Simultaneous Giant Virus and Virophage Quantification Using Droplet Digital PCR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model System
2.2. DNA Extraction
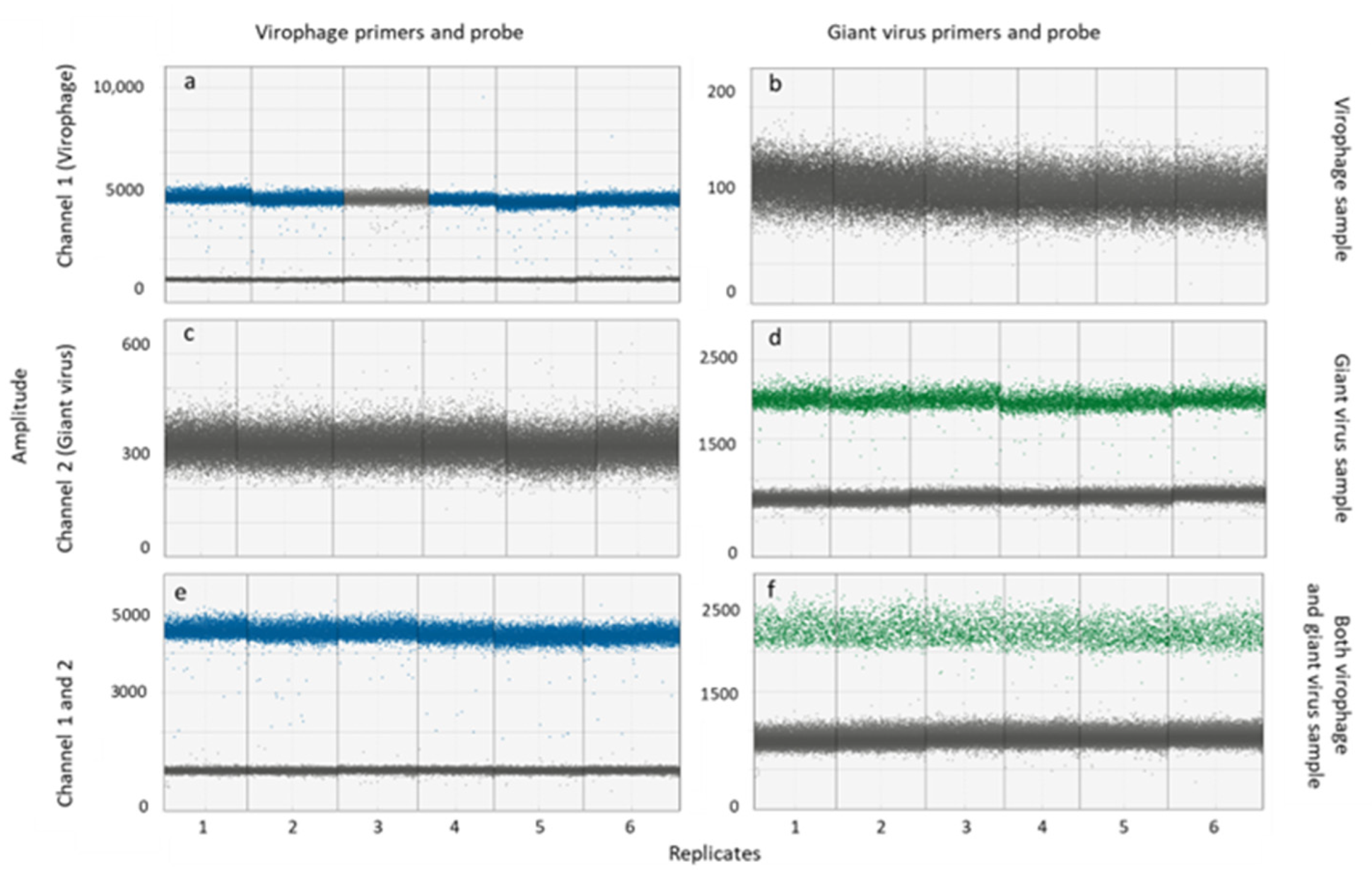
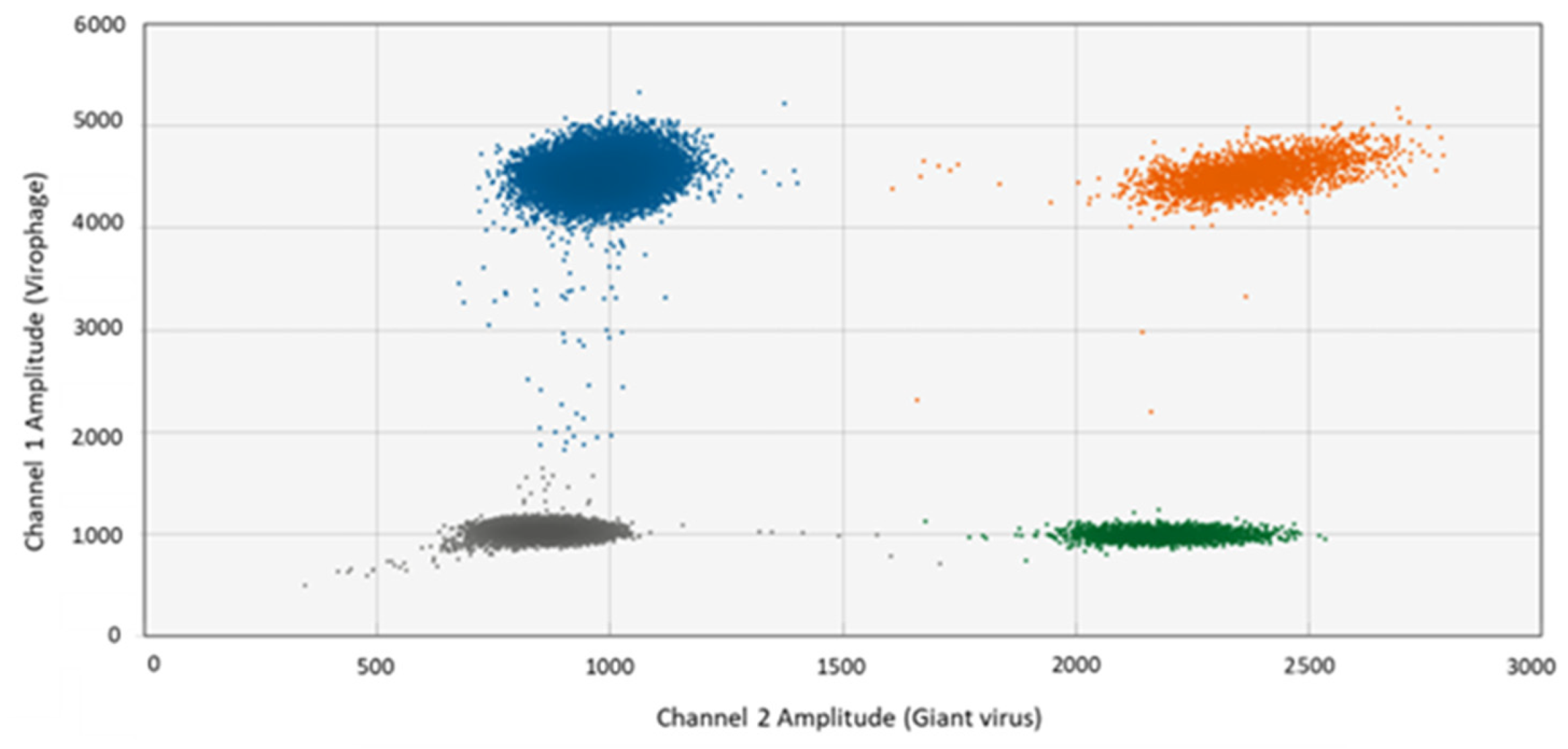
2.3. Evaluation of Primer and Probe Specificities in Singleplex and Multiplex ddPCR
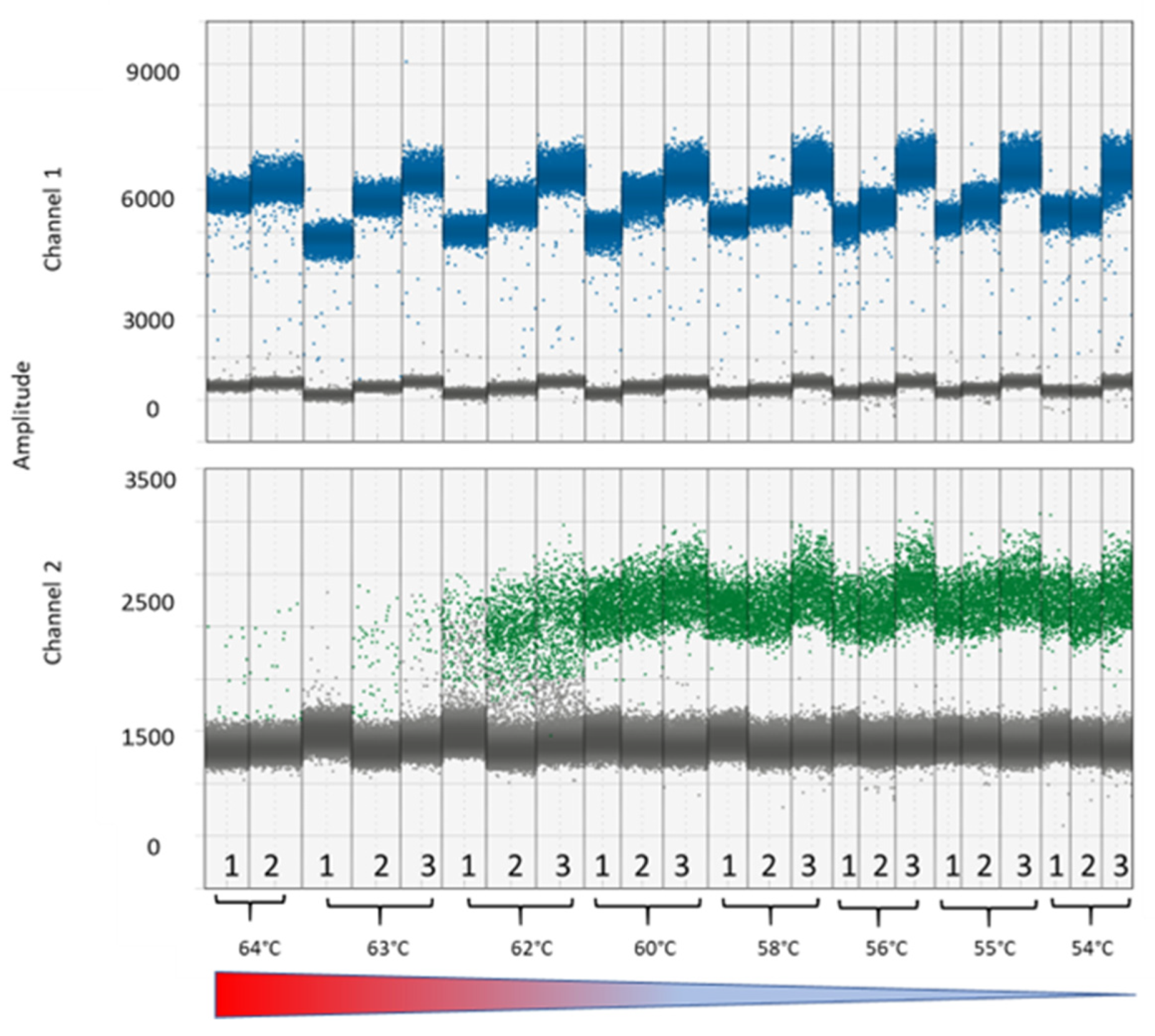
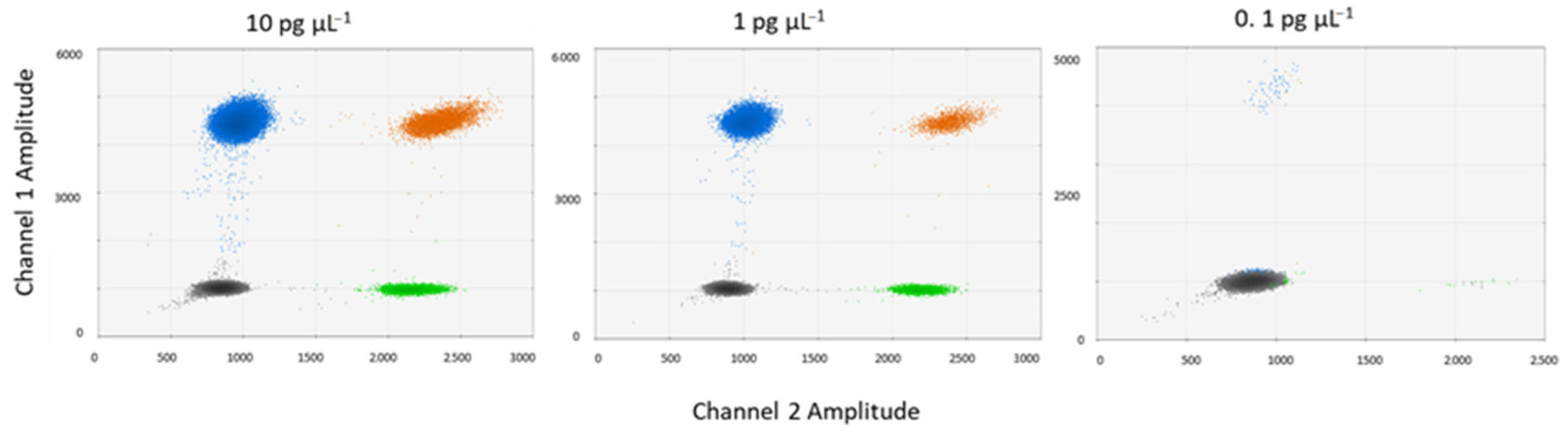
2.4. Optimization of PCR Conditions and DNA Concentrations
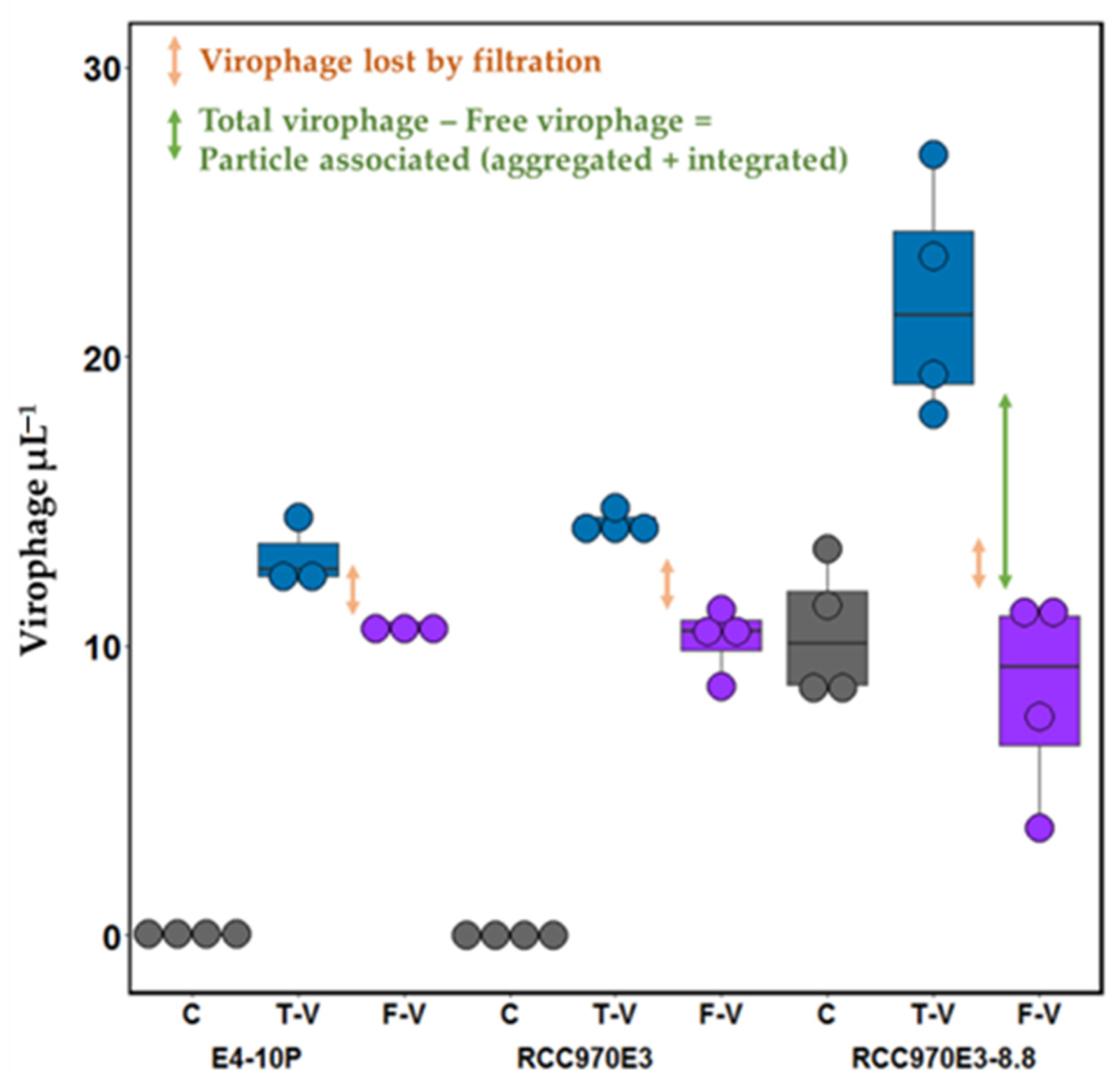
2.5. Discrimination between Free and Particle Associated Virophage
3. Results
3.1. Specificity of Primers and Probes in Singleplex and Multiplex ddPCR
3.2. PCR and DNA Conditions
3.3. Discrimination between Free and Particle-Associated Virophages
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cobián Güemes, A.G.; Youle, M.; Cantú, V.A.; Felts, B.; Nulton, J.; Rohwer, F. Viruses as Winners in the Game of Life. Annu. Rev. Virol. 2016, 3, 197–214. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M. Marine Viruses: Truth or Dare. Annu. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frickel, J.; Sieber, M.; Becks, L. Eco-Evolutionary Dynamics in a Coevolving Host–Virus System. Ecol. Lett. 2016, 19, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Suttle, C.A.; Chan, A.M.; Cottrel, M.T. Infection of Phytoplankton by Viruses and Reduction of Primary Productivity. J. Colloid Interface Sci. 1990, 374, 685–689. [Google Scholar] [CrossRef]
