Abstract
Viral infections cause a variety of acute and chronic human diseases, sometimes resulting in small local outbreaks, or in some cases spreading across the globe and leading to global pandemics. Understanding and exploiting virus–host interactions is instrumental for identifying host factors involved in viral replication, developing effective antiviral agents, and mitigating the severity of virus-borne infectious diseases. The diversity of CRISPR systems and CRISPR-based tools enables the specific modulation of innate immune responses and has contributed impressively to the fields of virology and immunology in a very short time. In this review, we describe the most recent advances in the use of CRISPR systems for basic and translational studies of virus–host interactions.
Keywords:
CRISPR/Cas; interferon effector proteins; interferon induction; pathogen recognition receptor; pathogen-associated molecular pattern; Toll-like receptor; cGAS/STING; DNA sensors; interferon stimulated genes; pooled libraries; epitranscriptomics; HBV; HDV; HCV; HIV; SARS-CoV-2; yellow fever virus; KSHV; HSV; EBOV; ZIKV; influenza A virus; CHIKV 1. Introduction
The development of antibacterial drugs in the first half of the 20th century provided the opportunity to control the most serious bacterial infections and markedly reduced the corresponding fatality and disability rates. However, compared to bacteria, viruses have a far more complicated life cycle and are remarkably more heterogeneous, which significantly complicates the development of broad-spectrum antivirals. The pandemic caused by the novel SARS-CoV-2 coronavirus has caused millions of deaths worldwide and resulted in the most severe economic recession since World War II [1]. Chronic viral infections by pathogens such as the human immunodeficiency virus (HIV) and the hepatitis B, C, and D viruses (HBV, HCV, and HDV, respectively) have been causing epidemics that kill millions of people for many decades [2,3]. For most viral diseases, there are no effective antiviral therapies or no therapies at all. A different approach is to activate specific or non-specific immune responses (immunotherapy or immunomodulation), which can contribute to the elimination or cessation of viral replication by adaptive and/or innate immunity.
The most important components of antiviral immune defense include innate immunity (mostly the interferon system [4] and natural killer cells [5]) and adaptive immunity (CD4+ and CD8+ T cells). Compared to CD8+ T-cytotoxic lymphocytes [6] and natural killer (NK) cells [7], which can destroy infected cells and thus contribute to viral clearance, activation of the interferon (IFN) system can upregulate expression of interferon-stimulated genes (ISGs), which directly inhibit viral replication [4].
Activating the IFN system is a complex, multistep process comprising (1) recognition of viral patterns (nucleic acids or proteins); (2) activation of adaptor kinases; (3) phosphorylation and nuclear translocation of transcriptional factors IRF3/IRF7; and (4) stimulation of IFN gene transcription. Secreted IFN molecules act as autocrine or paracrine mediators that stimulate IFN receptor complexes on the cell surface. The activation of IFN receptors stimulates the JAK/STAT signal transduction pathway and induces the expression of downstream ISGs [8,9]. Recombinant IFNs are approved for use as prophylactic or non-specific immunotherapy for a number of viral infections [10,11].
Notably, though quite effective in treating certain viral infections, therapeutic IFNs were developed several decades ago. Since then, immunology has made impressive progress, developing cytokine-based medications (e.g., recombinant IFNs [12]), next-generation chemical immunomodulators (agonists of Toll-like receptors [13,14] and antiviral sensors [15]) and biologics (therapeutic vaccines [15], chimeric antigen receptor (CAR) immune cells [16], retargeting and bispecific antibodies [17], etc.). However, in the never-ending evolutionary arms race between viruses and host immune responses, the latter is always at a disadvantage. Viruses have long been known to evade immunity, whilst the frequent mutations they acquire during replication in their host cells reduce the efficacy of antiviral therapies [18,19]. Due to the vast heterogeneity of viruses, the mechanisms of viral immune evasion are diverse and poorly described.
Modulating viral replication and modifying the immune response using novel molecular biology tools provide unprecedented means to study virus–host interactions and, possibly, build the foundation for new types of antivirals. In particular, adapting the bacterial defense system CRISPR–Cas for gene editing and beyond has already made these systems routine and very robust biological tools. CRISPR–Cas functions via binding of the Cas protein recruited to target DNA or RNA molecules by a short guiding RNA (sgRNA). Recognition of the target locus demands the presence of a short PAM sequence (two to seven nucleotides) immediately 3′ of the target site. Initially, pioneering studies of the recent Nobel Laureates J. Doudna and E. Charpentier demonstrated that the Cas9 protein can cleave target DNA by forming DNA double-strand breaks (DSB), so this technology can be used for programmed gene editing [20]. Less than 10 years later, the CRISPR toolkit has expanded dramatically and has been complemented with new CRISPR–Cas systems of different types, Cas proteins with modified PAM recognition (the PAMless Cas is possibly to be developed in the recent years), Cas proteins with increased or altered specificity, Cas nickases and related technologies (PrimeEditing, base editors), dead Cas proteins with additional functional domains (epigenetic modifiers, transcription activators/repressors), and other tools (reviewed in [21]). The invention of genome-wide CRISPR screens (CRISPRi, CRISPRa, CRISPRko) has allowed the examination of thousands of genes to identify their particular impact in human disease, including in infectious diseases [22]. Many CRISPR tools have been leveraged to develop novel antiviral approaches based on enhancing antiviral immune responses.
In this manuscript, we will review the main mechanisms of antiviral adaptive and innate immune responses and IFN systems, including recognition of viral patterns, activation of IFN secretion, and mechanisms by which viruses evade immunity. In the second part of the review, we will discuss the results and approaches used in recent studies to identify novel host factors, elucidate virus–host interactions, modulate antiviral immunity, and clarify the mechanisms of antiviral ISGs. Importantly, we also highlight recent developments in the use of CRISPR systems to enhance antiviral responses and their potential use as therapeutic agents in viral diseases.
2. The Role of Innate Immunity in Restricting Viral Replication
The innate immune system is the first line of defense against invading pathogens. Pathogen-associated molecular patterns (PAMP) can be sensed by pattern recognition receptors (PRR), resulting in the activation of signaling pathways that contribute to the elimination of these foreign agents [23]. One key player in these pathways is IFN. There are three families of IFN, with type I and III IFN serving as direct antivirals, and type II being the main regulator of antiviral innate and adaptive immune responses in different cell types [23]. Type I and III IFN activate the expression of ISGs, which have broad-spectrum antiviral activity [24].
2.1. Pattern Recognition Receptors (PRR)
Three main types of immune sensors for viral PAMPs (proteins and nucleic acids) have been discovered: Toll-like receptors (TLR), RIG-like receptors (RLR) (RIG-I, MDA5), cytosolic DNA sensors (DNA-PKcs, cGAS, AIM2 etc.), and nuclear DNA sensors (IFI16, hnRNPA2B1, cGAS etc.) [25,26,27,28]. TLR are localized on the cell surface (TLR1, TLR2, TLR4, TLR5, TLR6) and on the surface of endosomes (TLR3, TLR7, TLR8, TLR9). TLR2 and TLR4, anchored in the cell membrane, recognize viral proteins and induce downstream immune signaling. TLR3, TLR7/8, and TLR9 sense viral dsRNA (generated mostly as a by-product of viral replication), ssRNA, and CpG-rich DNA, correspondingly. Pathogen-derived RNA can also be detected by retinoic acid-inducible gene I (RIG-I)-like receptors (RIG-I and MDA-5) [29]. The main receptor that recognizes cytosolic DNA is cyclic GMP-AMP (cGAMP) synthase (cGAS) [30], but in various types of immune cells, foreign DNA can also be detected by the sensor-molecule “absent in melanoma 2” (AIM2) [28].
2.1.1. Viral Sensing by TLR and RLR: Concise Overview
TLRs are transmembrane proteins containing three domains: ectodomain recognizing PAMP, transmembrane domain, and cytosolic Toll/IL-1 receptor (TIR) domain [31]. Once bound to their ligands, TLRs oligomerize via their TIR domains, recruiting adaptor proteins and initiating signal transduction.
Depending on the adaptor proteins recruited to activated TLRs, downstream signaling occurs via “myeloid differentiation primary response 88” (MyD88)-dependent or TIR domain-containing adaptor-inducing interferon-β (TRIF)-dependent pathways (Figure 1A). Most TLRs recruit MyD88 adaptor, but TLR3 recruits TRIF [32]. MyD88 forms a signaling complex with kinases IRAK4 and IRAK1/2 [33], leading to IRAK4 trans-autophosphorylation followed by the activation of IRAK1/2. In turn, this complex phosphorylates and activates NF-κB, IRF5, and IRF7 [34]. TRIF interacts with TRAF, resulting either in TBK-1 activation with subsequent IRF-3 phosphorylation or in IkB degradation and the release and activation of NF-κB transcription factors. The main effect of NF-κB activation is the secretion of pro-inflammatory cytokines and activation of pro-IL-1β. IRF3 and IRF7 primarily result in induction of the IFN response, while IRF5 acts both as a pro-inflammatory factor and an IFN inducer.
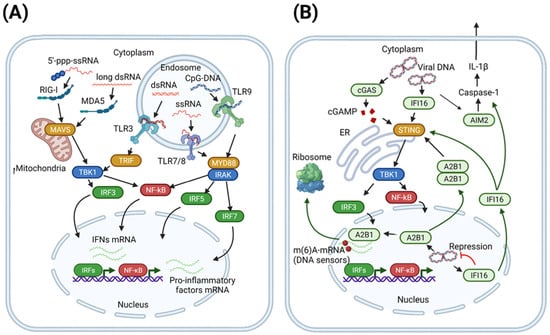
Figure 1.
Sensing of foreign nucleic acids. (A) TLR- and RLR-mediated sensing of foreign nucleic acids. Different types of cytoplasmic foreign RNA are recognized by RIG-I or MDA5 sensors followed by activation of MAVS and downstream TBK1-IRF signaling. In endosomes, RNA and CpG-DNA activate TLRs that result in one of the two signaling pathways involving TRIF or MYD88-IRAK. Activation of IRFs induces the expression of interferons and mRNA of pro-inflammatory factors. (B) Cytoplasmic and nuclear sensors of foreign DNA. Cytoplasmic DNA can be sensed by a number of sensors, including IFI16, cGAS, and AIM2. The first two factors activate the STING pathway that ultimately induces TBK1/IRF and interferon secretion. Upon recognition of cytoplasmic DNA, AIM2 induces caspase-1-dependent maturation of pro-IL-1β (pro-inflammatory response). A2B1 (hnRNPA2B1) and IFI16, among other factors, can participate in the sensing of foreign DNA in the nuclei of cells. IFI16 interferes with foreign nuclear DNA by epigenetic silencing, whereas A2B1 recognizes foreign DNA as well as activates and enhances innate antiviral responses. This picture was created in BioRender. Abbreviation: A2B1—hnRNPA2B1; m6a—methyl-6-adenine RNA; ER—endoplasmic reticulum.
Signaling through RIG-I and MDA5 receptors, which recognize cytosolic RNA, is essential for eliciting IFNα/β responses and, ultimately, clearing or inhibiting the incoming virus (Figure 1A). RIG-I and MDA5 signal through “mitochondrial antiviral-signaling protein” (MAVS; also known as IPS-1, VISA, and CARDIF) [29,35]. This increases the production of type I IFNs and pro-inflammatory cytokines. IFNs in turn induce ISG expression and suppress viral infection. In addition, the overactivation of mitochondrial MAVS adaptor protein leads to IFN-independent and Caspase-9-dependent activation of apoptosis in infected cells. The pro-apoptotic activity of MAVS can often be abrogated by different viral proteins as a mechanism of viral immune evasion [36].
