4. Bioinformatic Analysis of JC1
2.5. Mass Spectrometry Analysis of JC1
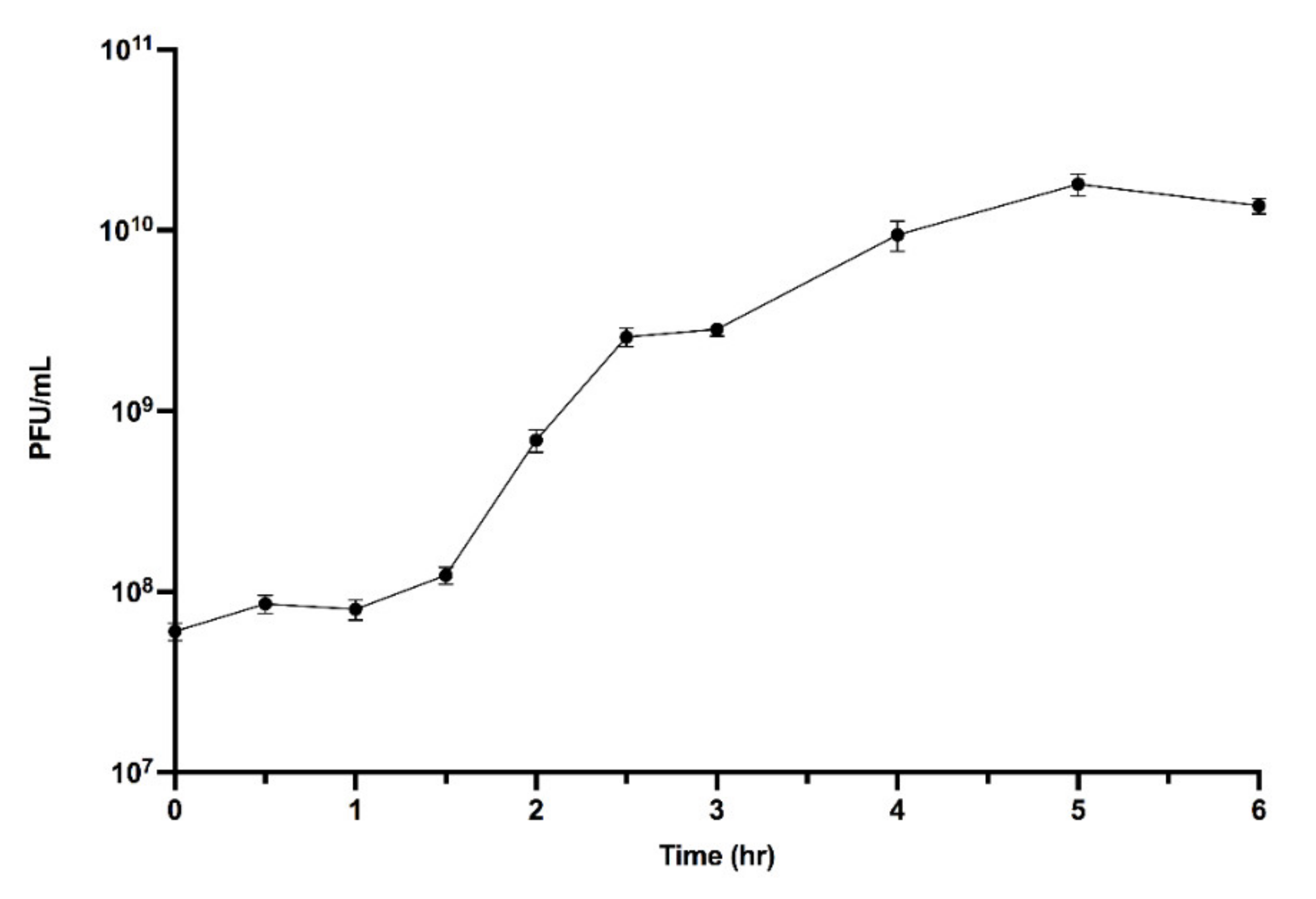
2.6. One-Step Growth Curve
2.7. Complementation of LPS Mutants
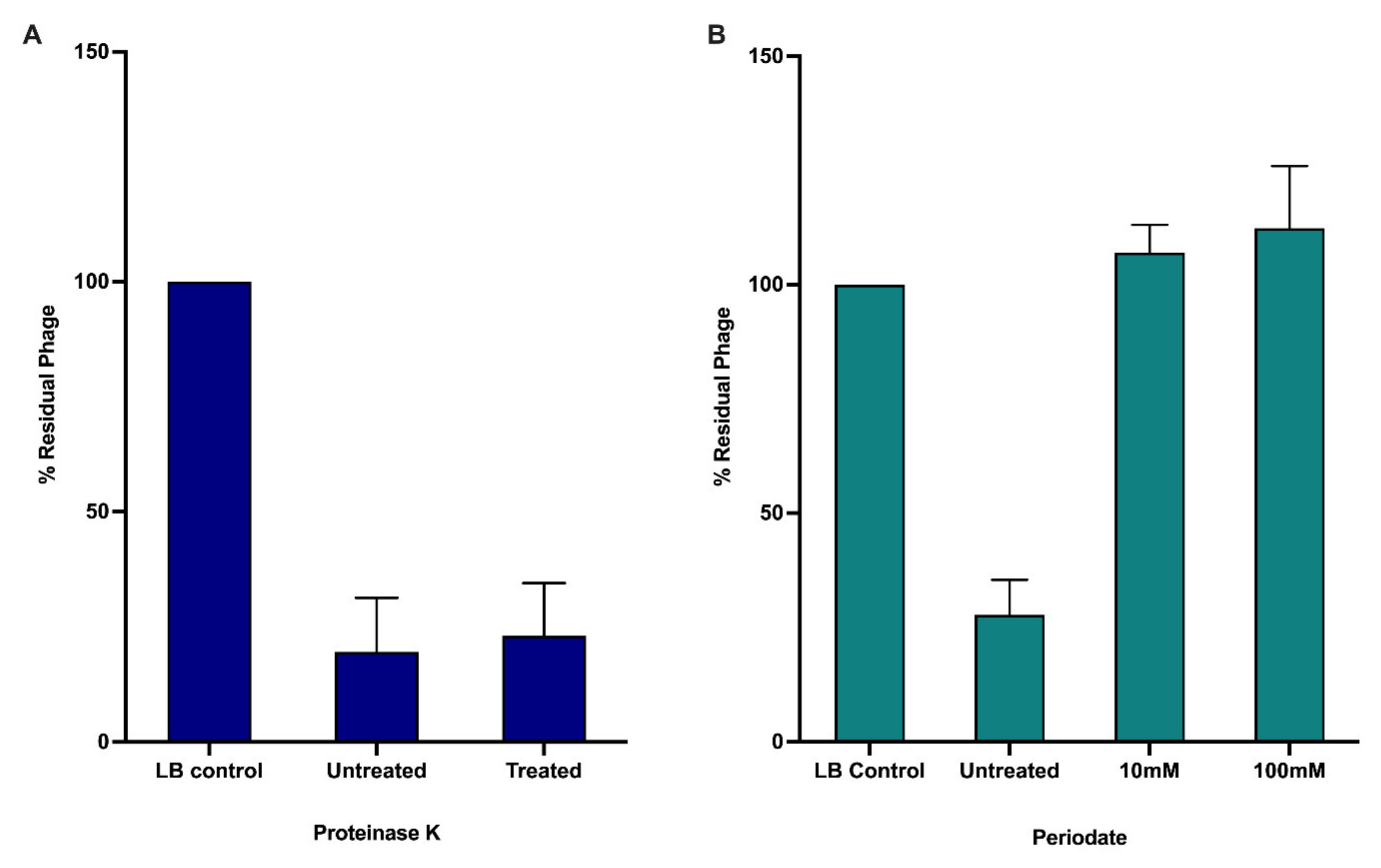
2.8. Identification of Phage Receptor
2.9. Determination of JC1 Lifestyle and Integration Site
2.10. Growth Analysis of Van1 vs. JC1 Lysogen and JC1 Virulence Index
2.11. Virulence Assay Using Galleria mellonella
2.12. Normal Human Serum (NHS) Percent Survival
3. Results and Discussion
3.1. Isolation, Morphology, and Host Range
3.2. Receptor Binding
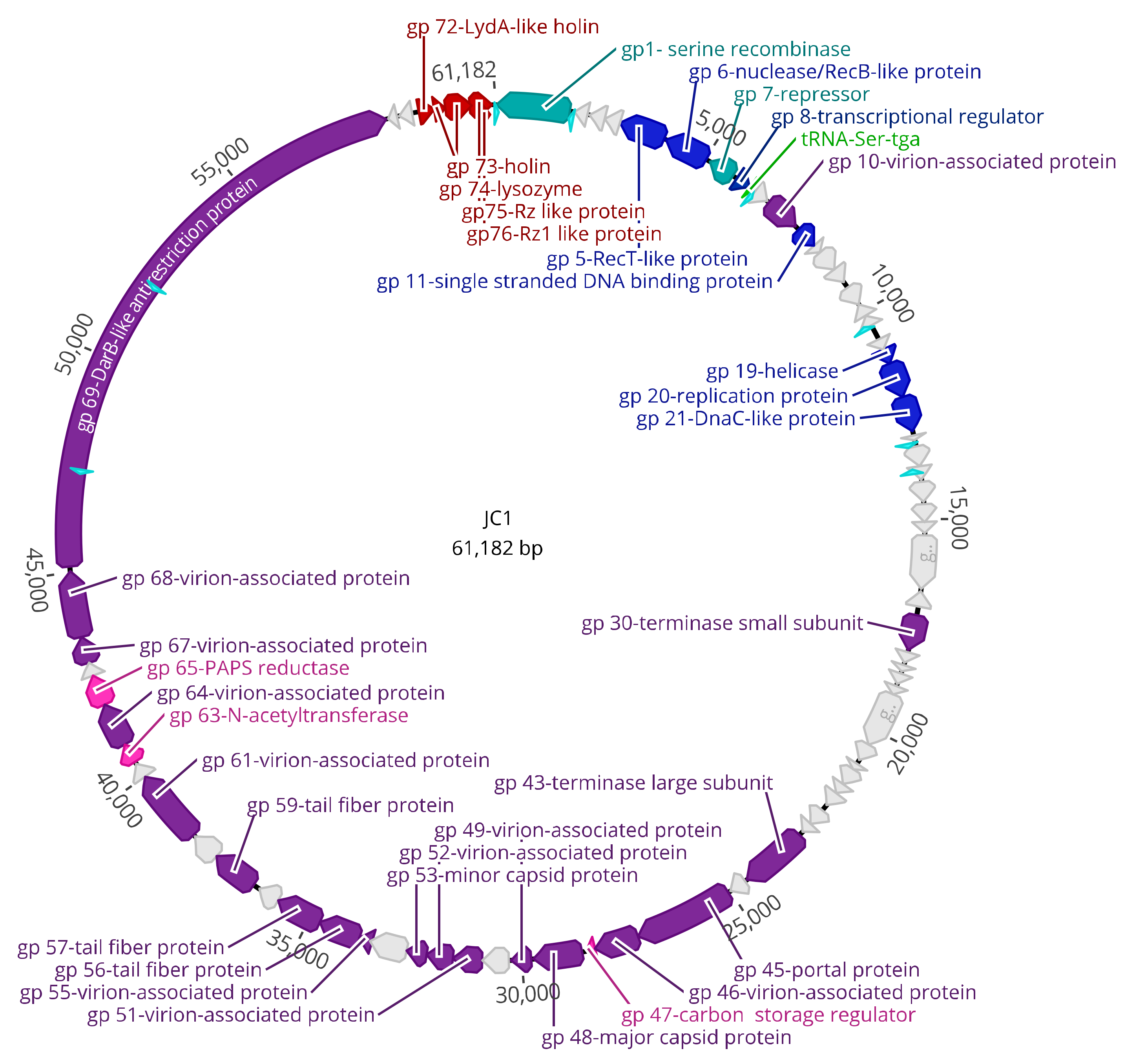
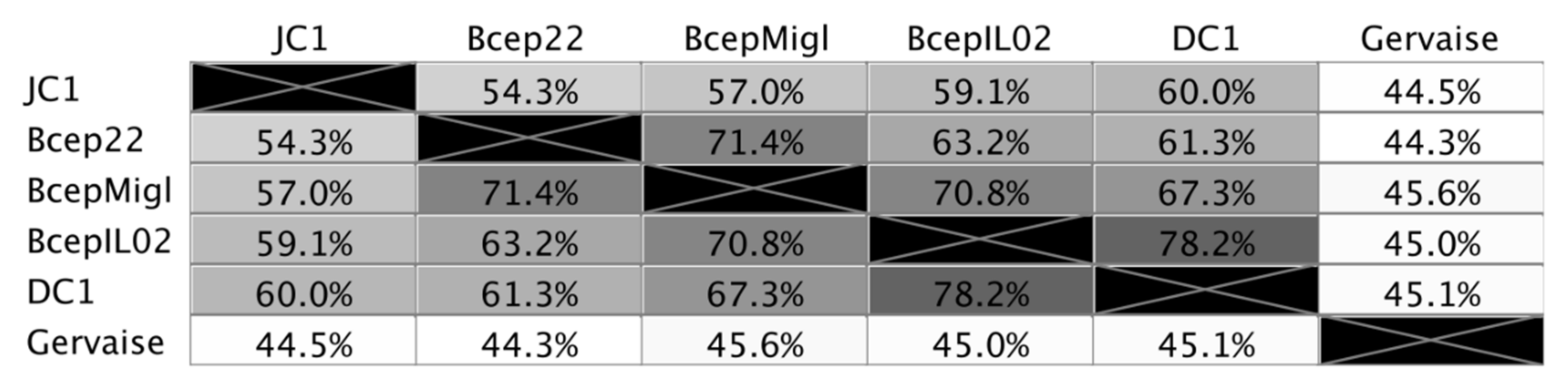
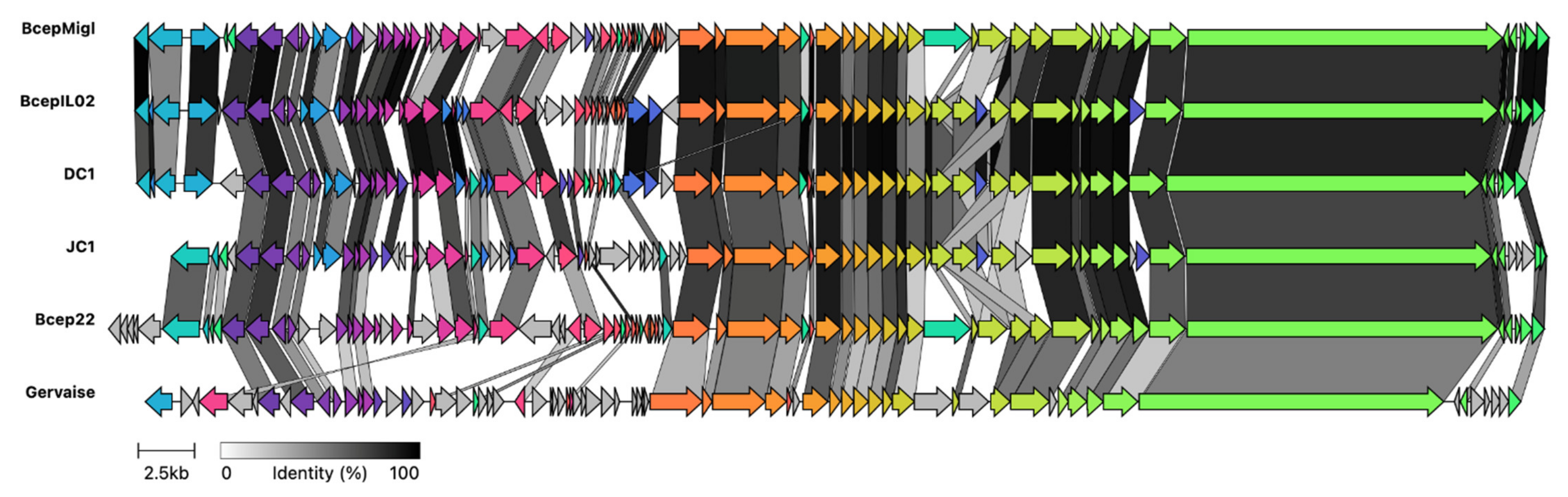