- Fischer, M.G.; Hackl, T. Host Genome Integration and Giant Virus-Induced Reactivation of the Virophage Mavirus. Nature 2016, 540, 288–291. [Google Scholar] [CrossRef]
- La Scola, B.; Desnues, C.; Pagnier, I.; Robert, C.; Barrassi, L.; Fournous, G.; Merchat, M.; Suzan-Monti, M.; Forterre, P.; Koonin, E.; et al. The Virophage as a Unique Parasite of the Giant Mimivirus. Nature 2008, 455, 100–104. [Google Scholar] [CrossRef]
- Duponchel, S.; Fischer, M.G. Viva Lavidaviruses! Five Features of Virophages That Parasitize Giant DNA Viruses. PLoS Pathog. 2019, 15, e1007592. [Google Scholar] [CrossRef]
- La Scola, B.; Campocasso, A.; N’Dong, R.; Fournous, G.; Barrassi, L.; Flaudrops, C.; Raoult, D. Tentative Characterization of New Environmental Giant Viruses by MALDI-TOF Mass Spectrometry. Intervirology 2010, 53, 344–353. [Google Scholar] [CrossRef]
- Fischer, M.G.; Allen, M.J.; Wilson, W.H.; Suttle, C.A. Giant Virus with a Remarkable Complement of Genes Infects Marine Zooplankton. Proc. Natl. Acad. Sci. USA 2010, 107, 19508–19513. [Google Scholar] [CrossRef] [Green Version]
- Sheng, Y.; Wu, Z.; Xu, S.; Wang, Y. Isolation and Identification of a Large Green Alga Virus (Chlorella Virus XW01) of Mimiviridae and Its Virophage (Chlorella Virus Virophage SW01) by Using Unicellular Green Algal Cultures. J. Virol. 2022, 96, e0211421. [Google Scholar] [CrossRef]