RIG-I recognizes mainly blunt-end short RNAs with 5-triphospate [37], and MDA5 senses long viral dsRNA. RNA binding changes the conformation of RIG-I and MDA5: RIG-I forms tetramers, whereas MDA-5 oligomerizes. RIG-I and MDA-5 activation depends on the binding of their CARD domain to K63 polyubiquitin chains [38]. TRIM25 and RIPLET ubiquitin ligases synthesize polyubiquitin chains, promoting RIG-I signaling. Activated RIG-I and MDA-5 recruit MAVS for further signaling. Upon activation, MAVS aggregates on the mitochondrial surface [39]. When small aggregates are assembled, they can recruit other MAVS molecules to form large aggregates. MAVS activates signaling through IKK and TBK1 kinases, leading to NF-κB and IRF3 activation. Ubiquitin ligases TRAF2, TRAF5, and TRAF6 also can be recruited to MAVS and activate downstream signaling [40]. It is well established that RIG-I triggers the innate immune response during infection by orthomyxoviruses, paramyxoviruses, rhabdoviruses, and other viruses [41]. RIG-I is the major PRR that initiates the host antiviral response against hepatitis C virus (HCV) via recognition of poly-U/UC motifs in HCV RNA [42]. RIG-I was shown to be important both for sensing hepatitis B virus (HBV) RNA (more specifically, the 5′-ε region of HBV pre-genomic RNA, the major form of HBV RNA), but it is also a direct antiviral factor that impairs interaction between HBV polymerase and pre-genomic RNA [43].
The induction of miR146a by HBV was shown to attenuate antiviral innate immune responses by targeting RIG-I and RIG-I enhancer. Impairment of RIG-I and RIG-I enhancer signaling by HBV may be one of the major mechanisms responsible for the evasion of host immunity by HBV [44]. Similarly, activation of RIG-I was shown to inhibit human immunodeficiency virus (HIV) replication in macrophages by inducing the expression of critical ISGs such as APOBECs, tetherin, and CC chemokines [45]. However, HIV also developed a protease that counteracts RIG-I signaling: the expression of HIV protease promotes the loss of cytoplasmic RIG-I by sequestering it in lysosomes [46].
2.1.2. Foreign DNA Recognition by Cytosolic and Nuclear DNA Sensors
The cytosolic sensor of DNA is cyclic GMP-AMP synthase (cGAS), which contains two major DNA-binding domains and a nucleotidyltransferase domain [30]. cGAS binds DNA independently of the nucleotide sequence [30], instead targeting its sugar-phosphate backbone or recognizing Y-shaped structures of ssDNAs [47,48]. Upon binding to DNA in the cytosol, cGAS synthesizes a second messenger cGAMP from ATP and GTP [30], which activates the adaptor “stimulator of interferon genes” (STING). STING protein contains four transmembrane domains and is localized on the endoplasmic reticulum. STING itself cannot bind DNA, but it undergoes a conformational change upon cGAS binding and translocates to the nuclear compartment for TBK1 and IKK complex activation [49]. That leads to the activation of transcription factors IRF3 and NF-κB, resulting in the expression of type I IFN and pro-inflammatory cytokines [50].
Cytosolic DNA can also initiate inflammasome formation, which is a platform for pro-inflammatory cytokine maturation, via recognition by AIM2-like receptors (ALRs) [28,51]. Nucleic acid recognition leads to AIM2 dimerization and further interaction with apoptosis-associated speck-like protein containing a CARD (ASC) [52] followed by the activation of caspases [53].
Although initially enigmatic, in recent years, a plethora of potential nuclear sensors of foreign DNA were identified, including cGAS, IFI16, hnRNPA2B1, DA/ZBP1, TLR7/9, ZCCHC3, RNA Pol III, etc. [54]. Among them, for cGAS (recognition of DNA double-strand breaks), IFI16 (functions as a transcriptional repressor of foreign DNA), and hnRNPA2B1 (activates and amplifies antiviral response), DNA-sensing activity was directly determined (Figure 1B). More detailed information about the functioning of nuclear DNA sensors can be found in recent reviews [54].
2.2. Restriction of Viral Replication by Interferon-Stimulated Genes (ISGs)
Upon activation, PRRs initiate a signaling cascade that leads to IFN production. In turn, IFNs activate the JAK–STAT signaling pathway, resulting in the subsequent expression of numerous ISGs with broad antiviral activity. ISGs can restrict virtually every step of the viral life cycle (Figure 2).

Figure 2.
Restriction of viral life cycle by different ISGs for viruses with nuclear replication. ISGs with antiviral activity are shown for different steps of viral replication, including binding and viral entry, capsid disassembly, nuclear import, reverse transcription, viral nucleic acid replication/transcription, nuclear export, translation, capsid assembly, budding and release of viral particles. This picture was created in BioRender.
2.2.1. Restriction of Viral Entry
Viral entry is commonly inhibited by such ISGs as CH25H, which converts cholesterol to 25-hydrocholesterol (25HC). 25HC changes the composition of the cell membrane, thus blocking membrane fusion between the virus and cell [55]. CH25H has broad antiviral activity and reduces infection by vesicular stomatitis virus (VSV), herpes simplex virus (HSV), HIV, HCV, Ebola virus (EBOV), Nipah virus, Zika virus (ZIKV), and other viruses [55,56,57]. Another factor that inhibits viral endocytosis is human nuclear receptor coactivator 7 (NCOA7), which binds vacuolar H+-ATPase, resulting in the degradation of viral particles [58]. Interferon-induced transmembrane protein (IFITM) family proteins have also been shown to block viral infection at the stage of viral fusion and cytosolic entry. IFITM proteins have broad antiviral tropism, including influenza A virus (IAV), dengue virus (DENV), West Nile virus (WNV), EBOV, Marburg virus, SARS-CoV, SARS-CoV-2, rhabdovirus, bunyavirus, HCV, HIV, and others [59,60,61,62,63,64,65,66,67,68]. Most recently, screening of the ISG library revealed that lymphocyte antigen 6 complex, locus E (LY6) potently restricts infection by multiple coronaviruses, including SARS-CoV-2 [69].
2.2.2. Restriction of Protein Translation
Many ISGs impair viral protein translation. Protein kinase R (PKR) is widely known to inhibit the production of viral proteins [70]. The family of IFIT proteins inhibits the translation of viral proteins by different mechanisms, for example, by blocking the initiation of translation [71]. ISG15 can co-translationally conjugate with viral proteins [72]. Recently discovered ISG Schlafen 11 (SLFN11) inhibits rare tRNA codons, which are sometimes used by viruses [73]. Another mechanism is employed by an ISG Shiftless, which inhibits the ribosomal frameshifting used by HIV to regulate the ratio of its proteins [74].
2.2.3. Restriction of Viral Replication
Many ISGs can restrict viral replication. Viperin blocks DNA and RNA viruses by binding viral proteins and thus preventing their replication [75]. The process of reverse transcription can be targeted by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC3G), which induces mutations in the viral genome [76]. IFI6 was shown to disrupt replication flavivirus organelles [77], while RBBP6 protein impairs transcription of EBOV [78].
Several ISGs can destabilize and destroy viral RNAs. Oligoadenylate synthetases (OAS) catalyze the formation of 2′–5′-linked oligoadenylates that activate cellular RNase L, resulting in the degradation of viral RNA genomes [79]. Endonuclease ZAP can inhibit viral replication by preventing the accumulation of mRNA in the cytoplasm [80]. APOBEC3A, APOBEC3B [81] and AID [82] have been shown to directly deaminate and destroy HBV nuclear depo, covalently closed circular DNA, thus paving the ways for developing novel anti-HBV therapeutics. ISG20 can interfere with viral replication by several mechanisms. ISG20 can impair mRNA synthesis and protein translation of RNA viruses [83,84]. Potentially, ISG20 can also contribute to the restriction of HBV replication and degradation of HBV cccDNA by APOBEC3A [85].
2.2.4. Induction of Inflammatory Response
TLR activation can result in the maturation and migration of immune cells (dendritic cells), enhanced phagocytosis and generation of reactive oxygen species (macrophages and neutrophils), overproduction of co-stimulatory molecules (e.g., B cells), and other immune responses [86]. TLR activation must be tightly regulated for adequate innate immune response to pathogen DNA, and dysregulated TLR signaling is associated with chronic inflammatory conditions and, in some cases, septic shock [87,88,89].
Overall, the effects of ISGs and their mechanisms of action are diverse, with some playing a major role in the restriction of viral agents, while some being dispensable (or with an incremental effect) for the antiviral response. It is noteworthy that the effects of ISG interaction networks in some cases may have cumulative effect that, in certain infections, could be more important for viral restriction than effects of individual ISGs.
2.3. Host Factors Targeted by Viruses for Immune Evasion
The limited capacity of host cells to sense pathogen nucleic acids is explained by the existence of elaborate immune evasion mechanisms utilized by viruses, which have evolved a myriad mechanism allowing them to avoid recognition by immune sensors; some of these mechanisms are summarized in Table 1 and depicted in Figure 3.

Figure 3.
Mechanisms of viral immune evasion. (A) Evasion of immune recognition. (B) Blockade of interferon signaling. This picture was created in BioRender. Abbreviations: NA—nucleic acids; ISG—interferon-stimulated genes; IFNs—interferons; PTM—post-transcriptional modifications; SOCS—suppressor of cytokine signaling.
Many groups of viruses can inhibit PRRs themselves. For example, vaccinia virus (VACV) was reported to block TLR9 signaling by binding viral protein A46R to the MyD88 adaptor [90]. Human cytomegalovirus (HCMV) disrupts TLR signaling pathways with its protein US7, which promotes the ubiquitination of TLR3 [91]. Kaposi’s sarcoma-associated herpesvirus (KSHV) and HBV can block TLR2, TLR4, and TLR9 expression, reducing the levels of pro-inflammatory cytokines [92,93].

Table 1.
Host factors targeted by viruses and mechanisms of immune evasion.
Table 1.