3.3. Genomic Characterization
3.4. DNA Replication, Repair, and Regulation Module
3.5. Virion Morphogenesis Module
3.6. Lysis Module
3.7. Moron Genes
3.8. Analysis of JC1 Structural Proteins
3.9. Integration Site Characterization
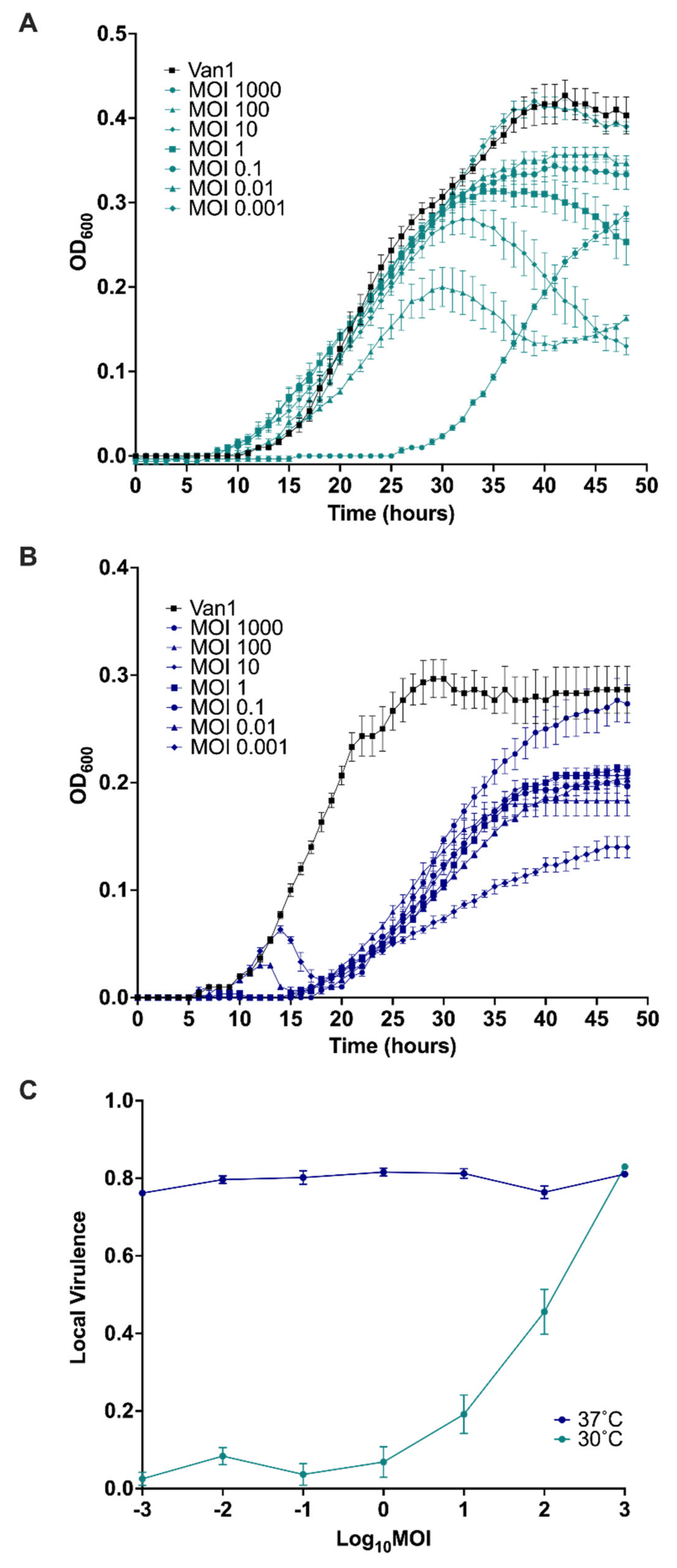
3.10. Virulence Index of JC1
3.11. Lysogenic Conversion
3.12. Comparison of Van1 & Van1::JC1 in Galleria mellonella and Normal Human Serum (NHS)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- O’Neil, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. The Review on Antimicrobial Resistance; Government of the United Kingdom: London, UK, 2016; Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 1 March 2020).