- Raoult, D.; Audic, S.; Robert, C.; Abergel, C.; Renesto, P.; Ogata, H.; La Scola, B.; Suzan, M.; Claverie, J.M. The 1.2-Megabase Genome Sequence of Mimivirus. Science 2004, 306, 1344–1351. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T. Giant Viruses in the Environment: Their Origins and Evolution. Curr. Opin. Virol. 2011, 1, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Gallot-Lavallée, L.; Maumus, F. Provirophages in the Bigelowiella Genome Bear Testimony to Past Encounters with Giant Viruses. Proc. Natl. Acad. Sci. USA 2015, 112, E5318–E5326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackl, T.; Duponchel, S.; Barenhoff, K.; Weinmann, A.; Fischer, M.G. Virophages and Retrotransposons Colonize the Genomes of a Heterotrophic Flagellate. eLife 2021, 10, e72674. [Google Scholar] [CrossRef]
- Krupovic, M.; Kuhn, J.H.; Fischer, M.G. A Classification System for Virophages and Satellite Viruses. Arch. Virol. 2016, 161, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Berjón-Otero, M.; Koslová, A.; Fischer, M.G. The Dual Lifestyle of Genome-Integrating Virophages in Protists. Ann. N. Y. Acad. Sci. 2019, 1447, 97–109. [Google Scholar] [CrossRef]
- Taylor, B.; Weitz, J.; Brussaard, C.; Fischer, M. Quantitative Infection Dynamics of Cafeteria Roenbergensis Virus. Viruses 2018, 10, 468. [Google Scholar] [CrossRef] [Green Version]
- Taylor, B.P.; Cortez, M.H.; Weitz, J.S. The Virus of My Virus Is My Friend: Ecological Effects of Virophage with Alternative Modes of Coinfection. J. Theor. Biol. 2014, 354, 124–136. [Google Scholar] [CrossRef] [Green Version]