Host factors targeted by viruses and mechanisms of immune evasion.
| Target | Virus | Viral Protein | Mechanism |
|---|---|---|---|
| RIG-I | Influenza virus | NS1 | Direct interaction, TRIM and RIPLET binding |
| Coxsackievirus B3 | 2Apro | Cleavage | |
| Epstein-Barr virus | LMP1 | Proteasomal degradation | |
| SARS-CoV | Nucleocapsid protein | Binding TRIM25 | |
| Respiratory syncytial virus | NS1 | Binding TRIM25 | |
| Human papilloma virus | E6 | Proteasomal degradation of TRIM25 | |
| WNV | NS1 | Proteasomal degradation of TRIM25 | |
| HCV | NS3-4A | Cleavage of RIPLET | |
| DENV/WNV | NS3 | Binding 14-3-3ε | |
| TLR9 | Vaccinia virus | A46R | MyD88 adaptor binding |
| TLR3 | Human cytomegalovirus (HCMV) | US7 | Resultant ubiquitination of TLR3 |
| HIV | No data | Inhibits phosphorylation of IRF3, IRF7, STAT1, STAT3 | |
| MDA5 | Measles virus | V | Prevention of MDA5 dephosphorylation |
| SARS-CoV-2 | NSP8 | Impairment of K63-linked polyubiquitination | |
| DENV/WNV | NS3 | Binding 14-3-3ε | |
| IFI16 | HSV1 | ICP0/Ul41 | Ubiquitinoylation/inhibition of expression |
| CMV | pUL83 | Direct interaction | |
| cGAS | HSV1 | Ul41/Ul37/Vp22 | Inhibition of expression or enzymatic activity |
| HCMV | UL31/pp65 | Direct interaction/enzymatic activity inhibition | |
| Kaposi’s sarcoma-associated herpesvirus (KSHV) | ORF52/LANA | Direct interaction/enzymatic activity inhibition | |
| ZIKV | NS1 | Stabilization of caspase1 | |
| TRIF | HCV | NS3-4A | Cleavage |
| MAVS | DENV | NS2B3 | Binding of MFN1 and MFN2 proteins |
| Rhinoviruses | 2A and 3C | Cleavage | |
| STING | Adenovirus | E1A | Binding of STING |
| HPV18 | E7 | Binding of STING | |
| KSHV | vIRF1 | Prevention of STING interaction with downstream factors | |
| HBV | Pol | Prevention of STING polyubiquitylation | |
| HCV | NS4B | Inhibition of downstream signaling | |
| HIV | Vpx | Antagonizes cGAS/STING-triggered NF-κB signaling [94] | |
| DENV | NS2B3 | Inhibition of downstream signaling | |
| Yellow fever virus | NS4B | Inhibition of downstream signaling | |
| IKKε | Lassa fever virus | Nucleoprotein | Inhibition of autocatalytic activity |
| EBOV | Vp53 | Direct binding | |
| TBK1 | HIV | Vpr and Vif | Direct binding |
| EBOV | Vp53 | Direct binding | |
| Human herpesvirus 8 | vIRF1 | Interaction with CBP/p300 | |
| HSV1 | ICP34.5 | Binding of TBK1 | |
| KSHV | ORF45 | Alternative substrate for TBK1 | |
| IRF7 | Enterovirus 68 | 3Cpro | Cleavage |
| STAT2 | DENV | NS5 | Ubiquitination |
| IRF3 | SARS-CoV | ORF3b, ORF6 and N | IRF-3 inhibition |
Other viruses have been shown to disrupt signaling by MDA5 and RIG-I, which are two crucial RNA sensors responsible for viral recognition in the cytoplasm. Enteroviruses (e.g., coxsackievirus B3) encode 2A and 3C proteases that cleave MDA5 and RIG-I [95]. Negative-sense, single-strand RNA measles virus (MV) can inhibit MDA5 activation by its V protein, which binds to phosphatases PP1α and PP1γ and prevents the dephosphorylation of MDA5 during infection [96]. Similarly, Epstein–Barr virus (EBV) encodes «latent membrane protein» (LMP1) that mediates RIG-I proteasomal degradation [97], whereas influenza virus NS1 protein can interact with RIG-I and impair its function [98]. One of the mechanisms whereby SARS-CoV-2 inhibits innate immunity is by suppressing of MDA-5 by nonstructural protein NSP8 [99].
Some viruses do not interact with MDA5 or RIG-I directly but rather counteract the activity of factors responsible for RIG-I activation, such as TRIM25 and RIPLET (mediate RIG-I ubiquitinoylation) and 14-3-3ε. Influenza virus protein NS1 binds both factors and decreases the production of IFNs [100], while SARS-CoV nucleocapsid protein and respiratory syncytial virus NS1 protein target TRIM25 and prevent the activation of RIG-I [101,102]. TRIM25 can also be targeted to proteasomal degradation by HPV’s E6 protein and WNV’s NS1 [103,104]. HCV proteases NS3-4A can cleave RIPLET and thus impair RIG-I activation [105]. The third factor, 14-3-3ε, can be bound by NS3 proteases of DENV and WNV that block subsequent immune response activation [106].
Viral DNA can be recognized and sensed not only by cytoplasmic (cGAS/STING), but also by nuclear DNA sensors, including IFI16, which can sense DNA viruses replicating in the nucleus [107]. Many viruses elaborated mechanisms to dampen IFI16 activity. For instance, immediate early protein ICP0 of HSV-1 ubiquitinylates IFI16, promoting its degradation [108,109]. The matrix protein pUL83 of HCMV blocks IFI16 signaling via direct interaction [110], while HCMV UL41 protein inhibits IFI16 and cGAS expression at the mRNA level [111,112].
ORF52 of KSHV and NS1 protein of ZIKV were likewise shown to inhibit cGas protein [113] and promote its degradation [114]. Similarly, Vp21 and the matrix protein UL37 of HSV1 inhibit cGAS [115,116]. HCMV UL31 interacts with cGAS, and its matrix protein pp65 binds cGAS to prevent its interaction with STING [117,118]. LANA protein of KSHV can also inhibit cGAS by direct interaction [119].
Viruses also can act on downstream factors of immune signaling pathways. In particular, NS proteases of HCV block several transcription factors involved in regulating innate immune responses. NS3-4A inhibits transcriptional factor IRF3 by binding MAVS [120] and cleaves the TRIF protein [121]. The NS2B3 protease of DENV binds and blocks MFN1 and MFN2 proteins, which are important regulators of MAVS [122]. MAVS is also targeted for cleavage and degradation by 2A and 3C proteases of rhinoviruses [123].
As already mentioned, STING is a crucial component of the cGAS–STING signaling axis that is responsible for detecting viral nucleic acids and deploying antiviral defense responses. Many DNA viruses interfere with the function of STING by cleaving it or impairing its ubiquitination (an important post-translational modification required for its function and for the regulation of innate immune responses). For example, adenoviral E1A protein and HPV18 E7 protein are known STING pathway inhibitors [124]. vIRF1 protein of KSHV prevents STING pathway activation by interacting with TBK1 and IRF3, which is the target of TBK1 and an important transcriptional factor required for IFN induction [125]. HBV polymerase prevents STING polyubiquitylation, reducing STING function [126] and impairing IFN-β induction. Surprisingly, RNA viruses also can affect the DNA-sensing STING pathway. This is significant, because many RNA viruses not directly detected by cytoplasmic or nuclear DNA sensors can induce the release of nuclear or mitochondrial DNA into the cytoplasm, followed by activation of the cGAS–STING axis and pro-inflammatory and antiviral innate responses. For instance, HCV NS4B protein, DENV NS2B3 protease, and yellow fever virus (YFV) NS4B protein all block STING downstream signaling and impair the IFN response [127].
IKKε and TBK1 kinases are signal transducers in MAVS and STING signaling pathways. IKKε is frequently targeted by viral proteins, including Lassa fever virus nucleoprotein [128], whereas TBK1 is inhibited by Vpr and Vif protein of HIV [129]. Both IKKε and TBK1 are blocked by EBOV Vp35 protein [130]. Herpesvirus ICP34.5 protein can bind TBK1, resulting in decreased IFN type I expression [131].
HHV-8 protein vIRF1 prevents the interaction of STING and TBK1, thereby inhibiting STING, TBK1, and IRF3 activation. This protein can also interact with transcriptional activator CBP/p300, impairing CBP/p300 and IRF3 association and reducing the efficacy of transcriptional activation from IRF3-dependent promoters [132]. A different strategy for TBK1 inactivation is used by KSHV protein ORF45, which competes with IRF3 as a TBK1 substrate, stopping innate immune signaling at the TBK1–IRF3 binding step [133].
Viruses can inhibit transcriptional factors that mediate IFN induction and ISG activation. Enterovirus 68 (EV-D68) 3Cpro cleaves IRF7 during infection [134]. DENV NS5 protein leads to the ubiquitination and degradation of STAT2 [135,136], which is a factor of signal transduction from receptor to IFN-I/III. NS5 can also inhibit IFN signaling by cleaving STAT2 [137]. SARS-CoV proteins ORF3b, ORF6, and N have been shown to antagonize the IFN pathway mostly by inhibiting IRF-3 protein [138]. ZIKV has been demonstrated to suppress IFNβ in vitro [139].
Chikungunya virus (CHIKV) effectively blocks the translation of ISG mRNAs, preventing the antiviral immune response [140]. One of the important mediators of this process may be the viral nsP2 protein, which could be involved in inhibiting STAT signaling [141]. Several studies reported that cellular nucleoporins are destroyed and mis-localized and that nucleo-cytoplasmic trafficking pathways are disrupted in cells infected with rhino- and polioviruses. The 2A protease of rhinoviruses (poliovirus or other enteroviruses) can cleave translation initiation factor elF4G, resulting in the translational shutdown of cellular mRNAs [142]. In addition, 2A protease cleaves nuclear pore proteins Nup62 and Nup98, while 3C protease cleaves Nup153 [135,143]. These cleavage events alter the efficacy of the host immune response signaling and promote immune evasion.
HIV Vif protein is widely known to block antiviral host responses by mediating the proteasomal degradation of APOBEC enzymes [144]. Tetherin, a host transmembrane IFN-induced protein, blocks the detachment and release of enveloped viruses. HIV can evade the antiviral activity of tetherin by expressing the Vpu protein, which binds tetherin and inhibits its activity [145].
To conclude, viruses develop elaborate mechanisms to evade or become resistant to innate antiviral response. Identifying factors able to abolish viral replication and contribute to viral clearance is important for developing novel therapeutics.
3. Emerging CRISPR–Cas Tools
Since their adaptation for genome editing, CRISPR–Cas systems have become one of the most widespread tools in molecular biology. The most commonly used CRISPR–Cas system is CRISPR–Cas type II, in which the Cas9 protein introduces a DSB in the target DNA site via nucleolytic activity of RuvC and HNH domains [20]. Resulting DSBs can be repaired predominantly by homologous recombination (HR) or non-homologous end joining (NHEJ), or occasionally by alternative pathways [146,147,148]. NHEJ is an error-prone DSB repair mechanism that results in out-of-frame mutations at the DSB site and, as a consequence, leads to gene inactivation. Alternatively, in the presence of a homologous DNA template, DSBs can be repaired by HR, which preserves the integrity of the genome [149]. The main disadvantages of CRISPR–Cas-mediated cleavage include (a) potential genotoxicity [150,151], the formation of large (up to several kb) deletions at the target site and extensive chromosome aberrations, including chromothripsis [152,153]; (b) low efficacy of DNA integration by the HR mechanism [154]; (c) intracellular responses to CRISPR–Cas components [155]; and (d) potential involvement of repair pathways other than NHEJ or HR that may yield unpredictable on-target mutations [146,156].
Inactivating Cas nucleolytic activity by single-point mutations in RuvC and HNH nuclease domains generates a nuclease-null or “dead” Cas (dCas) system [157]. The dCas protein retains its ability to bind to the target site but cannot generate DSBs. Developing chimeric dCas-X systems, in which X is any functional domain, endowed CRISPR–Cas with numerous additional functional modalities and enabled unprecedented manipulation of both the genome and the epigenome [21].
3.1. Modulation of Gene Transcription by CRISPR
CRISPR-activation (CRISPRa) or interference (CRISPRi) systems are based on fusing transcriptional activators/repressors to dCas [157]. The system is recruited to the regulatory regions of target genes (promoters or enhancers) to transactivate or suppress gene transcription. The major advantages of the CRISPRa approach over overexpressing cDNA are the precise, tunable control of target gene expression and ability to overexpress selected or all isoforms of a gene.
First-generation CRISPRa systems include dCas9–(VP16)n [158], dCas9–p65–HSF1 [159], and dCas9–p300 [160], and they are characterized by relatively low activation efficiency. Robust activation of target gene transcription is feasible when using several sgRNAs targeting an extended genomic region. Historically, one of the first activators was based on herpesvirus protein VP16 domains, which transactivate gene expression by recruiting pre-initiation complex factors to the dCas9–VP16-bound regions. The signal produced by VP16-based CRISPRa systems can be further amplified by fusing multiple VP16 domains (3–10 units) [158,161]. The transcription factor p65-HSF1 is another example of directly activated gene transcription. Another CRISPRa system takes advantage of p300 acetyltransferase, which modifies the epigenetic state of the promoter and enhancer regions by directly acetylating histone H3 at lysine K27 [160]. This acetylation induces gene transcription and may also result in the recruitment of additional transcription factors to the region of interest.