- Council of Canadian Academies. When Antibiotics Fail: The Expert Panel on the Potential Socio-Economic Impacts of Antimicrobial Resistance in Canada; Council of Canadian Academies: Ottawa, ON, Canada, 2019. [Google Scholar]
- Mahenthiralingam, E.; Urban, T.A.; Goldberg, J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005, 3, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Scoffone, V.C.; Chiarelli, L.R.; Trespidi, G.; Mentasti, M.; Riccardi, G.; Buroni, S. Burkholderia cenocepacia infections in cystic fibrosis patients: Drug resistance and therapeutic approaches. Front. Microbiol. 2017, 8, 1592. [Google Scholar] [CrossRef] [PubMed]
- Cystic Fibrosis Canada-The Canadian Cystic Fibrosis Registry 2019 Annual Data Report. Available online: https://www.cysticfibrosis.ca/registry/2019AnnualDataReport.pdf. (accessed on 20 January 2021).
- Gordillo Altamirano, F.L.; Barr, J.J. Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed]
- Davis, C.M.; McCutcheon, J.G.; Dennis, J.J. Aztreonam lysine increases the activity of phages E79 and PhiKZ against Pseudomonas aeruginosa PA01. Microorganisms 2021, 9, 152. [Google Scholar] [CrossRef]
- Kamal, F.; Dennis, J.J. Burkholderia cepacia complex phage-antibiotic synergy (PAS): Antibiotics stimulate lytic phage activity. Appl. Environ. Microbiol. 2015, 81, 1132–1138. [Google Scholar] [CrossRef]
- Comeau, A.M.; Tétart, F.; Trojet, S.N.; Prère, M.F.; Krisch, H.M. Phage-antibiotic synergy (PAS): β-lactam and quinolone antibiotics stimulate virulent phage growth. PLoS ONE 2007, 2, e799. [Google Scholar] [CrossRef]
- Chaudhry, W.N.; Concepción-Acevedo, J.; Park, T.; Andleeb, S.; Bull, J.J.; Levin, B.R. Synergy and order effects of antibiotics and phages in killing Pseudomonas aeruginosa biofilms. PLoS ONE 2017, 12, e0168615. [Google Scholar] [CrossRef]
- Seed, K.D.; Dennis, J.J. Isolation and characterization of bacteriophages of the Burkholderia cepacia complex. FEMS Microbiol. Lett. 2005, 251, 273–280. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Coenye, T.; Chung, J.W.; Speert, D.P.; Govan, J.R.W.; Taylor, P.; Vandamme, P. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 2000, 38, 910–913. [Google Scholar] [CrossRef]
- Gonzalez, C.F.; Pettit, E.A.; Valadez, V.A.; Provin, E.M. Mobilization, cloning, and sequence determination of a plasmid-encoded polygalacturonase from a phytopathogenic Burkholderia (Pseudomonas) cepacia. Mol. Plant Microbe Interact. 1997, 10, 840–851. [Google Scholar] [CrossRef] [PubMed]
- McKenney, D.; Brown, K.E.; Allison, D.G. Influence of Pseudomonas aeruginosa exoproducts on virulence factor production in Burkholderia cepacia: Evidence of interspecies communication. J. Bacteriol. 1995, 177, 6989. [Google Scholar] [CrossRef] [PubMed]
- Stanier, R.Y.; Palleroni, N.J.; Doudoroff, M. The Aerobic Pseudomonads: A taxonomic study. J. Gen. Microbiol. 1966, 43, 159–271. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.P.; Lessie, T.G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J. Bacteriol. 1994, 176, 4034–4042. [Google Scholar] [CrossRef] [PubMed]
- Gaffney, T.D.; Lessie, T.G. Insertion-sequence-dependent rearrangements of Pseudomonas cepacia plasmid PTGL1. J. Bacteriol. 1987, 169, 224–230. [Google Scholar] [CrossRef]
- Vandamme, P.; Holmes, B.; Vancanneyt, M.; Coenye, T.; Hoste, B.; Coopman, R.; Revets, H.; Lauwers, S.; Gillis, M.; Kersters, K.; et al. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 1997, 47, 1188–1200. [Google Scholar] [CrossRef]
- Chu, K.K.; Davidson, D.J.; Halsey, T.K.; Chung, J.W.; Speert, D.P. Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect. Immun. 2002, 70, 2715–2720. [Google Scholar] [CrossRef]
- Whiteford, M.L.; Wilkinson, J.D.; McColl, J.H.; Conlon, F.M.; Michie, J.R.; Evans, T.J.; Paton, J.Y. Outcome of Burkholderia (Pseudomonas) cepacia colonisation in children with cystic fibrosis following a hospital outbreak. Thorax 1995, 50, 1194–1198. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Campbell, M.E.; Henry, D.A.; Speert, D.P. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: Analysis by randomly amplified polymorphic DNA fingerprinting. J. Clin. Microbiol. 1996, 34, 2914–2920. [Google Scholar] [CrossRef]
- Revets, H.; Vandamme, P.; Van Zeebroeck, A.; De Boeck, K.; Struelens, M.J.; Verhaegen, J.; Ursi, J.P.; Verschraegen, G.; Franckx, H.; Malfroot, A.; et al. Burkholderia (Pseudomonas) cepacia and cystic fibrosis: The epidemiology in Belgium. Acta Clin. Belg. 1996, 51, 222–230. [Google Scholar] [CrossRef]
- Lynch, K.H.; Dennis, J.J. Development of a species-specific Fur gene-based method for identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 2008, 46, 447–455. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McKevitt, A.I.; Bajaksouzian, S.; Klinger, J.D.; Woods, D.E. Purification and characterization of an extracellular protease from Pseudomonas cepacia. Infect. Immun. 1989, 57, 771. [Google Scholar] [CrossRef] [PubMed]
- Mahenthiralingam, E.; Simpson, D.A.; Speert, D.P. Identification and characterization of a novel DNA marker associated with epidemic Burkholderia cepacia strains recovered from patients with cystic fibrosis. J. Clin. Microbiol. 1997, 35, 808. [Google Scholar] [CrossRef] [PubMed]
- Conway, B.A.D.; Chu, K.K.; Bylund, J.; Altman, E.; Speert, D.P. Production of exopolysaccharide by Burkholderia cenocepacia results in altered cell-surface interactions and altered bacterial clearance in mice. J. Infect. Dis. 2004, 190, 957–966. [Google Scholar] [CrossRef]
- Govan, J.R.W.; Doherty, C.J.; Nelson, J.W.; Brown, P.H.; Greening, A.P.; Maddison, J.; Dodd, M.; Webb, A.K. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 1993, 342, 15–19. [Google Scholar] [CrossRef]
- Hutchison, M.L.; Poxton, I.R.; Govan, J.R.W. Burkholderia cepacia produces a hemolysin that is capable of inducing apoptosis and degranulation of mammalian phagocytes. Infect. Immun. 1998, 66, 2033. [Google Scholar] [CrossRef]
- Lewenza, S.; Conway, B.; Greenberg, E.P.; Sokol, P.A. Quorum sensing in Burkholderia cepacia: Identification of the LuxRI homologs CepRI. J. Bacteriol. 1999, 181, 748. [Google Scholar] [CrossRef]
- Balandreau, J.; Viallard, V.; Cournoyer, B.; Coenye, T.; Laevens, S.; Vandamme, P. Burkholderia cepacia genomovar III is a common plant-associated bacterium. Appl. Environ. Microbiol. 2001, 67, 982–985. [Google Scholar] [CrossRef]
- LiPuma, J.J.; Spilker, T.; Coenye, T.; Gonzalez, C.F. An epidemic Burkholderia cepacia complex strain identified in soil. Lancet 2002, 359, 2002–2003. [Google Scholar] [CrossRef]
- LiPuma, J.J.; Mortensen, J.E.; Dasen, S.E.; Edlind, T.D.; Schidlow, D.V.; Burns, J.L.; Stull, T.L. Ribotype analysis of Pseudomonas cepacia from cystic fibrosis treatment centers. J. Pediatr. 1988, 113, 859–862. [Google Scholar] [CrossRef]
- Walsh, T.A.; Ballou, D.P. Halogenated protocatechuates as substrates for protocatechuate dioxygenase from Pseudomonas cepacia. J. Biol. Chem. 1983, 258, 14413–14421. [Google Scholar] [CrossRef]
- Gillis, M.; Van Van, T.; Bardin, R.; Goor, M.; Hebbar, P.; Willems, A.; Segers, P.; Kersters, K.; Heulin, T.; Fernandez, M.P. Polyphasic taxonomy in the genus Burkholderia leading to an emended description of the genus and proposition of Burkholderia vietnamiensis Sp. Nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 1995, 45, 274–289. [Google Scholar] [CrossRef]
- Burns, J.L.; Jonas, M.; Chi, E.Y.; Clark, D.K.; Berger, A.; Griffith, A. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 1996, 64, 4054–4059. [Google Scholar] [CrossRef] [PubMed]