- Fritz, A.; Hofmann, P.; Majda, S.; Dahms, E.; Dröge, J.; Fiedler, J.; Lesker, T.R.; Belmann, P.; DeMaere, M.Z.; Darling, A.E.; et al. CAMISIM: Simulating Metagenomes and Microbial Communities. Microbiome 2019, 7, 17. [Google Scholar] [CrossRef] [Green Version]
- Schulz, F.; Roux, S.; Paez-Espino, D.; Jungbluth, S.; Walsh, D.A.; Denef, V.J.; McMahon, K.D.; Konstantinidis, K.T.; Eloe-Fadrosh, E.A.; Kyrpides, N.C.; et al. Giant Virus Diversity and Host Interactions through Global Metagenomics. Nature 2020, 578, 432–436. [Google Scholar] [CrossRef]
- Brussaard, C.P.D.; Payet, J.P.; Winter, C.; Weinbauer, M.G. Quantification of Aquatic Viruses by Flow Cytometry. Man. Aquat. Viral Ecol. 2010, 11, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Suttle, C.A.; Fuhrman, J.A. Enumeration of Virus Particles in Aquatic or Sediment Samples by Epifluorescence Microscopy. Man. Aquat. Viral Ecol. 2010, 15, 145–153. [Google Scholar]
- Furness, G.; Youngner, J.S. One-Step Growth Curves for Vaccinia Virus in Cultures of Monkey Kidney Cells. Virology 1959, 9, 386–395. [Google Scholar] [CrossRef]
- McDermott, G.P.; Do, D.; Litterst, C.M.; Maar, D.; Hindson, C.M.; Steenblock, E.R.; Legler, T.C.; Jouvenot, Y.; Marrs, S.H.; Bemis, A.; et al. Multiplexed Target Detection Using DNA-Binding Dye Chemistry in Droplet Digital PCR. Anal. Chem. 2013, 85, 11619–11627. [Google Scholar] [CrossRef] [PubMed]