Second-generation CRISPRa systems include Scaffold [162], VPR [163], SunTag variants [164], SAM [165], and more recently developed dCas9–CBP [166] and SPH [167]. Compared to first-generation systems, these systems are characterized by improved on-target gene activation, and they allow the multiple overexpression of genes with a single sgRNA. The key property of second-generation CRISPRa systems is that they mobilize several activation domains. They rely on different strategies, such as dCas9 fused to various activation domains. The principle of second-generation activators is the recruitment of several activation domains to target sites. The VPR activator consists of VP64 herpes virus protein, p65 transcriptional factor, and Rta (transcription factor of Epstein–Barr virus). In the SunTag system, GCN4 peptide arrays are fused to dCas9. In the same cell, activation domains (VP64, p300, p65-HSF1, or others) fused to GCN4-scFv are produced. GCN4-scFv is a single-chain antibody fragment with a high affinity for GCN4 arrays. As a result, up to 10 copies of hybrid Activator-scFv domains are recruited to the dCas9 binding site to activate transcription. The scaffold uses unmodified dCas9 protein and sgRNA modified with two special RNA aptamers (e.g., MS2 aptamers). Additionally, activators fused with the aptamer-specific protein are produced (MCP protein in case of MS2). Four activator–MCP domains are recruited to sgRNA aptamers in the dCas9 binding site for transcriptional induction. In SAM, dCas9 protein is modified with VP64 activator and attracts four p65–HSF1 activation domains to aptamer-modified sgRNA for pronounced transcriptional activation (reviewed in [21]).
The most widely used CRISPRi system is dCas9-KRAB [168]. The KRAB inhibitory domain attracts epigenetic enzymes that mediate the deposition of inactive chromatin marks (H3K9Me3 and H3K27Me3) to the target site (gene promoters or enhances), subsequently repressing transcription. Alternatively, the EZH2 inhibitory domain may be used [169].
3.2. CRISPR-Based DNA- and RNA-Editing Tools
Base editors (BE) are molecular tools that introduce pinpoint nucleotide replacements into the target DNA site. Functional domains of BE are cytidine and adenosine deaminases. Instead of dCas proteins, nCas9 with only one inactivated nuclease domain is used in the latest BEs. After binding to the target site, deaminases catalyze the editing of nucleotides in a narrow nucleotide window of one DNA strand and nicking of the complementary strand. Cytidine BEs catalyze cytidine deamination and generation on uridine (U). After the nicked strand and deaminated nucleotides are repaired, U is changed to a thymine (T) residue. Adenine BEs generate inosine (I) instead of adenine, resulting in guanine (G) after repair. Thus, cytosine BEs mediate C→T/G→A editing and adenine BEs mediate A→G/T→C editing. Base editing results in very infrequent insertion/deletion (indel) mutations and thus a safer analog of the gene editing systems. CRISPR-STOP and iSTOP technologies use cytidine BE to create stop codons in early exons of genes, knocking out gene expression without introducing DSB. More extensive descriptions and characterization of different CRISPR–Cas tools can be found in other reviews [170].
In the recent years, several RNA editing tools based on RNA-specific CRISPR–Cas13 were developed. Cas13 proteins are specific to RNA, interacting with target moieties by CRISPR RNA (crRNA), and they exhibit ssRNA-specific nuclease activity. Cas13-based approaches have already been used to target viral RNA and inhibit the replication of ssRNA viruses, including LCMV, IAV, VSV, SARS-CoV-2, etc. [171,172]. Nucleolytically null dead Cas13 proteins (dCas13) were generated and used as platforms to develop RNA base editors, such as REPAIR (utilizes an evolved variant of ADAR2 adenine deaminase to deaminate nucleotides). dCas13 serves as a vehicle to recruit ADAR2 to the crRNA:target RNA duplex. Next, ADAR2 mediates A→I conversion in target RNA [173].
Evolved ADAR2 protein exhibits dual A→I and C→U deaminase activity (RESCUE system) [174]. By replacing ADAR2 deaminase domain with APOBEC3A deaminase, it was possible to generate a C→U RNA-specific editase (CURE system) [175]. Additionally, truncated variants of Cas13 RNA editing tools (<1000 aa) that can be easily packaged into common AAV were engineered [176]. These advancements have become a prominent milestone on the path to create CRISPR-based therapeutics.
Differences in the properties and applications of DNA and RNA base editors were reviewed elsewhere [21]. A short summary about CRISPRa/CRISPRi and base editing systems is listed in Table 2.

Table 2.
Comparison of most common CRISPRa, CRISPRi, and base editing systems.
Different variants of CRISPR–Cas systems are widely used in basic and translational research. Current directions of CRISPR–Cas applications in antiviral immunity can be divided into three areas: screening for potent antiviral genes, basic research into antiviral immunity, and development of antiviral approaches, which we review below.
5. Modulating Epitranscriptomics in Viral Infections
Epitranscriptomics is a dynamically developing research field. The main RNA epigenetic mark is methyl-6-adenosine (m6A) [263]. While its function is not completely understood, its role in different RNA metabolism processes has been established, including translation, degradation, splicing, export, and folding [264]. The regulation of m6A deposition and functions is mediated by three classes of enzymes: m6A writers, m6a erasers, and m6A readers [265]. Writers are part of the RNA methyltransferase complex, which catalyzes the methylation of adenosine residues on RNA and establishes m6A markers (for example, METTL3, METTL14, and other proteins). Erasers perform the opposite function, removing m6A residues. Among m6A erasers are ALKBH5 and FTO proteins. m6A readers bind m6A residues on RNA and enhance translation, induce degradation, or perform other functions. Examples of m6A readers are YTHDC1/2 and YTHDF1/2/3 proteins (m6A metabolism reviewed in [266,267]).
To date, m6A metabolism is known to significantly impact the life cycle of pathogenic viruses (including HIV, EV71, IAV, KSHV, HBV, HCV, HPV, ZIKV, and EBV). In viral biology, m6A can regulate viral replication and/or viral RNA stability, latency state, cell cycle, and other functions. m6A is also involved in the progression of virus-related tumors (including HBV- and HCV-related hepatocellular carcinoma and HPV-related cervical carcinoma) (reviewed in [268]).
The participation of m6A in regulating antiviral immunity has been described as well [269]. hnRNPA2B1, a recently identified nuclear sensor of viral DNA, amplifies type I IFN response after HSV-1 infection by inducing cGAS, IFI16, and STING proteins via m6A-dependent binding of hnRNPA2B1 to corresponding mRNAs; this subsequently increases the nuclear export and translation of these proteins [270]. Using a model of human metapneumovirus, Lu et al. (2020) demonstrated that m6A deposition on viral RNA reduces its recognition by RIG-I RNA sensors and decreases the activation of IFN secretion. Viral RNA may be recognized as self RNA by RIG-I sensors after m6A deposition, allowing immune evasion [271]. Similarly, Chen et al. (2019) reported that m6A can serve as a marker of self RNA to prevent the activation of sensors such as RIG-I, MDA5, OAS, OASL, and PKR [272]. Additionally, Winkler et al. (2019) demonstrated that IFNB mRNA can be targeted for methylation by m6A deposition, blocking the IFN response. Depleting m6A writers and readers activates the IFN response and depletes viral propagation in models of different viral infections, including CMV, IAV, adenovirus, and VSV. Moreover, CMV infection leads to an increased expression of m6A writers and readers, potentially increasing the degradation of IFN mRNA to allow immune evasion [269]. At the same time, DDX46 factor has been demonstrated to mobilize the m6A eraser ALKBH5 to induce the retention of Mavs, Traf3, and Traf6 mRNAs in the nucleus and prevent their translation. Thus, DDX46 negatively regulates IFN response by inducing m6A depletion (not deposition) on mRNA of immune factors.
Investigations of epitranscriptomics were initially based on CRISPR–Cas mediated knockout of m6A modulators (writers, readers, or erasers) [269]. Such an approach only increased or decreased the overall levels of m6A methylation, but some m6A-related effects have been shown to be dependent on RNA localization [273]. Creating systems for site-specific m6A methylation/demethylation has become a new goal of epitranscriptomics. Systems for RNA methylation are based on using a dCas9 protein fused to single-chain METTL14-METTL3 methyltransferase complex (dCas9-based writer). The target site for RNA methylation is determined by sgRNA and short PAM-containing antisense DNA oligonucleotide (PAMmer). PAMmer is necessary for the system to function, as Cas9-based proteins cannot bind RNA independently of it. m6A demethylation systems are dCas9 proteins fused to ALKBH5 or FTO m6A demethylation enzymes (dCas9-based erasers) [274]. Replacing the dCas9 protein in this system with a type VI dCas13 protein eliminated the necessity for PAMmers, because type VI proteins, including dCas13, are specific to ssRNA and can bind it independently. These modified m6A writer and eraser systems have become simpler and more convenient than dCas9-based systems [275].
6. Conclusions
Innate immunity is the first line of defense against infectious pathogens. Vertebrate hosts have evolved multiple mechanisms to restrict viral infection, suppress viral replication, and ultimately clear the virus from the body. However, under such selective pressure, viruses efficiently adapt to evade immunity and establish productive infection. Viruses can block the expression and/or function of various host proteins, become resilient to antiviral factors, or develop a stealth mode to avoid the recognition and triggering of immune responses. The interaction of viruses with infected cells is an area of active research that is promising not only for fundamental discoveries but also for translational studies and clinical practice.
Although potentially very promising, CRISPR–Cas approaches suffer from a number of limitations that may perturb their application in vivo, in clinical practice. First, the off-target activity of CRISPR–Cas systems is one of the most important challenges that is yet to be overcome despite the recent development of high-fidelity Cas variants. The generation of megabase-long indels, chromothripsis, etc. as a result of on-target or off-target nucleolytic cleavage of DNA may compromise the attempts to use Cas nucleases for therapeutic purposes [151,276]. Similar to Cas nucleases, the off-target activity of RNA-nucleases and RNA base editors was described. A reduction in the off-target activity of CRISPR-based approaches is possible by using evolved Cas proteins and functional domains with improved properties, orthologous nucleases, modified sgRNAs, or by introducing CRISPR–Cas as ribonucleoprotein complexes [277,278,279,280,281]. Second, Cas proteins are derived from bacteria and archaea, so that many enzymes are immunogenic and can be rapidly eliminated from the body by pre-existing immunity [282]. Among other approaches, the engineering of Cas proteins with immunosilenced or masked epitopes [283], and the application of Cas proteins from non-pathogenic organisms could become potential solutions for the safe, repeated administration of CRISPR–Cas in vivo. Last, but not least, is the issue of CRISPR–Cas in vivo targeted delivery, which is hampered by the large molecular weight of Cas proteins and dCas-based proteins as well as opportunities to deliver CRISPR–Cas as DNA, RNA, or in the form of ribonucleoprotein complexes. Pros and cons of these strategies are reviewed in [284].
To conclude, novel CRISPR and CRISPR-based tools are undoubtedly the most robust and efficient means to investigate virus–host interactions, identify and overcome mechanisms of viral immune evasion, and develop novel antiviral approaches. CRISPR–Cas has already revolutionized fundamental and translational research, as well as the field of molecular diagnostics. Clinical application of CRISPR–Cas techniques has recently provided promising early in vivo results to cure human genetic [285] and infectious diseases [286]. Further progress in this field will profoundly depend on the safety of CRISPR–Cas technologies for particular purposes and the effective delivery of gene editing complexes to the desired organs.