- Larsen, G.Y.; Stull, T.L.; Burns, J.L. Marked phenotypic variability in Pseudomonas cepacia isolated from a patient with cystic fibrosis. J. Clin. Microbiol. 1993, 31, 788. [Google Scholar] [CrossRef] [PubMed]
- Yeager, C.M.; Bottomley, P.J.; Arp, D.J.; Hyman, M.R. Inactivation of toluene 2-monooxygenase in Burkholderia cepacia G4 by alkynes. Appl. Environ. Microbiol. 1999, 65, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T.; Vandamme, P.; LiPuma, J.J.; Govan, J.R.W.; Mahenthiralingam, E. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 2003, 41, 2797. [Google Scholar] [CrossRef]
- Mahenthiralingam, E.; Baldwin, A.; Dowson, C.G. Burkholderia cepacia complex bacteria: Opportunistic pathogens with important natural biology. J. Appl. Microbiol. 2008, 104, 1539–1551. [Google Scholar] [CrossRef]
- Palleroni, N.J.; Ballard, R.W.; Ralston, E.; Doudoroff, M. Deoxyribonucleic acid homologies among some Pseudomonas species. J. Bacteriol. 1972, 110, 1–11. [Google Scholar] [CrossRef]
- Yabannavar, A.V.; Zylstra, G.J. Cloning and characterization of the genes for p-nitrobenzoate degradation from Pseudomonas pickettii YH105. Appl. Environ. Microbiol. 1995, 61, 4284–4290. [Google Scholar] [CrossRef]
- McCutcheon, J.; Peters, D.; Dennis, J. Identification and characterization of type IV pili as the cellular receptor of broad host range Stenotrophomonas maltophilia bacteriophages DLP1 and DLP2. Viruses 2018, 10, 338. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Delcher, A.L.; Bratke, K.A.; Powers, E.C.; Salzberg, S.L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 2007, 23, 673–679. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinf. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; Von Heijne, G.; Sonnhammer, E.L.L. Predicting transmembrane protein topology with a hidden Markov model:Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gíslason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 2022, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.; Sulakvelidze, A. (Eds.) Working with Bacteriophages: Common Techniques and Methodological Approaches; Bacteriophages; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Kiljunen, S.; Datta, N.; Dentovskaya, S.V.; Anisimov, A.P.; Knirel, Y.A.; Bengoechea, J.A.; Holst, O.; Skurnik, M. Identification of the lipopolysaccharide core of Yersinia pestis and Yersinia pseudotuberculosis as the receptor for bacteriophage ΦA1122. J. Bacteriol. 2011, 193, 4963. [Google Scholar] [CrossRef]
- Williams, R.; Meader, E.; Mayer, M.; Narbad, A.; Roberts, A.P.; Mullany, P. Determination of the AttP and AttB sites of phage ΦCD27 from Clostridium difficile NCTC 12727. J. Med. Microbiol. 2013, 62, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Peters, D.L.; McCutcheon, J.G.; Dennis, J.J. Characterization of novel broad-host-range bacteriophage DLP3 specific to Stenotrophomonas maltophilia as a potential therapeutic agent. Front. Microbiol. 2020, 11, 1358. [Google Scholar] [CrossRef] [PubMed]
- Duong-Nu, T.M.; Jeong, K.; Hong, S.H.; Puth, S.; Kim, S.Y.; Tan, W.; Lee, K.H.; Lee, S.E.; Rhee, J.H. A stealth adhesion factor contributes to Vibrio vulnificus pathogenicity: Flp pili play roles in host invasion, survival in the bloodstream and resistance to complement activation. PLoS Pathog. 2019, 15, e1007767. [Google Scholar] [CrossRef] [PubMed]
- McKay, G.A.; Woods, D.E.; MacDonald, K.L.; Poole, K. Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect. Immun. 2003, 71, 3068. [Google Scholar] [CrossRef]
- Ackermann, H.-W. Frequency of morphological phage descriptions in the year 2000-Brief Review. Arch. Virol. 2001, 146, 843–857. [Google Scholar] [CrossRef]
- Gill, J.J.; Summer, E.J.; Russell, W.K.; Cologna, S.M.; Carlile, T.M.; Fuller, A.C.; Kitsopoulos, K.; Mebane, L.M.; Parkinson, B.N.; Sullivan, D.; et al. Genomes and characterization of phages Bcep22 and BcepIL02, founders of a novel phage type in Burkholderia cenocepacia. J. Bacteriol. 2011, 193, 5300–5313. [Google Scholar] [CrossRef]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Characterization of DC1, a broad-host-range Bcep22-like podovirus. Appl. Environ. Microbiol. 2012, 78, 889–891. [Google Scholar] [CrossRef]
- Ragupathi, N.K.D.; Veeraraghavan, B. Accurate identification and epidemiological characterization of Burkholderia cepacia complex: An update. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 7. [Google Scholar] [CrossRef]
- Lauman, P.; Dennis, J.J. Advances in phage therapy: Targeting the Burkholderia cepacia complex. Viruses. 2021, 13, 1331. [Google Scholar] [CrossRef] [PubMed]
- Loutet, S.A.; Flannagan, R.S.; Kooi, C.; Sokol, P.A.; Valvano, M.A. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J. Bacteriol. 2006, 188, 2073–2080. [Google Scholar] [CrossRef] [PubMed]
- Ortega, X.; Silipo, A.; Saldfas, M.S.; Bates, C.C.; Molinaro, A.; Valvano, M.A. Biosynthesis and structure of the Burkholderia cenocepacia K56-2 lipopolysaccharide core oligosaccharide: Truncation of the core oligosaccharide leads to increased binding and sensitvity to polymyxin B. J. Biol. Chem. 2009, 284, 21738–21751. [Google Scholar] [CrossRef] [PubMed]
- Holden, M.T.G.; Seth-Smith, H.M.B.; Crossman, L.C.; Sebaihia, M.; Bentley, S.D.; Cerdeño-Tárraga, A.M.; Thomson, N.R.; Bason, N.; Quail, M.A.; Sharp, S.; et al. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J. Bacteriol. 2009, 91, 261–277. [Google Scholar] [CrossRef]
- Gautheret, D.; Lambert, A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001, 313, 1003–1011. [Google Scholar] [CrossRef]
- Macke, T.J.; Ecker, D.J.; Gutell, R.R.; Gautheret, D.; Case, D.A.; Sampath, R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001, 29, 4724–4735. [Google Scholar] [CrossRef]
- Lesnik, E.A.; Sampath, R.; Levene, H.B.; Henderson, T.J.; McNeil, J.A.; Ecker, D.J. Prediction of Rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001, 29, 3583. [Google Scholar] [CrossRef]
- Hofacker, I.L.; Fontana, W.; Stadler, P.F.; Bonhoeffer, L.S.; Tacker, M.; Schuster, P. Fast folding and comparison of RNA secondary structures. Monatshefte für Chem./Chem. Mon. 1994, 125, 167–188. [Google Scholar] [CrossRef]
- Weigel, C.; Seitz, H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006, 30, 321–381. [Google Scholar] [CrossRef]
- Summer, E.J.; Gonzalez, C.F.; Carlisle, T.; Mebane, L.M.; Cass, A.M.; Savva, C.G.; LiPuma, J.J.; Young, R. Burkholderia cenocepacia phage BcepMu and a family of Mu-like phages encoding potential pathogenesis factors. J. Mol. Biol. 2004, 340, 49–65. [Google Scholar] [CrossRef]
- Casjens, S.R. Diversity among the tailed-bacteriophages that infect the Enterobacteriaceae. Res. Microbiol. 2008, 159, 340. [Google Scholar] [CrossRef] [PubMed]
- Goudie, A.D.; Lynch, K.H.; Seed, K.D.; Stothard, P.; Shrivastava, S.; Wishart, D.S.; Dennis, J.J. Genomic sequence and activity of KS10, a transposable phage of the Burkholderia cepacia complex. BMC Genom. 2008, 9, 615. [Google Scholar] [CrossRef] [PubMed]
- Lynch, K.H.; Abdu, A.H.; Schobert, M.; Dennis, J.J. Genomic characterization of JG068, a novel virulent Podovirus active against Burkholderia cenocepacia. BMC Genom. 2013, 14, 574. [Google Scholar] [CrossRef] [PubMed]
- Roszniowski, B.; Latka, A.; Maciejewska, B.; Vandenheuvel, D.; Olszak, T.; Briers, Y.; Holt, G.S.; Valvano, M.A.; Lavigne, R.; Smith, D.L.; et al. The temperate Burkholderia phage AP3 of the Peduovirinae shows efficient antimicrobial activity against B. cenocepacia of the IIIA Lineage. Appl. Microbiol. Biotechnol. 2017, 101, 1203–1216. [Google Scholar] [CrossRef]
- Lynch, K.H.; Liang, Y.; Eberl, L.; Wishart, D.S.; Dennis, J.J. Identification and characterization of ΦH111-1. Bacteriophage 2013, 3, e26649. [Google Scholar] [CrossRef][Green Version]