- Rački, N.; Dreo, T.; Gutierrez-Aguirre, I.; Blejec, A.; Ravnikar, M. Reverse Transcriptase Droplet Digital PCR Shows High Resilience to PCR Inhibitors from Plant, Soil and Water Samples. Plant Methods 2014, 10, 42. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.; et al. The Need for Transparency and Good Practices in the QPCR Literature. Nat. Methods 2013, 10, 1063–1067. [Google Scholar] [CrossRef]
- Koch, H.; Jeschke, A.; Becks, L. Use of DdPCR in Experimental Evolution Studies. Methods Ecol. Evol. 2016, 7, 340–351. [Google Scholar] [CrossRef] [Green Version]
- Morella, N.M.; Yang, S.C.; Hernandez, C.A.; Koskella, B. Rapid Quantification of Bacteriophages and Their Bacterial Hosts in Vitro and in Vivo Using Droplet Digital PCR Norma. J. Virol. Methods 2018, 259, 18–24. [Google Scholar] [CrossRef]
- Martinez-hernandez, F.; Garcia-heredia, I.; Gomez, M.L.; Maestre-carballa, L.; Martínez, J.M.; Martinez-garcia, M.; Martinez-garcia, M. Droplet Digital PCR for Estimating Absolute Abundances of Widespread Pelagibacter Viruses. Front. Microbiol. 2019, 10, 1226. [Google Scholar] [CrossRef] [Green Version]
- Koskella, B.; Brockhurst, M.A. Bacteria–Phage Coevolution as a Driver of Ecological and Evolutionary Processes in Microbial Communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [Green Version]
- Lievens, E.J.P.; Agarkova, I.; Dunigan, D.D.; Etten, J.L.V.; Becks, L. Life History Diversity and Signals of Trade-Offs in a Large Group of Chloroviruses. bioRxiv 2022. [Google Scholar] [CrossRef]