Author Contributions
Conceptualization, S.B., A.K., D.K.; writing—original draft preparation, S.B., A.K., D.K.; writing—review and editing, S.B., E.B., E.V., V.G., I.G., D.G., D.K.; supervision, D.K., V.C.; project administration, D.K.; funding acquisition, D.G., D.K., V.C. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by RFBR-DFG grant 20-515-12010 and GL 595/9-1; and RFBR grant 20-015-00442. The National Reference Center for Hepatitis B Viruses and Hepatitis D Viruses at the Justus Liebig University Giessen is supported by the German Ministry of Health via the Robert Koch Institute, Berlin, Germany.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| HIV | Human immunodeficiency virus |
| HBV | Hepatitis B virus |
| HCV | Hepatitis C virus |
| HDV | Hepatitis D virus |
| NK | Natural killer cells |
| IFN | Interferon |
| ISGs | Interferon-stimulated genes |
| JAK/STAT | Janus kinase/signal transducers and activators of transcription |
| DSB | Double-strand breaks |
| PAMP | Pathogen-associated molecular patterns |
| PRR | Pattern recognition receptors |
| TLR | Toll-like receptors |
| ssRNA | Single-stranded RNA |
| dsRNA | Double-stranded RNA |
| RIG-I | Retinoic acid-inducible gene I |
| cGAMP | Cyclic GMP-AMP |
| cGAS | Cyclic GMP-AMP synthase |
| AIM2 | Absent in melanoma 2 |
| TIR | Toll/IL-1 receptor |
| TRIF | TIR domain-containing adaptor-inducer interferon-β |
| MAVS | Mitochondrial antiviral-signaling protein |
| ssDNA | Single-stranded DNA |
| STING | Stimulator of interferon genes |
| ALRs | AIM2-like receptors |
| ASC | Apoptosis-associated speck-like protein containing a CARD |
| 25HC | 25-hydrocholesterol |
| VSV | Vesicular stomatitis virus |
| HSV | Herpes simplex virus |
| EBOV | Ebola virus |
| ZIKV | Zika virus |
| NCOA7 | Nuclear receptor coactivator 7 |
| IAV | Influenza A virus |
| DENV | Dengue virus |
| WNV | West Nile virus |
| PKR | Protein kinase R |
| SLFN11 | Schlafen 11 |
| APOBEC | Apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like |
| IFI16 | Gamma-interferon-inducible protein 16 |
| IFITM1 | Interferon-induced transmembrane protein 1 |
| OAS | Oligoadenylate synthetase |
| VACV | Vaccinia virus |
| HCMV | Human cytomegalovirus |
| KSHV | Kaposi’s sarcoma-associated herpesvirus |
| MV | Measles virus |
| LMP1 | Latent membrane protein 1 |
| YFV | Yellow fever virus |
| HDAC | Histone deacetylase |
| SINV | Sin Nombre virus |
| ONNV | o’nyong’nyong virus |
| SINV | Sindbis virus |
| SVV | Seneca Valley virus |
| VEEV | Venezuelan encephalitis virus |
| CHIKV | Chikungunya virus |
| RV | Rotavirus |
| MVA | Modified vaccinia virus Ankara |
| CMV | Cytomegalovirus |
| SINV-EEEV | chimeric eastern equine encephalitis virus |
| SeV | Sendai virus |
| m6A | methyl-6-adenosine |
| TTLL12 | Tubulin Tyrosine Ligase Like 12 |
References
- Sayed, A.; Peng, B. Pandemics and income inequality: A historical review. SN Bus. Econ. 2021, 1, 1–17. [Google Scholar] [CrossRef]
- World Health Organization. Global Hepatitis Report; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- GBD 2016 Mortality Collaborators. Global, regional, and national under-5 mortality, adult mortality, age-specific mortality, and life expectancy, 1970–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017, 390, 1084–1150. [Google Scholar] [CrossRef] [Green Version]
- Malmgaard, L. Induction and regulation of IFNs during viral infections. J. Interf. Cytokine Res. 2004, 24, 439–454. [Google Scholar] [CrossRef]
- Biron, C.A.; Nguyen, K.B.; Pien, G.C.; Cousens, L.P.; Salazar-Mather, T.P. Natural killer cells in antiviral defense: Function and regulation by innate cytokines. Annu. Rev. Immunol. 1999, 17, 189–220. [Google Scholar] [CrossRef]
- Uzhachenko, R.V.; Shanker, A. CD8+ T lymphocyte and NK cell network: Circuitry in the cytotoxic domain of immunity. Front. Immunol. 2019, 10, 1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waggoner, S.N.; Reighard, S.D.; Gyurova, I.E.; Cranert, S.A.; Mahl, S.E.; Karmele, E.P.; McNally, J.P.; Moran, M.T.; Brooks, T.R.; Yaqoob, F.; et al. Roles of natural killer cells in antiviral immunity. Curr. Opin. Virol. 2016, 16, 15–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akira, S.; Uematsu, S.; Takeuchi, O. Pathogen recognition and innate immunity. Cell 2006, 124, 783–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Platanias, L.C. Mechanisms of type-I-and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005, 5, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Antonelli, G.; Scagnolari, C.; Moschella, F.; Proietti, E. Twenty-five years of type I interferon-based treatment: A critical analysis of its therapeutic use. Cytokine Growth Factor Rev. 2015, 26, 121–131. [Google Scholar] [CrossRef]
- Park, A.; Iwasaki, A. Type I and type III interferons–induction, signaling, evasion, and application to combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Isorce, N.; Lucifora, J.; Zoulim, F.; Durantel, D. Immune-modulators to combat hepatitis B virus infection: From IFN- D to novel investigational immunotherapeutic strategies. Antiviral Res. 2015, 122, 69–81. [Google Scholar] [CrossRef]
- Durantel, D.; Zoulim, F. Review New antiviral targets for innovative treatment concepts for hepatitis B virus and hepatitis delta virus. J. Hepatol. 2016, 64, S117–S131. [Google Scholar] [CrossRef] [PubMed]
- Martinsen, J.T.; Gunst, J.D.; Højen, J.F.; Tolstrup, M.; Søgaard, O.S. The use of Toll-like receptor agonists in HIV-1 cure strategies. Front. Immunol. 2020, 11, 1112. [Google Scholar] [CrossRef] [PubMed]
- Fanning, G.C.; Zoulim, F.; Hou, J.; Bertoletti, A. Therapeutic strategies for hepatitis B virus infection: Towards a cure. Nat. Rev. Drug Discov. 2019, 18, 827–844. [Google Scholar] [CrossRef] [PubMed]
- Zhen, A.; Carrillo, M.A.; Kitchen, S.G. Chimeric antigen receptor engineered stem cells: A novel HIV therapy. Immunotherapy 2017, 9, 401–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nyakatura, E.K.; Soare, A.Y.; Lai, J.R. Bispecific antibodies for viral immunotherapy. Hum. Vaccin. Immunother. 2017, 13, 836–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finlay, B.B.; McFadden, G. Anti-immunology: Evasion of the host immune system by bacterial and viral pathogens. Cell 2006, 124, 767–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowie, A.G.; Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Brezgin, S.; Kostyusheva, A.; Kostyushev, D.; Chulanov, V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019, 20, 6041. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [Green Version]
- Feng, J.; Wickenhagen, A.; Turnbull, M.L.; Rezelj, V.V.; Kreher, F.; Tilston-Lunel, N.L.; Slack, G.S.; Brennan, B.; Koudriakova, E.; Shaw, A.E.; et al. Interferon-Stimulated Gene (ISG)-Expression Screening Reveals the Specific Antibunyaviral Activity of ISG20. J. Virol. 2018, 92, e02140-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burleigh, K.; Maltbaek, J.H.; Cambier, S.; Green, R.; Gale, M., Jr.; James, R.C.; Stetson, D.B. Human DNA-PK activates a STING-independent DNA sensing pathway. Sci. Immunol. 2020, 5, eaba4219. [Google Scholar] [CrossRef] [Green Version]
- Akira, S.; Takeda, K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004, 4, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Barral, P.M.; Sarkar, D.; Su, Z.-Z.; Barber, G.N.; DeSalle, R.; Racanello, V.R.; Fisher, P.B. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: Key regulators of innate immunity. Pharmacol. Ther. 2009, 124, 219–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burckstummer, T.; Baumann, C.; Blüml, S.; Dixit, E.; Dürnberger, G.; Jahn, H.; Planyavsky, M.; Bilban, M.; Colinge, J.; Bennet, K.L.; et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 2009, 10, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, M.; Kikuchi, M.; Natsukawa, T.; Shinobu, N.; Imaizumi, T.; Miyagushi, M.; Taira, K.; Akira, S.; Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004, 5, 730–737. [Google Scholar] [CrossRef]
- Sun, L.; Wu, J.; Du, F.; Chen, X.; Chen, Z.J. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 2013, 339, 786–791. [Google Scholar] [CrossRef] [Green Version]
- Kawai, T.; Akira, S. The role of pattern-recognition receptors in innate immunity: Update on Toll-like receptors. Nat. Immunol. 2010, 11, 373–384. [Google Scholar] [CrossRef]
- Sato, S.; Sugiyama, M.; Yamamoto, M.; Watanabe, Y.; Kawai, T.; Takeda, K.; Akira, S. Toll/IL-1 receptor domain-containing adaptor inducing IFN-beta (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-kappa B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 2003, 171, 4304–4310. [Google Scholar]
- Lin, S.-C.; Lo, Y.-C.; Wu, H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawai, T.; Sato, S.; Ishii, K.J.; Coban, C.; Hemmi, H.; Yamamoto, M.; Terai, K.; Matsuda, M.; Inoue, J.-i.; Uematsu, S.; et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004, 5, 1061–1068. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Moore, C.B.; Leisman, R.M.; O’Connor, B.P.; Berhstralh, D.T.; Chen, Z.J.; Pickles, R.J.; Ting, J.P.-Y. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS ONE 2009, 4, e5466. [Google Scholar] [CrossRef]
- Hornung, V.; Ellegast, J.; Kim, S.; Brzózka, K.; Jung, A.; Kato, H.; Poeck, H.; Akira, S.; Conzelmann, K.-K.; Schlee, M.; et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science 2006, 314, 994–997. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Kinch, L.N.; Brautigam, C.A.; Chen, X.; Du, F.; Grishin, N.V.; Chen, Z.J. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 2012, 36, 959–973. [Google Scholar] [CrossRef] [Green Version]
- Hou, F.; Sun, L.; Zheng, H.; Skaug, B.; Jiang, Q.-X.; Chen, Z.J. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 2011, 146, 448–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Chen, J.; Cai, X.; Wu, J.; Chen, X.; Wu, Y.-T.; Sun, L.; Chen, Z.J. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2013, 2, e00785. [Google Scholar] [CrossRef]
- Kato, H.; Takeuchi, O.; Sato, S.; Yoneyama, M.; Yamamoto, M.; Matsui, K.; Uematsu, S.; Jung, A.; Kawai, T.; Ishii, K.J.; et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006, 441, 101–105. [Google Scholar] [CrossRef]