- Lynch, K.H.; Stothard, P.; Dennis, J.J. Genomic analysis and relatedness of P2-like phages of the Burkholderia cepacia complex. BMC Genom. 2010, 11, 599. [Google Scholar] [CrossRef]
- Summer, E.J.; Gonzalez, C.F.; Bomer, M.; Carlile, T.; Embry, A.; Kucherka, A.M.; Lee, J.; Mebane, L.; Morrison, W.C.; Mark, L.; et al. Divergence and mosaicism among virulent soil phages of the Burkholderia cepacia complex. J. Bacteriol. 2006, 188, 255. [Google Scholar] [CrossRef]
- Iida, S.; Streiff, M.B.; Bickle, T.A.; Arber, W. Two DNA antirestriction systems of bacteriophage P1, DarA, and DarB: Characterization of DarA- phages. Virology 1987, 157, 156–166. [Google Scholar] [CrossRef]
- Piya, D.; Vara, L.; Russell, W.K.; Young, R.; Gill, J.J. The multicomponent antirestriction system of phage P1 is linked to capsid morphogenesis. Mol. Microbiol. 2017, 105, 399. [Google Scholar] [CrossRef]
- Summer, E.J.; Berry, J.; Tran, T.A.T.; Niu, L.; Struck, D.K.; Young, R. Rz/Rz1 lysis gene equivalents in phages of gram-negative hosts. J. Mol. Biol. 2007, 373, 1098–1112. [Google Scholar] [CrossRef]
- Reddy, B.L.; Saier, M.H. Topological and phylogenetic analyses of bacterial holin families and superfamilies. Biochim. Biophys. Acta 2013, 1828, 2654. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Velleman, M.; Arber, W. Three functions of bacteriophage P1 involved in cell lysis. J. Bacteriol. 1996, 178, 1099. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Vukov, N.; Moll, I.; Bläsi, U.; Scherer, S.; Loessner, M.J. Functional regulation of the Listeria monocytogenes bacteriophage A118 holin by an intragenic inhibitor lacking the first transmembrane domain. Mol. Microbiol. 2003, 48, 173–186. [Google Scholar] [CrossRef]
- Sheehan, M.M.; Stanley, E.; Fitzgerald, G.F.; Van Sinderen, D. Identification and characterization of a lysis module present in a large proportion of bacteriophages infecting Streptococcus thermophilus. Appl. Environ. Microbiol. 1999, 65, 569. [Google Scholar] [CrossRef]
- Vermassen, A.; Leroy, S.; Talon, R.; Provot, C.; Popowska, M.; Desvaux, M. Cell wall hydrolases in bacteria: Insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 2019, 10, 331. [Google Scholar] [CrossRef]
- Palmer, T.; Finney, A.J.; Saha, C.K.; Atkinson, G.C.; Sargent, F. A holin/peptidoglycan hydrolase-dependent protein secretion system. Mol. Microbiol. 2021, 115, 345–355. [Google Scholar] [CrossRef]
- Park, T.; Struck, D.K.; Dankenbring, C.A.; Young, R. The pinholin of lambdoid phage 21: Control of lysis by membrane depolarization. J. Bacteriol. 2007, 189, 9135. [Google Scholar] [CrossRef]
- Pourciau, C.; Lai, Y.J.; Gorelik, M.; Babitzke, P.; Romeo, T. Diverse mechanisms and circuitry for global regulation by the RNA-binding protein CsrA. Front. Microbiol. 2020, 11, 2709. [Google Scholar] [CrossRef]
- Tanaka, S.; Matsushita, Y.; Yoshikawa, A.; Isono, K. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol. Gen. Genet. 1989, 217, 289–293. [Google Scholar] [CrossRef]
- Vetting, M.W.; Magnet, S.; Nieves, E.; Roderick, S.L.; Blanchard, J.S. A bacterial acetyltransferase capable of regioselective N-acetylation of antibiotics and histones. Chem. Biol. 2004, 11, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Chartron, J.; Shiau, C.; Stout, C.D.; Carroll, K.S. 3′-phosphoadenosine-5′-phosphosulfate reductase in complex with thioredoxin: A structural snapshot in the catalytic cycle. Biochemistry 2007, 46, 3942. [Google Scholar] [CrossRef] [PubMed]
- Silverstein, J.L.; Goldberg, E.B. T4 DNA Injection. II. Protection of Entering DNA from Host Exonuclease, V. Virology 1976, 72, 212–223. [Google Scholar] [CrossRef]
- Silverstein, J.L.; Goldberg, E.B. T4 DNA Injection. I. Growth cycle of a gene 2 mutant. Virology 1976, 72, 195–211. [Google Scholar] [CrossRef]
- Anton, B.P.; Saleh, L.; Benner, J.S.; Raleigh, E.A.; Kasif, S.; Roberts, R.J. RimO, a MiaB-like enzyme, methylthiolates the universally conserved Asp88 residue of ribosomal protein S12 in Escherichia Coli. Proc. Natl. Acad. Sci. USA 2008, 105, 1826–1831. [Google Scholar] [CrossRef]
- Kurland, C.; Hughes, D.; Ehrenberg, M. Escherichia Coli and Salmonella: Cellular and Molecular Biology; American Association for the Advancement of Science: Washington, DC, USA, 1996; Volume 240, pp. 979–1004. [Google Scholar]
- Carr, J.F.; Hamburg, D.M.; Gregory, S.T.; Limbach, P.A.; Dahlberg, A.E. Effects of streptomycin resistance mutations on posttranslational modification of ribosomal protein S12. J. Bacteriol. 2006, 188, 2020–2023. [Google Scholar] [CrossRef]
- Storms, Z.J.; Teel, M.R.; Mercurio, K.; Sauvageau, D. The virulence index: A metric for quantitative analysis of phage virulence. Ther. Appl. Res. 2020, 1, 27–36. [Google Scholar] [CrossRef]
- Shan, J.; Korbsrisate, S.; Withatanung, P.; Adler, N.L.; Clokie, M.R.J.; Galyov, E.E. Temperature dependent bacteriophages of a tropical bacterial pathogen. Front. Microbiol. 2014, 5, 599. [Google Scholar] [CrossRef]
- Lynch, K.H.; Seed, K.D.; Stothard, P.; Dennis, J.J. Inactivation of Burkholderia cepacia complex phage KS9 gp41 identifies the phage repressor and generates lytic virions. J. Virol. 2010, 84, 1276–1288. [Google Scholar] [CrossRef]
- Boyd, E.F.; Brüssow, H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002, 10, 521–529. [Google Scholar] [CrossRef]
- Summer, E.J.; Gill, J.J.; Upton, C.; Gonzalez, C.F.; Young, R. Role of phages in the pathogenesis of Burkholderia or “Where are the toxin genes in Burkholderia phages?”. Curr. Opin. Microbiol. 2007, 10, 410. [Google Scholar] [CrossRef] [PubMed]
- Nzula, S.; Vandamme, P.; Govan, J.R.W. Sensitivity of the Burkholderia cepacia complex and Pseudomonas aeruginosa to transducing bacteriophages. FEMS Immunol. Med. Microbiol. 2000, 28, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Langley, R.; Kenna, D.T.; Vandamme, P.; Ure, R.; Govan, J.R.W. Lysogeny and bacteriophage host range within the Burkholderia cepacia complex. J. Med. Microbiol. 2003, 52, 483–490. [Google Scholar] [CrossRef]
- Hens, D.K.; Ghosh, A.N.; Kumar, R. A new small temperate DNA phage BcP15 isolated from Burkholderia cepacia DR11. Arch. Virol. 2005, 150, 2421–2428. [Google Scholar] [CrossRef] [PubMed]
- Holt, G.S.; Lodge, J.K.; McCarthy, A.J.; Graham, A.K.; Young, G.; Bridge, S.H.; Brown, A.K.; Veses-Garcia, M.; Lanyon, C.V.; Sails, A.; et al. Shigatoxin encoding bacteriophage Φ24B modulates bacterial metabolism to raise antimicrobial tolerance. Sci. Rep. 2017, 7, 40424. [Google Scholar] [CrossRef]
- Seed, K.D.; Dennis, J.J. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect. Immun. 2008, 76, 1267–1275. [Google Scholar] [CrossRef]
- Seed, K.D.; Dennis, J.J. Experimental bacteriophage therapy increases survival of Galleria mellonella larvae infected with clinically relevant strains of the Burkholderia cepacia complex. Antimicrob. Agents Chemother. 2009, 53, 2205–2208. [Google Scholar] [CrossRef]
- Kamal, F.; Peters, D.L.; McCutcheon, J.G.; Dunphy, G.B.; Dennis, J.J. Use of greater wax moth larvae (Galleria mellonella) as an alternative animal infection model for analysis of bacterial pathogenesis. Methods Mol. Biol. 2019, 1898, 163–171. [Google Scholar]
- Dennis, J.J. Burkholderia cenocepacia virulence microevolution in the CF lung: Variations on a theme. Virulence 2017, 8, 618–620. [Google Scholar] [CrossRef]
- Govan, J.R.W.; Brown, A.R.; Jones, A.M. Evolving epidemiology of Pseudomonas aeruginosa and the Burkholderia cepacia complex in cystic fibrosis lung infection. Future Microbiol. 2007, 2, 153–164. [Google Scholar] [CrossRef]