- Retel, C.; Kowallik, V.; Huang, W.; Werner, B.; Künzel, S.; Becks, L.; Feulner, P.G.D. The Feedback between Selection and Demography Shapes Genomic Diversity during Coevolution. Sci. Adv. 2019, 5, eaax0530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wodarz, D. Evolutionary Dynamics of Giant Viruses and Their Virophages. Ecol. Evol. 2013, 3, 2103–2115. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Chan, L.-K.; Egan, R.; Malmstrom, R.R.; McMahon, K.D.; Sullivan, M.B. Ecogenomics of Virophages and Their Giant Virus Hosts Assessed through Time Series Metagenomics. Nat. Commun. 2017, 8, 858. [Google Scholar] [CrossRef]
- Yau, S.; Lauro, F.M.; DeMaere, M.Z.; Brown, M.V.; Thomas, T.; Raftery, M.J.; Andrews-Pfannkoch, C.; Lewis, M.; Hoffman, J.M.; Gibson, J.A.; et al. Virophage Control of Antarctic Algal Host–Virus Dynamics. Proc. Natl. Acad. Sci. USA 2011, 108, 6163–6168. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.C.; Carbonneau, J.; Shelton, D.N.; Boivin, G. Optimization of Droplet Digital PCR from RNA and DNA Extracts with Direct Comparison to RT-QPCR: Clinical Implications for Quantification of Oseltamivir-Resistant Subpopulations. J. Virol. Methods 2015, 224, 58–66. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Yao, Z.; Liu, J.; Zhang, H.; Din, G.M.U.; Zhao, S.; Chen, W.; Liu, T.; Gao, L. Development of Droplet Digital PCR for the Detection of Tilletia Laevis, Which Causes Common Bunt of Wheat, Based on the SCAR Marker Derived from ISSR and Real-Time PCR. Sci. Rep. 2020, 10, 16106. [Google Scholar] [CrossRef]
| Name | Description | Sequence (5′ to 3′) |
|---|---|---|
| Mavirus_forward | Forward primer | GAATGTCTCGCGGTTTAGGT |