- Saito, T.; Owen, D.M.; Jiang, F.; Marcotrigiano, J.; Gale, M., Jr. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 2008, 454, 523–527. [Google Scholar] [CrossRef] [Green Version]
- Sato, S.; Li, K.; Kameyama, T.; Hayashi, T.; Ishida, Y.; Murakami, S.; Watanabe, T.; Lijima, S.; Sakurai, Y.; Watashi, K.; et al. The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for Hepatitis B Virus Article The RNA Sensor RIG-I Dually Functions as an Innate Sensor and Direct Antiviral Factor for Hepatitis B Virus. Immunity 2015, 42, 123–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, Z.; Zhang, J.; Han, Q.; Su, C.; Qu, J.; Xu, D.; Zhang, C.; Tian, Z. Hepatitis B virus inhibits intrinsic RIG-I and RIG-G immune signaling via inducing miR146a. Sci. Rep. 2016, 6, 26150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, X.; Li, J.; Zhou, Y.; Ho, W. RIG-I activation inhibits HIV replication in macrophages. J. Leukoc. Biol. 2013, 94, 337–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solis, M.; Nakhaei, P.; Jalalirad, M.; Lacoste, J.; Douville, R.; Arguello, M.; Zhao, T.; Laughrea, M.; Wainberg, M.A.; Hiscott, J. RIG-I-mediated antiviral signaling is inhibited in HIV-1 infection by a protease-mediated sequestration of RIG-I. J. Virol. 2011, 85, 1224–1236. [Google Scholar] [CrossRef] [Green Version]
- Civril, F.; Deimling, T.; de Oliviera Mann, C.C.; Ablasser, A.; Moldt, M.; Witte, G.; Hornung, V.; Hopfner, K.-P. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wu, J.; Du, F.; Xu, H.; Sun, L.; Chen, Z.; Brautigam, C.A.; Zhang, X.; Chen, Z.J. The cytosolic DNA sensor cGAS forms an oligomeric complex with DNA and undergoes switch-like conformational changes in the activation loop. Cell Rep. 2014, 6, 421–430. [Google Scholar] [CrossRef] [Green Version]
- Gao, P.; Ascano, M.; Zillinger, T.; Wang, W.; Dai, P.; Serganov, A.A.; Gaffney, B.L.; Shuman, S.; Jones, R.A.; Deng, L.; et al. Structure-function analysis of STING activation by c[G(2′,5′)pA(3′,5′)p] and targeting by antiviral DMXAA. Cell 2013, 154, 748–762. [Google Scholar] [CrossRef] [Green Version]
- Ishikawa, H.; Barber, G.N. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 2008, 455, 674–678. [Google Scholar] [CrossRef]
- Tschopp, J.; Martinon, F.; Burns, K. NALPs: A novel protein family involved in inflammation. Nat. Rev. Mol. Cell Biol. 2003, 4, 95–104. [Google Scholar] [CrossRef]
- Fernandes-Alnemri, T.; Yu, J.-W.; Datta, P.; Wu, J.; Alnemri, E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009, 458, 509–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamkanfi, M.; Dixit, V.M. Mechanisms and functions of inflammasomes. Cell 2014, 157, 1013–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, F.; Yuan, Y.; Ma, F. Function and regulation of nuclear DNA sensors during viral infection and tumorigenesis. Front. Immunol. 2020, 11, 624556. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Y.; Aliyari, R.; Chikere, K.; Li, G.; Marsden, M.D.; Smith, J.K.; Pernet, O.; Guo, H.; Nusbaum, R.; Zack, J.A.; et al. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 2013, 38, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Deng, Y.-Q.; Wang, S.; Ma, F.; Aliyari, R.; Huang, X-Y.; Zhang, N-N.; Watanabe, M.; Dong, H-L.; Liu, P.; et al. 25-Hydroxycholesterol protects host against Zika virus infection and its associated microcephaly in a mouse model. Immunity 2017, 46, 446–456. [Google Scholar] [CrossRef] [Green Version]
- Romero-Brey, I.; Romero-Brey, I.; Berger, C.; Colpitts, C.C.; Boldanova, T.; Engelmann, M.; Todt, D.; Perin, P.M.; Behrendt, P.; Vondran, F.W.R.; et al. Interferon-inducible cholesterol-25-hydroxylase restricts hepatitis C virus replication through blockage of membranous web formation. Hepatology 2015, 62, 702–714. [Google Scholar]
- Doyle, T.; Moncorgé, O.; Bonaventure, B.; Pollpeter, D.; Lussignol, M.; Tauziet, M.; Apolonia, L.; Catanese, M.-T.; Goujon, C.; Malim, M.H. The interferon-inducible isoform of NCOA7 inhibits endosome-mediated viral entry. Nat. Microbiol. 2018, 3, 1369–1376. [Google Scholar] [CrossRef] [PubMed]
- Kühnl, A.; Musiol, A.; Heitzig, N.; Johnson, D.; Ehrhardt, C.; Grewal, T.; Gerke, V.; Ludwig, S.; Rescher, U. Late endosomal/lysosomal cholesterol accumulation is a host cell-protective mechanism inhibiting endosomal escape of influenza A virus. MBio 2018, 9, e01345-18. [Google Scholar] [CrossRef] [Green Version]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.H.; Everitt, A.R.; Vora, M.; et al. The CD225 domain of IFITM3 is required for both IFITM protein association and inhibition of influenza A virus and dengue virus replication. J. Virol. 2013, 87, 7837–7852. [Google Scholar] [CrossRef] [Green Version]
- Tartour, K.; Nguyen, X.-N.; Approurchaux, R.; Assil, S.; Barateau, V.; Bloyet, L-M.; Gaillard, J.B.; Confort, M-P.; Escudero-Perez, B.; Gruffat, H.; et al. Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog. 2017, 13, e1006610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wrensch, F.; Karsten, C.B.; Gnirß, K.; Hoffmann, M.; Lu, K.; Takada, A.; Winkler, M.; Simmons, G.; Pöhlmann, S. Interferon-Induced Transmembrane Protein–Mediated Inhibition of Host Cell Entry of Ebolaviruses. J. Infect. Dis. 2015, 212, S210–S218. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Sehgal, M.; Hou, Z.; Cheng, J.; Shu, S.; Wu, S.; Guo, F.; Le Marchand, S.J.; Lin, H.; Chang, J.; et al. Identification of residues controlling restriction versus enhancing activities of IFITM proteins on entry of human coronaviruses. J. Virol. 2018, 92, e01535-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, G.; Kenney, A.D.; Kudryashova, E.; Zani, A.; Zhang, L.; Lai, K.K.; Hall-Stoodley, L.; Robinson, R.; Kudryashov, D.S.; Compton, A.A.; et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021, 40, e106501. [Google Scholar] [CrossRef]
- Weidner, J.M.; Jiang, D.; Pan, X.B.; Chang, J.; Block, T.M.; Guo, J.T. Interferon-induced cell membrane proteins, IFITM3 and tetherin, inhibit vesicular stomatitis virus infection via distinct mechanisms. J. Virol. 2010, 84, 12646–12657. [Google Scholar] [CrossRef] [Green Version]
- Mudhasani, R.; Tran, J.P.; Retterer, C.; Radoshitzky, S.R.; Kota, K.P.; Altamura, L.A.; Smith, J.M.; Packard, B.Z.; Luhn, J.H.; Constantino, J.; et al. IFITM-2 and IFITM-3 but not IFITM-1 restrict Rift Valley fever virus. J. Virol. 2013, 87, 8451–8464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Pan, Q.; Rong, L.; Liu, S.-L.; Liang, C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011, 85, 2126–2137. [Google Scholar] [CrossRef] [Green Version]
- Pfaender, S.; Mar, K.B.; Michailidis, E.; Kratzel, A.; Boys, I.N.; V’kovski, P.; Fan, W.; Kelly, J.N.; Hirt, D.; Ebert, N.; et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol. 2020, 5, 1330–1339. [Google Scholar] [CrossRef] [PubMed]
- Pindel, A.; Sadler, A. The role of protein kinase R in the interferon response. J. Interf. cytokine Res. Off. J. Int. Soc. Interf. Cytokine Res. 2011, 31, 59–70. [Google Scholar] [CrossRef]
- Vladimer, G.I.; Górna, M.W.; Superti-Furga, G. IFITs: Emerging Roles as Key Anti-Viral Proteins. Front. Immunol. 2014, 5, 94. [Google Scholar] [CrossRef] [Green Version]
- Durfee, L.A.; Lyon, N.; Seo, K.; Huibregtse, J.M. The ISG15 conjugation system broadly targets newly synthesized proteins: Implications for the antiviral function of ISG15. Mol. Cell 2010, 38, 722–732. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoggins, J.W.; Wilson, S.J.; Panis, M.; Murphy, M.Y.; Jones, C.T.; Beiniasz, P.; Rice, C.M.; et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 2011, 472, 481–485. [Google Scholar] [CrossRef] [PubMed]
- Helbig, K.J.; Beard, M.R. The role of viperin in the innate antiviral response. J. Mol. Biol. 2014, 426, 1210–1219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malim, M.H.; Bieniasz, P.D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb. Perspect. Med. 2012, 2, a006940. [Google Scholar] [CrossRef]
- Richardson, R.B.; Ohlson, M.B.; Eitson, J.L.; Kumar, A.; McDougal, M.B.; Boys, I.N.; Mar, K.B.; De La Cruz-Rivera, P.C.; Douglas, C.; Konopka, G.; et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 2018, 3, 1214–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batra, J.; Hultquist, J.F.; Liu, D.; Shtanko, O.; Dollen, J.V.; Satkamp, L.; Dollen, J.V.; Satkamp, L.; Jang, G.M.; Luthra, P.; et al. Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication. Cell 2018, 175, 1917–1930. [Google Scholar] [CrossRef] [Green Version]
- Chakrabarti, A.; Jha, B.K.; Silverman, R.H. New insights into the role of RNase L in innate immunity. J. Interf. Cytokine Res. Off. J. Int. Soc. Interf. Cytokine Res. 2011, 31, 49–57. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Ma, J.; Sun, J.; Gao, G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. USA 2007, 104, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Lucifora, J.; Xia, Y.; Reisinger, F.; Zhang, K.; Stadler, D.; Cheng, X.; Sprinzl, M.F.; Koppemsteiner, H.; Makowska, Z.; Volz, T.; et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014, 343, 1221–1228. [Google Scholar] [CrossRef]
- Qiao, Y.; Han, X.; Guan, G.; Wu, N.; Sun, J.; Pak, V.; Liang, G. TGF-beta triggers HBV cccDNA degradation through AID-dependent deamination. FEBS Lett. 2016, 590, 419–427. [Google Scholar] [CrossRef]
- Espert, L.; Degols, G.; Gongora, C.; Blondel, D.; Williams, B.R.; Silverman, R.H.; Mechti, N. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 2003, 278, 16151–16158. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Nguyen, X.-N.; Wang, L.; Approurchaux, R.; Zhang, C.; Panthu, B.; Gruffat, H.; Journo, C.A.; lais, S.; Qin, J.; et al. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLoS Pathog. 2019, 15, e1008093. [Google Scholar] [CrossRef] [PubMed]
- Stadler, D.; Kächele, M.; Jones, A.N.; Hess, J.; Urban, C.; Schneider, J.; Xia, Y.; Oswald, A.; Nebioglu, F.; Bester, R.; et al. Interferon-induced degradation of the persistent hepatitis B virus cccDNA form depends on ISG20. EMBO Rep. 2021, 22, e49568. [Google Scholar] [CrossRef] [PubMed]