- Walport, M.J. Complement. First of two parts. N. Engl. J. Med. 2001, 344, 1058–1066. [Google Scholar] [CrossRef] [PubMed]
- Jander, G.; Rahme, L.G.; Ausubel, F.M. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J. Bacteriol. 2000, 182, 3843–3845. [Google Scholar] [CrossRef] [PubMed]
- Brennan, M.; Thomas, D.Y.; Whiteway, M.; Kavanagh, K. Correlation between virulence of Candida albicans mutants in mice and Galleria mellonella larvae. FEMS Immunol. Med. Microbiol. 2002, 34, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.; Loh, J.M.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef]



| Burkholderia Species | Strain | Efficiency of Plating (EOP) | Source/Reference |
|---|---|---|---|
| B. cepacia | ATCC 25416 T | ND | Onion/[13,14,15] |
| ATCC 17759 | ND | Soil, Trinidad/[13,14,15,16] | |
| CEP509/LMG 18821 | 6.7 × 10−7 | CF patient, Australia/[13] | |
| CEP521 | 7.3 × 10−7 | CF patient, Canada/CBCCRRR * | |
| B. multivorans | ATCC 17616 | +++ | Soil, USA/[13,16,17,18,19] |
| C3430 | ND | CF patient, Canada/[20] | |
| C1576, LMG 16660 | ND | CF-e patient, UK/[13,19,21] | |
| C5274 | 8.0 × 10−6 | CF patient, Canada/[20] | |
| C5393 | ND | CF patient, Canada/[13,20] | |
| C5568 | ++ | CF patient, Canada/[20] | |
| JC1 | ++ | CF patient, Canada/[20] | |
| LMG 13010 T | ++ | CF patient, Belgium/[13,19,20,21,22,23] | |
| M1512 | ND | CF patient, Canada/[24] | |
| M1865 | ND | CF patient, Canada/[24] | |
| R810 | ND | CF patient, Canada/[24] | |
| R1159 | ND | CF patient, Canada/[24] | |
| B. cenocepacia | AU1054 | 0.93 | CF patient/[18] |
| 715j | ND | CF patient, USA/[25] | |
| BS1 | ++ | CF patient, Canada/This study | |
| BS2 | ++ | CF patient, Canada/This study | |
| BS3 | ++ | CF patient, Canada/This study | |
| C1257 | ++ | CF-e patient, USA/[20] | |
| C4455 | ++ | CF-e patient, Canada/[20] | |
| C5424 | 0.25 | CF-e patient, Canada/[13,21] | |
| C6433 | ND | CF-e patient, Canada/[13,21,26] | |
| C8963 | 0.3 | CF patient, Canada/[27] | |
| C9343 | ND | CF patient, Canada/[27] | |
| CEP511 | ++ | CF-e patient, Australia/[13,22] | |
| CEP0868 | 0.002 | CF patient, Argentina/[25] | |
| D1 | ND | Soil, USA/[24] | |
| HI2424 | ND | Soil, USA/[24] | |
| J2315 | 0.14 | CF-e patient, UK/[13,28,29] | |
| K56-2 | +++ | CF-e patient, Canada/[13,30] | |
| K63-3 | ND | CF-e patient, Canada/[30] | |
| LMG 19240 | ND | Wheat soil, Australia/[31] | |
| MCO-3 | ND | Maize soil, USA/[32] | |
| PC184 | 0.19 | CF-e patient, USA/[13,33] | |
| R161 | + | CF patient, Canada/[24] | |
| R452 | + | CF patient, Canada/[24] | |
| R750 | 0.9 | CF patient, Canada/[24] | |
| R1284 | 0.022 | CF patient, Canada/[24] | |
| R1285 | 0.6 | CF patient, Canada/[24] | |
| R1314 | ++ | CF patient, Canada/[24] | |
| R1434 | 0.86 | CF patient, Canada/[24] | |
| R1619 | ++ | CF patient, Canada/[24] | |
| R1882 | 0.53 | CF patient, Canada/[24] | |
| R1883 | 0.47 | CF patient, Canada/[24] | |
| R1884 | 1 | CF patient, Canada/[24] | |
| R2314 | 0.73 | CF patient, Canada/[24] | |
| RK1b | 0.31 | CF patient, Canada/[24] | |
| S11528 | 0.8 | CF patient, Canada/[24] | |
| Van1 | 1 | CF patient, Canada/This study | |
| B. stabilis | LMG 14294 | + | CF patient, Belgium/[13,23] |
| C7322/LMG 18870 | 6.0 × 10−4 | CF patient, Canada/[13,22] | |
| R450 | ND | CF patient, Canada/[24] | |
| R2140 | + | CF patient, Canada/[24] | |
| R2339 | ND | CF patient, Canada/[24] | |
| B. vietnamiensis | DBO1 | ND | Soil, USA/[34] |
| LMG 10929 T | ND | Rice, Vietnam/[13,19,35] | |
| PC259/LMG 18835 | + | CF patient, USA/[13,36,37] | |
| G4 | ND | Soil, USA/[38] | |
| B. dolosa | AU0158 | ++ | CF patient, USA/[39] |
| CEP021 | + | CF patient, USA/[39] | |
| E12 | 8.6 × 10−5 | CF patient, UK/[39] | |
| STM1441 | ND | Soil, Senegal/[39] | |
| B. ambifaria | AMMD T | ND | Soil, USA/[39] |
| ATCC 53266 | 2.3 × 10−5 | Soil, USA/[39] | |
| CEP996 | 0.31 | CF patient, Australia/[39] | |
| M53 | ND | Soil, USA/[24] | |
| B. anthina | AU1293 | 0.8 | CF patient, USA/[39] |
| C1765 | 4.3 × 10−3 | CF patient, UK/[39] | |
| J2552 | ND | Soil, UK/[39] | |
| W92T | ND | Soil, USA/[39] | |
| B. pyrrocinia | ATCC 15958 | ND | Soil, Japan/[39] |
| ATCC 39277 | ND | Soil, USA/[39] | |
| BC011 | ND | Water, USA/[39] | |
| C1469 | ND | CF patient, UK/[39] | |
| Bcc Group K | CEP0964 | ND | CF patient, Canada/[24] |
| CEP1056 | ++ | CF patient, Canada/[24] | |
| R445 | 3.1 × 10−5 | CF patient, Canada/[24] | |
| B. lata | 383 | ND | Soil, Trinidad/[40] |
| Burkholderia sp. | JS150 | 1 | Soil, USA/[24] |
| Pandoraea sp. | R1717 | ND | CF patient, Canada/[24] |
| Ralstonia pickettii | ATCC 27511 | 0.7 | Patient isolate, USA/[41] |
| YH105 | ND | Soil, USA/[42] |
| Bacterial Strain | Phenotype | pSCRhaB2 | pSCRhaB2-Complement |
|---|---|---|---|
| K56-2 | Wildtype LPS | + | + |
| SAL1 | K56-2 hldA:: pSL5, Lacks inner and outer core | – | + |
| CCB1 | K56-2 waaC::pGPΩTp, Lacks inner and outer core | – | + |
| XOA8 | K56-2 wabO::pGPΩTp, Lacks inner and outer core | – | + |
| XOA7 | K56-2 waaL::pGPΩTp, Lacks outer core | + | + |
| XOA15 | K56-2 wabR::pGPΩTp, Lacks outer core | + | + |
| XOA17 | K56-2 wabS::pGPΩTp, Lacks outer core | + | + |
| RSF19 | K56-2 wbxE:: pRF201, Lacks O-antigen | + | + |
| Gene | Start | End | Strand | Length(aa) | Putative Function | BLASTp Hit | Species | Coverage (%) | E-Value | Identity (%) | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1660 | 5 | – | 551 aa | serine recombinase | serine recombinase-like protein | Burkholderia phage Bcep22 | 100 | 0 | 70.29 | NP_944235.2 |
| 2 | 2081 | 1794 | – | 95 aa | hypothetical protein | hypothetical protein Bcep22_gp07 | Burkholderia phage Bcep22 | 72 | 2 × 10−27 | 65.22 | YP_009173769.1 |
| 3 | 2493 | 2146 | – | 115 aa | hypothetical protein | hypothetical protein Bcep22_gp09 | Burkholderia phage Bcep22 | 98 | 2 × 10−21 | 40.35 | NP_944237.1 |
| 4 | 2867 | 2490 | – | 125 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 3 × 10−84 | 96.80 | WP_217093966.1 |
| 5 | 3895 | 2864 | – | 343 aa | RecT-like protein | RecT-like protein | Burkholderia phage Bcep22 | 100 | 1 × 10−178 | 74.16 | NP_944238.1 |
| 6 | 4976 | 3945 | – | 343 aa | nuclease/RecB-like protein | nuclease/RecB-like protein | Burkholderia phage Bcepmigl | 98 | 0 | 84.32 | YP_007236753.1 |
| 7 | 5697 | 5104 | – | 197 aa | repressor | transcriptional regulator | Burkholderia phage Bcepmigl | 100 | 7 × 10−81 | 59.90 | YP_007236754.1 |
| 8 | 5821 | 6165 | + | 114 aa | transcriptional regulator | transcriptional regulator | Burkholderia phage DC1 | 78 | 8 × 10−38 | 66.29 | YP_006589939.1 |
| 9 | 6335 | 6685 | + | 116 aa | hypothetical protein | hypothetical protein G167_gp75 | Burkholderia phage Bcepmigl | 96 | 4 × 10−67 | 87.50 | YP_007236756.1 |
| 10 | 6737 | 7546 | + | 269 aa | virion-associated protein a | hypothetical protein BcepIL02_gp11 | Burkholderia phage Bcepil02 | 99 | 2 × 10−143 | 73.98 | YP_002922683.1 |
| 11 | 7642 | 8142 | + | 166 aa | single stranded DNA binding protein | single stranded DNA binding protein | Burkholderia phage DC1 | 100 | 4 × 10−82 | 81.33 | YP_006589943.1 |
| 12 | 8151 | 8369 | + | 72 aa | hypothetical protein | hypothetical protein B862_gp69 | Burkholderia phage DC1 | 100 | 3 × 10−40 | 88.89 | YP_006589944.1 |
| 13 | 8366 | 8842 | + | 158 aa | hypothetical protein | hypothetical protein UAM5_00057 | Ralstonia phage UAM5 | 99 | 2 × 10−68 | 65.61 | CAH0532174.1 |
| 14 | 8839 | 9201 | + | 120 aa | hypothetical protein | hypothetical protein KMC44_gp61 | Ralstonia phage Cimandef | 98 | 1 × 10−59 | 70.87 | YP_010078217.1 |
| 15 | 9352 | 9807 | + | 151 aa | hypothetical protein | hypothetical protein B862_gp66 | Burkholderia phage DC1 | 84 | 7 × 10−68 | 85.94 | YP_006589947.1 |