| Mavirus _reverse | Reverse primer | TGGCTACAAGTGCTTCATCTAC |
| Mavirus _probe | FAM Probe (Ch 1) | 56-FAM/ATTATATCCACCCACGGGCAGCAG/3BHQ_1 |
| CroV_forward | Forward primer | GAAACTGGAAATGCTCGTGTTAT |
| CroV_reverse | Reverse primer | GGGAAAGAACCTGGTCGTAATAC |
| CroV_probe | HEX Probe (Ch 2) | 5HEX/ACTGCAACACCTGCAATCAATCAACC/3BHQ_1 |
| Parameter | Tested Range | Optimal Value |
|---|---|---|
| PCR annealing temperature | 64–54 °C | 58 °C |
| DNA template | 0.1–1–10 pg µL−1 | 1 pg µL−1 |
| Primer/Probe CroV | Recommended concentrations | Primer 375 nM Probe 125 nM |
| Primer/Probe mavirus | Recommended concentrations | Primer 375 nM Probe 125 nM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
del Arco, A.; Fischer, M.; Becks, L. Simultaneous Giant Virus and Virophage Quantification Using Droplet Digital PCR. Viruses 2022, 14, 1056. https://doi.org/10.3390/v14051056
del Arco A, Fischer M, Becks L. Simultaneous Giant Virus and Virophage Quantification Using Droplet Digital PCR. Viruses. 2022; 14(5):1056. https://doi.org/10.3390/v14051056
Chicago/Turabian Styledel Arco, Ana, Matthias Fischer, and Lutz Becks. 2022. "Simultaneous Giant Virus and Virophage Quantification Using Droplet Digital PCR" Viruses 14, no. 5: 1056. https://doi.org/10.3390/v14051056
APA Styledel Arco, A., Fischer, M., & Becks, L. (2022). Simultaneous Giant Virus and Virophage Quantification Using Droplet Digital PCR. Viruses, 14(5), 1056. https://doi.org/10.3390/v14051056