- Kawai, T.; Akira, S. TLR Signaling. In Seminars in Immunology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 19, pp. 24–32. [Google Scholar]
- Könner, A.C.; Brüning, J.C. Toll-like receptors: Linking inflammation to metabolism. Trends Endocrinol. Metab. 2011, 22, 16–23. [Google Scholar] [CrossRef]
- Oka, T.; Hikoso, S.; Yamaguchi, O.; Taneike, M.; Takeda, T.; Tamai, T.; Oyabu, J.; Murakawa, T.; Nakayama, H.; Nishida, K.; et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature 2012, 485, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Vogl, T.; Tenbrock, K.; Ludwig, S.; Leukert, N.; Ehrhardt, C.; van Zoelen, M.A.D.; Nacken, W.; Foell, D.; van der Poll, T.; Sorg, C.; et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat. Med. 2007, 13, 1042–1049. [Google Scholar] [CrossRef]
- Kalamvoki, M.; Roizman, B. HSV-1 degrades, stabilizes, requires, or is stung by STING depending on ICP0, the US3 protein kinase, and cell derivation. Proc. Natl. Acad. Sci. USA 2014, 111, E611–E617. [Google Scholar] [CrossRef] [Green Version]
- Park, A.; Ra, E.A.; Lee, T.A.; Choi, H.J.; Lee, E.; Kang, S.; Seo, J.-Y.; Lee, S.; Parl, B. HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways. Nat. Commun. 2019, 10, 4670. [Google Scholar] [CrossRef]
- Bussey, K.A.; Reimer, E.; Todt, H.; Denker, B.; Gallo, A.; Konrad, A.; Ottinger, M.; Adler, H.; Stürzl, M.; Brune, W.; et al. The gammaherpesviruses Kaposi’s sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response. J. Virol. 2014, 88, 9245–9259. [Google Scholar] [CrossRef] [Green Version]
- Vincent, I.E.; Zannnetti, C.; Lucifora, J.; Norder, H.; Protzer, U.L.; Hainaut, P.; Zoulim, F.; Tommasino, M.; Trépo, C.; Hasan, U.; et al. Hepatitis B virus impairs TLR9 expression and function in plasmacytoid dendritic cells. PLoS ONE 2011, 6, e26315. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Rui, Y.; Lou, M.; Yin, L.; Xiong, H.; Zhou, Z.; Shen, S.; Chen, T.; Zhang, Z.; Zhao, N.; et al. HIV-2/SIV Vpx targets a novel functional domain of STING to selectively inhibit cGAS–STING-mediated NF-κB signalling. Nat. Microbiol. 2019, 4, 2552–2564. [Google Scholar] [CrossRef]
- Feng, Q.; Langereis, M.A.; Lork, M.; Nguyen, M.; Hato, S.V.; Lanke, K.; Emdad, L.; Bhoopathi, P.; Fisher, P.B.; Lloyd, R.E.; et al. Enterovirus 2Apro targets MDA5 and MAVS in infected cells. J. Virol. 2014, 88, 3369–3378. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.E.; Wang, M.K.; Rennick, L.J.; Full, F.; Gableske, S.; Mesman, A.W.; Gringhuis, S.I.; Geijtenbeek, T.B.H.; Duprex, W.P.; Gack, M.U. Antagonism of the phosphatase PP1 by the measles virus V protein is required for innate immune escape of MDA5. Cell Host Microbe 2014, 16, 19–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Sun, L.; Liu, W.; Duan, Z. Latent Membrane Protein 1 of Epstein-Barr Virus Promotes RIG-I Degradation Mediated by Proteasome Pathway. Front. Immunol. 2018, 9, 1446. [Google Scholar] [CrossRef]
- Jureka, A.S.; Kleinpeter, A.B.; Cornilescu, G.; Cornilescu, C.C.; Petit, C.M. Structural Basis for a Novel Interaction between the NS1 Protein Derived from the 1918 Influenza Virus and RIG-I. Structure 2015, 23, 2001–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Zhang, X.; Wang, F.; Wang, P.; Kuang, E.; Li, X. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. bioRxiv 2020, 2020.08.12.247767. [Google Scholar] [CrossRef]
- Rajsbaum, R.; Albrecht, R.A.; Wang, M.K.; Maharaj, N.P.; Versteeg, G.A.; Nistal-Villán, E.; García-Sastre, A.; Gack, M.U. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog. 2012, 8, e1003059. [Google Scholar] [CrossRef]
- Hu, Y.; Li, W.; Gao, T.; Cui, Y.; Jin, Y.; Ma, Q.; Liu, X.; Cao, C. SARS coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J. Virol. 2017, 91, e02143-16. [Google Scholar] [CrossRef] [Green Version]
- Ban, J.; Lee, N.-R.; Lee, N.-J.; Lee, J.K.; Quan, F.-S.; Inn, K.-S. Human Respiratory Syncytial Virus NS 1 Targets TRIM25 to Suppress RIG-I Ubiquitination and Subsequent RIG-I-Mediated Antiviral Signaling. Viruses 2018, 10, 716. [Google Scholar] [CrossRef] [Green Version]
- Chiang, C.; Pauli, E.-K.; Biryukov, J.; Feister, K.F.; Meng, M.; White, E.A.; Münger, K.; Howley, P.M.; Meyers, C.; Gack, M.U. The human papillomavirus E6 oncoprotein targets USP15 and TRIM25 to suppress RIG-I-mediated innate immune signaling. J. Virol. 2018, 92, e01737-17. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-L.; Ye, H.-Q.; Liu, S.-Q.; Deng, C.-L.; Li, X.-D.; Shi, P.-Y.; Zhang, B. West Nile Virus NS1 Antagonizes Interferon Beta Production by Targeting RIG-I and MDA5. J. Virol. 2017, 91, e02396-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oshiumi, H.; Miyashita, M.; Matsumoto, M.; Seya, T. A distinct role of Riplet-mediated K63-Linked polyubiquitination of the RIG-I repressor domain in human antiviral innate immune responses. PLoS Pathog. 2013, 9, e1003533. [Google Scholar] [CrossRef] [Green Version]
- Chan, Y.K.; Gack, M.U. A phosphomimetic-based mechanism of dengue virus to antagonize innate immunity. Nat. Immunol. 2016, 17, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Kerur, N.; Veettil, M.V.; Sharma-Walia, N.; Bottero, V.; Sadagopan, S.; Otageri, P.; Chandran, B. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi Sarcoma-associated herpesvirus infection. Cell Host Microbe 2011, 9, 363–375. [Google Scholar] [CrossRef] [Green Version]
- Orzalli, M.H.; Broekema, N.M.; Diner, B.A.; Hancks, D.C.; Elde, N.C.; Cristea, I.M.; Knipe, D.M. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl. Acad. Sci. USA 2015, 112, E1773–E1781. [Google Scholar] [CrossRef] [Green Version]
- Orzalli, M.H.; DeLuca, N.A.; Knipe, D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci. USA 2012, 109, E3008–E3017. [Google Scholar] [CrossRef] [Green Version]
- Cristea, I.M.; Moorman, N.J.; Terhune, S.S.; Cuevas, C.D.; O’Keefe, E.S.; Rout, M.P.; Chait, B.T.; Shenk, T. Human cytomegalovirus pUL83 stimulates activity of the viral immediate-early promoter through its interaction with the cellular IFI16 protein. J. Virol. 2010, 84, 7803–7814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orzalli, M.H.; Broekema, N.M.; Knipe, D.M. Relative Contributions of Herpes Simplex Virus 1 ICP0 and vhs to Loss of Cellular IFI16 Vary in Different Human Cell Types. J. Virol. 2016, 90, 8351–8359. [Google Scholar] [CrossRef] [Green Version]
- Su, C.; Zheng, C. Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J. Virol. 2017, 91, e02414-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.J.; Li, W.; Shao, Y.; Avey, D.; Fu, B.; Gillen, J.; Hand, T.; Ma, S.; Liu, X.; Miley, W.; et al. Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 2015, 18, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Liu, Q.; Wu, Y.; Ma, L.; Zhang, Z.; Liu, T.; Jin, S.; She, Y.; Li, Y.-P.; Cui, J. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 2018, 37, e99347. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, J.; Xu, J.; He, S.; Zeng, Y.; Xie, L.; Xie, N.; Liu, T.; Lee, K.; et al. Species-Specific Deamidation of cGAS by Herpes Simplex Virus UL37 Protein Facilitates Viral Replication. Cell Host Microbe 2018, 24, 234–248. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; You, H.; Su, C.; Li, Y.; Chen, S.; Zheng, C. Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J. Virol. 2018, 92, e00841-18. [Google Scholar] [CrossRef] [Green Version]
- Biolatti, M.; Dell’Oste, V.; Pautasso, S.; Gugliesi, F.; von Eineem, J.; Krapp, C.; Jakobsen, M.R.; Borgogna, C.; Gariglio, M.; De Andrea, M.; et al. Human Cytomegalovirus Tegument Protein pp65 (pUL83) Dampens Type I Interferon Production by Inactivating the DNA Sensor cGAS without Affecting STING. J. Virol. 2018, 92, e01774-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Z.-F.; Zou, H.-M.; Liao, B.-W.; Zhang, H.-Y.; Yang, Y.; Fu, Y.-Z.; Wang, S.-Y.; Luo, M.-H.; Wang, Y.-Y. Human Cytomegalovirus Protein UL31 Inhibits DNA Sensing of cGAS to Mediate Immune Evasion. Cell Host Microbe 2018, 24, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Chan, B.; Samarina, N.; Abere, B.; Weidner-Glunde, M.; Buch, A.; Pich, A.; Brinkmann, M.M.; Schulz, T.F. Cytoplasmic isoforms of Kaposi sarcoma herpesvirus LANA recruit and antagonize the innate immune DNA sensor cGAS. Proc. Natl. Acad. Sci. USA 2016, 113, E1034–E1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meylan, E.; Joseph, C.; Hofmann, K.; Moradpour, D.; Binder, M.; Bartenschlager, R.; Tschopp, J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 2005, 437, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Foy, E.; Ferreon, J.C.; Nakamura, M.; Ferreon, A.C.M.; Ikeda, M.; Ray, S.C.; Gale Jr, M.; Lemon, S.M. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. USA 2005, 102, 2992–2997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, C.-Y.; Liang, J.-J.; Li, J.-K.; Lee, Y.-L.; Chang, B.-L.; Su, C.-I.; Huang, W.-J.; Lai, M.M.C.; Lin, Y.-L. Dengue Virus Impairs Mitochondrial Fusion by Cleaving Mitofusins. PLoS Pathog. 2015, 11, e1005350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drahos, J.; Racaniello, V.R. Cleavage of IPS-1 in cells infected with human rhinovirus. J. Virol. 2009, 83, 11581–11587. [Google Scholar] [CrossRef] [Green Version]
- Lau, L.; Gray, E.E.; Brunette, R.L.; Stetson, D.B. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 2015, 350, 568–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Jacobs, S.R.; West, J.A.; Stopford, C.; Zhang, Z.; Davis, Z.; Barber, G.N.; Glaunsinger, B.A.; Dittmer, D.P.; Damania, B. Modulation of the cGAS-STING DNA sensing pathway by gammaherpesviruses. Proc. Natl. Acad. Sci. USA 2015, 112, E4306–E4315. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Li, J.; Chen, J.; Li, Y.; Wang, W.; Du, X.; Song, W.; Zhang, W.; Lin, L.; Yuan, Z. Hepatitis B Virus Polymerase Disrupts K63-Linked Ubiquitination of STING To Block Innate Cytosolic DNA-Sensing Pathways. J. Virol. 2015, 89, 2287–2300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, S.; Maestre, A.M.; Pagni, S.; Patel, J.R.; Savage, T.; Gutman, D.; Maringer, K.; Bernal-Rubio, D.; Shabman, R.S.; Simon, V.; et al. DENV inhibits type I IFN production in infected cells by cleaving human STING. PLoS Pathog. 2012, 8, e1002934. [Google Scholar] [CrossRef] [Green Version]