| 16 | 10,127 | 9801 | – | 108 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 2 × 10−75 | 100 | WP_217093979.1 |
| 17 | 10,389 | 10,132 | – | 85 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 2 × 10−53 | 100 | WP_217093980.1 |
| 18 | 10,744 | 10,974 | + | 76 aa | hypothetical protein | hypothetical protein B862_gp65 | Burkholderia phage DC1 | 100 | 3 × 10−39 | 84.21 | YP_006589948.1 |
| 19 | 10,971 | 11,357 | + | 128 aa | helicase | TPA: MAG TPA: hypothetical protein | Siphoviridae sp. | 61 | 6 × 10−18 | 44.30 | DAT31939.1 |
| 20 | 11,354 | 12,178 | + | 274 aa | eplication initiator protein | replication protein | Burkholderia phage Bcepmigl | 39 | 2 × 10−48 | 76.85 | YP_007236768.1 |
| 21 | 12,175 | 12,975 | + | 266 aa | DnaC-like protein | DnaC-like protein | Burkholderia phage Bcepil02 | 99 | 4 × 10−150 | 76.60 | YP_002922693.1 |
| 22 | 13,057 | 13,182 | + | 41 aa | hypothetical protein | hypothetical protein G167_gp61 | Burkholderia phage Bcepmigl | 100 | 1 × 10−9 | 56.10 | YP_007236770.1 |
| 23 | 13,337 | 13,765 | + | 142 aa | hypothetical protein | hypothetical protein B862_gp60 | Burkholderia phage DC1 | 98 | 5 × 10−51 | 60.28 | YP_006589953.1 |
| 24 | 13,771 | 14,106 | + | 111 aa | hypothetical protein | hypothetical protein BcepIL02_gp24 | Burkholderia phage Bcepil02 | 85 | 1 × 10−16 | 43.75 | YP_002922696.1 |
| 25 | 14,166 | 14,609 | + | 147 aa | hypothetical protein | TPA: MAG TPA_asm: hypothetical protein | Myoviridae sp. | 95 | 7 × 10−44 | 49.29 | DAL29776.1 |
| 26 | 14,652 | 15,041 | + | 129 aa | hypothetical protein | TPA: MAG TPA: hypothetical protein | Myoviridae sp. | 95 | 1 × 10−9 | 34.35 | DAP81611.1.1 |
| 27 | 15,044 | 15,325 | + | 93 aa | hypothetical protein | hypothetical protein B862_gp58 | Burkholderia phage DC1 | 100 | 3 × 10−40 | 70.83 | YP_006589955.1 |
| 28 | 15,364 | 16,602 | + | 412 aa | hypothetical protein | hypothetical protein Bcep22_gp31 | Burkholderia phage Bcep22 | 100 | 6 × 10−156 | 62.42 | NP_944260.2 |
| 29 | 17,017 | 16,658 | – | 119 aa | hypothetical protein | hypothetical protein KMC50_gp40 | Ralstonia phage Claudette | 85 | 2 × 10−25 | 49.02 | YP_010078630.1 |
| 30 | 17,210 | 18,010 | + | 266 aa | terminase small subunit | terminase small subunit | Burkholderia phage DC1 | 93 | 8 × 10−161 | 83.53 | YP_006589958.1 |
| 31 | 18,090 | 18,338 | + | 82 aa | hypothetical protein | hypothetical protein B862_gp53 | Burkholderia phage DC1 | 92 | 2 × 10−39 | 82.89 | YP_006589960.1 |
| 32 | 18,389 | 18,517 | + | 42 aa | hypothetical protein | hypothetical protein B862_gp49 | Burkholderia phage DC1 | 97 | 2 × 10−17 | 87.80 | YP_006589964.1 |
| 33 | 18,562 | 18,840 | + | 92 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 1 × 10−59 | 97.83 | WP_217093993.1 |
| 34 | 18,872 | 19,066 | + | 64 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 9 × 10−36 | 96.88 | WP_217093994.1 |
| 35 | 19,059 | 20,366 | + | 435 aa | hypothetical protein | hypothetical protein phiE131_040 | Burkholderia phage phiE131 | 47 | 9 × 10−23 | 41.40 | AYJ74306.1 |
| 36 | 20,356 | 20,766 | + | 136 aa | hypothetical protein | hypothetical protein HOT12_gp34 | Burkholderia phage vB_BmuP_KL4 | 84 | 5 × 10−32 | 68.10 | YP_009800723.1 |
| 37 | 20,851 | 21,033 | + | 60 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 8 × 10−33 | 100 | WP_217093996.1 |
| 38 | 21,030 | 21,416 | + | 128 aa | hypothetical protein | TPA: MAG TPA: Protein of unknown function (DUF2591) | Caudovirales sp. | 99 | 2 × 10−13 | 36.76 | DAH87964.1 |
| 39 | 21,413 | 21,700 | + | 95 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 9 × 10−63 | 100 | WP_217093998.1 |
| 40 | 21,697 | 22,035 | + | 112 aa | hypothetical protein | hypothetical protein Bcep22_gp48 | Burkholderia phage Bcep22 | 99 | 2 × 10−65 | 85.59 | NP_944277.1 |
| 41 | 22,172 | 22,582 | + | 136 aa | DUF2778 domain-containing protein | TPA: MAG TPA: Protein of unknown function (DUF2778) | Myoviridae sp. | 98 | 8 × 10−19 | 39.57 | DAO56318.1 |
| 42 | 22,579 | 22,860 | + | 93 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 7 × 10−57 | 98.92 | WP_217094000.1 |
| 43 | 22,948 | 24,549 | + | 533 aa | terminase large subunit | terminase large subunit | Burkholderia phage DC1 | 98 | 0 | 83.11 | YP_006589971.1 |
| 44 | 24,560 | 24,991 | + | 143 aa | hypothetical protein | hypothetical protein BcepIL02_gp45 | Burkholderia phage Bcepil02 | 99 | 2 × 10−82 | 82.39 | YP_002922717.1 |
| 45 | 25,013 | 27,310 | + | 765 aa | portal protein | phage portal protein | Burkholderia phage Bcepil02 | 95 | 0 | 77.53 | YP_002922718.1 |
| 46 | 27,318 | 28,340 | + | 340 aa | virion-associated proteina | hypothetical protein Bcep22_gp52 | Burkholderia phage Bcep22 | 98 | 3 × 10−110 | 57.82 | NP_944281.1 |
| 47 | 28,367 | 28,561 | + | 64 aa | carbon storage regulator | carbon storage regulator | Burkholderia phage DC1 | 100 | 1 × 10−30 | 89.06 | YP_006589976.1 |
| 48 | 28,660 | 29,754 | + | 364 aa | major capsid protein | major capsid protein | Burkholderia phage Bcepmigl | 100 | 0 | 92.03 | YP_007236797.1 |
| 49 | 29,821 | 30,288 | + | 155 aa | virion-associated protein | virion associated protein | Burkholderia phage Bcepmigl | 100 | 2 × 10−77 | 74.84 | YP_007236798.1 |
| 50 | 30,346 | 30,942 | + | 198 aa | hypothetical protein | hypothetical protein B862_gp33 | Burkholderia phage DC1 | 99 | 5 × 10−80 | 63.41 | YP_006589980.1 |
| 51 | 30,946 | 31,593 | + | 215 aa | virion-associated protein a | hypothetical protein B862_gp32 | Burkholderia phage DC1 | 100 | 7 × 10−147 | 92.09 | YP_006589981.1 |
| 52 | 31,590 | 32,219 | + | 209 aa | virion-associated protein a | hypothetical protein G167_gp30 | Burkholderia phage Bcepmigl | 100 | 6 × 10−117 | 77.14 | YP_007236801.1 |
| 53 | 32,229 | 32,648 | + | 139 aa | virion-associated protein | major capsid protein | Burkholderia phage DC1 | 100 | 6 × 10−93 | 94.24 | YP_006589983.1 |
| 54 | 32,653 | 33,528 | + | 291 aa | hypothetical protein | hypothetical protein B862_gp29 | Burkholderia phage DC1 | 97 | 9 × 10−147 | 74.39 | YP_006589984.1 |
| 55 | 33,510 | 33,788 | + | 92 aa | virion-associated protein | virion-associated phage protein | Burkholderia phage Bcepil02 | 100 | 2 × 10−53 | 90.22 | YP_002922729.1 |
| 56 | 33,790 | 34,740 | + | 316 aa | tail fiber protein | putative tail fiber protein | Burkholderia phage Bcepil02 | 100 | 4 × 10−177 | 76.90 | YP_002922730.1 |
| 57 | 34,744 | 35,814 | + | 356 aa | tail fiber protein | putative tail fiber protein | Burkholderia phage Bcepil02 | 100 | 2 × 10−145 | 64.54 | YP_002922731.1 |
| 58 | 35,811 | 36,323 | + | 170 aa | hypothetical protein | hypothetical protein BcepIL02_gp60 | Burkholderia phage Bcepil02 | 98 | 7 × 10−39 | 45.29 | YP_002922732.1 |
| 59 | 36,484 | 37,530 | + | 348 aa | tail fiber protein | TPA: MAG TPA: Endo N acetylneuraminidase | Siphoviridae sp. | 59 | 9 × 10−84 | 63.59 | DAM52127.1 |
| 60 | 37,532 | 38,221 | + | 229 aa | hypothetical protein | hypothetical protein | Pseudomonas phage Dolphis | 100 | 6 × 10−18 | 44.92 | QNJ57341.1 |
| 61 | 38,276 | 40,018 | + | 580 aa | head closure protein | virion-associated phage protein | Burkholderia phage Bcepil02 | 100 | 0 | 90.00 | YP_002922735.1 |
| 62 | 40,020 | 40,373 | + | 117 aa | hypothetical protein a | virion-associated phage protein | Burkholderia phage Bcepil02 | 97 | 2 × 10−62 | 86.09 | YP_002922736.1 |
| 63 | 40,424 | 40,864 | + | 146 aa | acetyltransferase | acetyltransferase | Burkholderia phage DC1 | 97 | 6 × 10−92 | 89.51 | YP_006589993.1 |
| 64 | 40,857 | 41,855 | + | 332 aa | virion-associated protein a | hypothetical protein B862_gp19 | Burkholderia phage DC1 | 100 | 0 | 91.27 | YP_006589994.1 |
| 65 | 41,867 | 42,589 | + | 240 aa | phosphoadenosine phosphosulfate reductase | phosphoadenosine phosphosulfate reductase | Burkholderia phage DC1 | 100 | 2 × 10−167 | 94.17 | YP_006589995.1 |
| 66 | 42,589 | 42,888 | + | 99 aa | hypothetical protein | hypothetical protein | Burkholderia multivorans | 100 | 2 × 10−59 | 100 | WP_217094022.1 |
| 67 | 42,905 | 43,495 | + | 196 aa | virion-associated protein | virion-associated phage protein | Burkholderia phage Bcepil02 | 30 | 1 × 10−4 | 49.21 | YP_002922740.1 |
| 68 | 43,506 | 45,038 | + | 510 aa | virion-associated protein a | hypothetical protein B862_gp17 | Burkholderia phage DC1 | 100 | 0 | 80.30 | YP_006589996.1 |
| 69 | 45,123 | 58,670 | + | 4515 aa | DarB-like antirestriction protein | DarB-like antirestriction protein | Burkholderia phage Bcep22 | 100 | 0 | 79.33 | NP_944303.1 |
| 70 | 58,940 | 58,701 | – | 79 aa | hypothetical protein | hypothetical protein BcepIL02_gp71 | Burkholderia phage Bcepil02 | 97 | 1 × 10−46 | 89.61 | YP_002922743.1 |
| 71 | 59,287 | 58,991 | – | 98 aa | hypothetical protein | hypothetical protein G167_gp14 | Burkholderia phage Bcepmigl | 100 | 2 × 10−41 | 67.35 | YP_007236817.1 |
| 72 | 59,443 | 59,790 | + | 115 aa | LydA-like holin | LydA-like holin | uncultured Caudovirales phage | 82 | 4 × 10−27 | 54.74 | CAB4121548.1 |
| 73 | 59,787 | 60,059 | + | 90 aa | holin | holin | Burkholderia phage vB_BceS_AH2 | 91 | 6 × 10−32 | 68.29 | YP_006561127.1 |
| 74 | 60,056 | 60,634 | + | 192 aa | lysozyme | hypothetical protein AXJ08_gp22 | Rhodoferax phage P26218 | 94 | 7 × 10−57 | 49.45 | YP_009222572.1 |
| 75 | 60,631 | 61,140 | + | 169 aa | Rz | Rz-like phage lysis protein | Burkholderia phage Bcep22 | 95 | 7 × 10−65 | 66.27 | NP_944308.1 |
| 76 | 60,866 | 61,087 | + | 73 aa | Rz1 | Rz1 | Burkholderia phage DC1 | 100 | 9 × 10−16 | 83.56 | YP_006590003.1 |
| Start | Program | Strand | Sequence | −ΔG |
|---|---|---|---|---|
| 1753 | Both | – | ATCGACTCCAACGGCACCCTCGCGGTGCCGTTTTTATTGCCC | −13.20 |
| 6258 | Rnamotif | + | CCAGCTGTTGAGCCTCCCGTTTCAGGGAGGCTTTTTGCCCGTA | −15.70 |
| 10,407 | Rnamotif | – | AGAGCGTCGTCGGCGGCCCGCACGGCCGCCaTTTTTTTCGATC | −16.00 |
| 13,228 | Rnamotif | + | GGCGACTTTGGTGGGCGGCTCGTACAGCGCCCGTTTTTTTTCACC | −9.60 |
| 13,893 | Rnamotif | – | CCGATGCGCACCGGCCGGATGTGGCTGATCCGGTTGTTGTATTCGCGG | −10.50 |
| 47,347 | Rnamotif | – | TCGGCCGACACCTTGCGGCGCTCGGCCGTGAGcaTCTTGTTCCAGC | −12.10 |
| 51,986 | Rnamotif | – | CCTCCTGAATCGCGCGCCAGATGGCGCGCTTCTGGTTCGGG | −15.60 |
| 61,154 | Both | + | GGCTGAGACTTCCCCGGCGCGAGCCGGGGTTTTTTATGCCG | −16.40 |
| Gp | Hit Type | PSSM-ID | Interval | E-Value | Accession | Short Name | Superfamily |
|---|---|---|---|---|---|---|---|
| 1 | specific | 238206 | 9–159 | 1.68 × 10−24 | cd00338 | Ser_Recombinase | cl02788 |
| 5 | superfamily | 413988 | 31–245 | 2.47 × 10−48 | cl04285 | RecT superfamily | - |
| 6 | superfamily | 415607 | 14–168 | 6.92 × 10−17 | cl09232 | YqaJ superfamily | - |
| 7 | specific | 238045 | 11–63 | 1.31 × 10−5 | cd00093 | HTH_XRE | cl22854 |
| 10 | superfamily | 413281 | 20–268 | 2.41 × 10−72 | cl02338 | DUF2303 superfamily | - |
| 14 | superfamily | 377777 | 34–98 | 6.08 × 10−8 | cl06229 | DUF1364 superfamily | - |
| 18 | specific | 404897 | 5–74 | 3.76 × 10−19 | pfam14090 | HTH_39 | cl16606 |
| 19 | specific | 214947 | 15–88 | 3.71 × 10−19 | smart00974 | T5orf172 | cl15257 |
| 20 | superfamily | 237940 | 106–141 | 2.12 × 10−3 | cl36477 | PRK15313 superfamily | - |
| 21 | superfamily | 422963 | 72–263 | 1.95 × 10−39 | cl38936 | P-loop_NTPase superfamily | - |
| 30 | specific | 397583 | 19–236 | 7.93 × 10−24 | pfam03592 | Terminase_2 | cl01513 |
| 38 | superfamily | 416328 | 2–120 | 1.32 × 10−21 | cl11584 | DUF2591 superfamily | - |
| 40 | superfamily | 404162 | 3–86 | 7.90 × 10−21 | cl16173 | DUF4031 superfamily | - |
| 41 | specific | 402478 | 1–125 | 6.70 × 10−48 | pfam10908 | DUF2778 | cl12489 |
| 43 | superfamily | 222858 | 51–252 | 3.53 × 10−09 | cl28557 | 17 superfamily | - |
| 45 | superfamily | 293119 | 57–616 | 9.79 × 10−10 | cl24922 | P22_portal superfamily | - |
| 45 | superfamily | 135173 | 668–765 | 8.02 × 10−5 | cl31366 | PRK04654 superfamily | - |
| 47 | specific | 396934 | 1–44 | 7.10 × 10−8 | pfam02599 | CsrA | cl00670 |
| 48 | specific | 404189 | 39–358 | 8.21 × 10−100 | pfam13252 | DUF4043 | cl22542 |
| 53 | superfamily | 412204 | 29–130 | 4.16 × 10−3 | cl00184 | CAS_like superfamily | - |
| 57 | specific | 404724 | 255–306 | 3.00 × 10−13 | pfam13884 | Peptidase_S74 | cl16452 |
| 59 | specific | 404724 | 248–304 | 2.58 × 10−12 | pfam13884 | Peptidase_S74 | cl16452 |
| 63 | specific | 224584 | 9–134 | 6.86 × 10−4 | COG1670 | RimL | cl34333 |
| 65 | specific | 238846 | 10–181 | 5.36 × 10−24 | cd01713 | PAPS_reductase | cl00292 |
| 68 | superfamily | 180240 | 299–400 | 9.02 × 10−7 | cl32090 | PRK05759 superfamily | - |
| 69 | superfamily | 226993 | 1810–2677 | 6.59 × 10−99 | cl18793 | COG4646 superfamily | |
| 69 | specific | 408627 | 4195–4401 | 1.56 × 10−36 | pfam18857 | LPD38 | cl40138 |
| 69 | specific | 408569 | 3170–3270 | 9.95 × 10−19 | pfam18798 | LPD3 | cl40093 |
| 69 | specific | 381594 | 80–192 | 7.10 × 10−14 | cd00254 | LT-like | cl00222 |
| 69 | specific | 223897 | 1537–1783 | 3.77 × 10−11 | COG0827 | YtxK | cl28092 |
| 69 | superfamily | 237171 | 1358–1500 | 3.31 × 10−7 | cl36163 | PRK12678 superfamily | - |
| 69 | superfamily | 223627 | 2412–2868 | 2.48 × 10−5 | cl33945 | HepA superfamily | - |
| 69 | superfamily | 235334 | 1071–1287 | 1.59 × 10−3 | cl35279 | PRK05035 superfamily | - |
| 72 | specific | 406481 | 24–103 | 3.77 × 10−16 | pfam16083 | Phage_holin_3_3 | cl24062 |
| 74 | superfamily | 226439 | 2–186 | 2.17 × 10−28 | cl34694 | ZliS superfamily | - |
| 75 | superfamily | 419854 | 54–163 | 2.39 × 10−10 | cl22701 | Phage_lysis superfamily | - |
| Protein | Score | Coverage | Unique Peptides (#) | Putative Function |
|---|---|---|---|---|
| gp48 | 298.44 | 56.04 | 27 | Major capsid protein |
| gp69 | 88.92 | 15.61 | 51 | DarB-like antirestriction protein |
| gp49 | 82.92 | 50.32 | 6 | Virion-associated protein |
| gp64 | 45.94 | 45.18 | 9 | Hypothetical protein |
| gp53 | 30.63 | 58.99 | 5 | Virion-associated protein |
| gp68 | 27.46 | 17.06 | 9 | Hypothetical Protein |
| gp45 | 25.85 | 22.22 | 14 | Portal protein |
| gp56 | 22.88 | 41.14 | 7 | Tail fiber protein |
| gp52 | 19.47 | 46.41 | 8 | Hypothetical protein |
| gp61 | 16.01 | 11.21 | 6 | Virion-associated phage protein |
| gp10 | 13.4 | 21.56 | 5 | Hypothetical protein |
| gp55 | 13.16 | 48.91 | 4 | Virion-associated phage protein |
| gp51 | 12.97 | 25.58 | 5 | Hypothetical protein |
| gp67 | 9.35 | 42.86 | 7 | Virion-associated phage protein |
| gp46 | 4.14 | 13.82 | 4 | Hypothetical protein |
| gp47 | 2.61 | 18.75 | 1 | Carbon storage regulator |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Davis, C.M.; Ruest, M.K.; Cole, J.H.; Dennis, J.J. The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1. Viruses 2022, 14, 938. https://doi.org/10.3390/v14050938
Davis CM, Ruest MK, Cole JH, Dennis JJ. The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1. Viruses. 2022; 14(5):938. https://doi.org/10.3390/v14050938
Chicago/Turabian StyleDavis, Carly M., Marta K. Ruest, Jamie H. Cole, and Jonathan J. Dennis. 2022. "The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1" Viruses 14, no. 5: 938. https://doi.org/10.3390/v14050938
APA StyleDavis, C. M., Ruest, M. K., Cole, J. H., & Dennis, J. J. (2022). The Isolation and Characterization of a Broad Host Range Bcep22-like Podovirus JC1. Viruses, 14(5), 938. https://doi.org/10.3390/v14050938