- Pythoud, C.; Shanaka I Rodrigo, W.W.; Pasqual, G.; Rothenberger, S.; Martínez-Sobrido, L.; Calros de la Torre, J.; Kunz, S. Arenavirus nucleoprotein targets interferon regulatory factor-activating kinase IKKε. J. Virol. 2012, 86, 7728–7738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harman, A.N.; Nasr, N.; Feetham, A.; Galoyan, A.; Alshehri, A.A.; Rambukwelle, D.; Botting, R.A.; Heiner, B.M.; Diefenbach, E.; Diefenbach, R.J.; et al. HIV Blocks Interferon Induction in Human Dendritic Cells and Macrophages by Dysregulation of TBK1. J. Virol. 2015, 89, 6575–6584. [Google Scholar] [CrossRef] [Green Version]
- Prins, K.C.; Cárdenas, W.B.; Basler, C.F. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J. Virol. 2009, 83, 3069–3077. [Google Scholar] [CrossRef] [Green Version]
- Verpooten, D.; Ma, Y.; Hou, S.; Yan, Z.; He, B. Control of TANK-binding kinase 1-mediated signaling by the gamma(1)34.5 protein of herpes simplex virus 1. J. Biol. Chem. 2009, 284, 1097–1105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, R.; Genin, P.; Mamane, Y.; Sgarbanti, M.; Battistini, A.; Harrington, W.J., Jr.; Barber, G.N.; Hiscott, J. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 2001, 20, 800–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Q.; Fu, B.; Wu, F.; Li, X.; Yuan, Y.; Zhu, F. ORF45 of Kaposi’s sarcoma-associated herpesvirus inhibits phosphorylation of interferon regulatory factor 7 by IKKε and TBK1 as an alternative substrate. J. Virol. 2012, 86, 10162–10172. [Google Scholar] [CrossRef] [Green Version]
- Xiang, Z.; Liu, L.; Lei, X.; Zhou, Z.; He, B.; Wang, J. 3C Protease of Enterovirus D68 Inhibits Cellular Defense Mediated by Interferon Regulatory Factor 7. J. Virol. 2015, 90, 1613–1621. [Google Scholar] [CrossRef] [Green Version]
- Watters, K.; Palmenberg, A.C. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J. Virol. 2011, 85, 10874–10883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, J.; Laurent-Rolle, M.; Maestre, A.M.; Rajsbaum, R.; Pisanelli, G.; Simon, V.; Mulder, L.C.F.; Fernandez-Sesma, A.; García-Sastre, A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013, 9, e1003265. [Google Scholar] [CrossRef] [Green Version]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.-Y.; Garcia-Sastre, A. NS5 of Dengue Virus Mediates STAT2 Binding and Degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, H.; Leo, H.; Shan, C.; Muruato, A.E.; Nunes, B.T.D.; Medeiros, D.B.A.; Zou, J.; Xie, X.; Giraldo, M.I.; Vasconcelos, P.F.C.; et al. An evolutionary NS1 mutation enhances Zika virus evasion of host interferon induction. Nat. Commun. 2018, 9, 414. [Google Scholar] [CrossRef]
- Fros, J.J.; Pijlman, G.P. Alphavirus Infection: Host Cell Shut-Off and Inhibition of Antiviral Responses. Viruses 2016, 8, 166. [Google Scholar] [CrossRef]
- White, L.K.; Sali, T.; Alvarado, D.; Gatti, E.; Pierre, P.; Streblow, D.; Defilippis, V.R. Chikungunya virus induces IPS-1-dependent innate immune activation and protein kinase R-independent translational shutoff. J. Virol. 2011, 85, 606–620. [Google Scholar] [CrossRef] [Green Version]
- Lamphear, B.J.; Yan, R.; Yang, F.; Waters, D.; Liebig, H.D.; Klump, H.; Kuechler, E.; Skern, T.; Rhoads, R.E. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human Coxsackievirus and rhinovirus. J. Biol. Chem. 1993, 268, 19200–19203. [Google Scholar] [CrossRef]
- Walker, E.J.; Younessi, P.; Fulcher, A.J.; McCuaig, R.; Thomas, B.J.; Bardin, P.G.; Jans, D.A.; Ghildyal, R. Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS ONE 2013, 8, e71316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conticello, S.G.; Harris, R.S.; Neuberger, M.S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003, 13, 2009–2013. [Google Scholar] [CrossRef] [Green Version]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Kostyushev, D.; Kostyusheva, A.; Brezgin, S.; Zarifyan, D.; Utkina, A.; Goptar, I.; Chulanov, V. Suppressing the NHEJ pathway by DNA-PKcs inhibitor NU7026 prevents degradation of HBV cccDNA cleaved by CRISPR/Cas9. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.H.Y.; Pannunzio, N.R.; Adachi, N.; Lieber, M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017, 18, 495–506. [Google Scholar] [CrossRef]
- Brandsma, I.; van Gent, D.C. Pathway choice in DNA double strand break repair: Observations of a balancing act. Genome Integr. 2012, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Overbeek, M.; Capurso, D.; Carter, M.M.; Thhompson, M.S.; Frias, E.; Russ, C.; Reece-Hoyes, J.S.; Nye, C.; Gradia, S.; Vidal, B.; et al. DNA repair profiling reveals nonrandom outcomes at Cas9-mediated breaks. Mol. Cell 2016, 63, 633–646. [Google Scholar] [CrossRef] [Green Version]
- Ihry, R.J.; Worringer, K.A.; Salick, M.R.; Frias, E.; Ho, D.; Theriault, K.; Kommineni, S.; Chen, J.; Sondey, M.; Ye, C.; et al. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018, 24, 939. [Google Scholar] [CrossRef]
- Zuccaro, M.V.; Xu, J.; Mitchell, C.; Marin, D.; Zimmerman, R.; Rana, B.; Weinstein, E.; King, R.T.; Palmerola, K.L.; Smith, M.E.; et al. Allele-specific chromosome removal after Cas9 cleavage in human embryos. Cell 2020, 183, 1650–1664. [Google Scholar] [CrossRef] [PubMed]
- Kosicki, M.; Tomberg, K.; Bradley, A. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018, 36, 765–771. [Google Scholar] [CrossRef]
- Leibowitz, M.L.; Papathanasiou, S.; Doerfler, P.A.; Blaine, L.; Sun, L.; Yao, Y.; Zhang, C.-Z.; Weiss, M.J.; Pellman, D. Chromothripsis as an on-target consequence of CRISPR–Cas9 genome editing. Nat. Genet. 2021, 53, 895–905. [Google Scholar] [CrossRef]
- Liu, M.; Rehman, S.; Tang, X.; Gu, K.; Fan, Q.; Chen, D.; Ma, W. Methodologies for improving HDR efficiency. Front. Genet. 2019, 9, 691. [Google Scholar] [CrossRef]
- Kim, S.; Koo, T.; Cho, H.-Y.; Lee, G.; Lim, D.-G.; Shin, H.S.; Kim, J.-S. CRISPR RNAs trigger innate immune responses in human cells. Genome Res. 2018, 28, 367–373. [Google Scholar] [CrossRef] [Green Version]
- Bae, S.; Kweon, J.; Kim, H.S.; Kim, J.-S. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 2014, 11, 705–706. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Cheng, A.W.; Wang, H.; Yang, H.; Shi, L.; Katz, Y.; Theunissen, T.W.; Rangarajan, S.; Shivalia, C.S.; Dadon, D.B.; Jaenisch, R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013, 23, 1163–1171. [Google Scholar] [CrossRef]
- Baeumler, T.A.; Ahmed, A.A.; Fulga, T.A. Engineering Synthetic Signaling Pathways with Programmable dCas9-Based Chimeric Receptors. Cell Rep. 2017, 20, 2639–2653. [Google Scholar] [CrossRef] [Green Version]
- Hilton, I.B.; D’ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Pinera, P.; Kocak, D.D.; Vockley, C.M.; Adler, A.F.; Kabadi, A.M.; Polstein, L.R.; Thakore, P.I.; Glass, K.A.; Ousterout, D.G.; Leong, K.W.; et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat. Methods 2013, 10, 973. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Rocha, P.P.; Luo, V.M.; Raviram, R.; Deng, Y.; Mazzoni, E.O.; Skok, J.A. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat. Commun. 2016, 7, 1–8. [Google Scholar] [CrossRef]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Lyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar] [CrossRef] [Green Version]
- Pflueger, C.; Tan, D.; Swain, T.; Nguyen, T.; Pflueger, J.; Nefzger, C.; Polo, J.M.; Ford, E.; Lister, R. A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res. 2018, 28, 1193–1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sajwan, S.; Mannervik, M. Gene activation by dCas9-CBP and the SAM system differ in target preference. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Liu, J.; Zhou, C.; Gao, N.; Rao, Z.; Li, H.; Hu, X.; Li, C.; Yao, X.; Shen, X.; et al. In vivo simultaneous transcriptional activation of multiple genes in the brain using CRISPR-dCas9-activator transgenic mice. Nat. Neurosci. 2018, 21, 440. [Google Scholar] [CrossRef]
- Thakore, P.I.; D’Ippolito, A.M.; Song, L.; Safi, A.; Shivakumar, N.K.; Kabadi, A.M.; Reddy, T.E.; Crawford, G.E.; Gersbach, C.A. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- O’Geen, H.; Bates, S.L.; Carter, S.S.; Nisson, K.A.; Halmai, J.; Fink, K.D.; Rhie, S.K.; Farnham, P.J.; Segal, D.J. Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics Chromatin 2019, 12, 26. [Google Scholar] [CrossRef]
- Rees, H.A.; Liu, D.R. Base editing: Precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 2018, 19, 770–788. [Google Scholar] [CrossRef]
- Abbott, T.R.; Dhamdhere, G.; Liu, Y.; Lin, X.; Goudy, L.; Zeng, L.; Chemparathy, A.; Chmura, S.; Heaton, N.S.; Debs, R.; et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell 2020, 181, 865–876. [Google Scholar] [CrossRef]
- Freije, C.A.; Myhrvold, C.; Boehm, C.K.; Lin, A.E.; Welch, N.L.; Carter, A.; Metsky, H.C.; Luo, C.Y.; Abudayyeh, O.O.; Gootenberg, J.S.; et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell 2019, 76, 826–837. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.T.; Zhang, F. A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Lv, J.; Li, Y.; Mao, S.; Li, Z.; Jing, Z.; Sun, Y.; Zhang, X.; Shen, S.; Wang, X.; et al. Programmable C-to-U RNA editing using the human APOBEC 3A deaminase. EMBO J. 2020, 39, e104741. [Google Scholar] [CrossRef]
- Xu, C.; Zhou, Y.; Xiao, Q.; He, B.; Geng, G.; Wang, Z.; Cao, B.; Dong, X.; Bai, W.; Wang, Y.; et al. Programmable RNA editing with compact CRISPR–Cas13 systems from uncultivated microbes. Nat. Methods 2021, 18, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Yeo, N.C.; Chavez, A.; Lance-Byrne, A.; Chan, Y.; Menn, D.; Milanova, D.; Kuo, C.-C.; Guo, X.; Sharma, S.; Tung, A.; et al. An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat. Methods 2018, 15, 611. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420. [Google Scholar] [CrossRef] [Green Version]
- Komor, A.C.; Zhao, K.T.; Packer, M.S.; Gaudelli, N.M.; Waterbury, A.L.; Koblan, L.W.; Kim, Y.B.; Badran, A.H.; Liu, D.R. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C: G-to-T: A base editors with higher efficiency and product purity. Sci. Adv. 2017, 3, eaao4774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Li, J.; Wang, Y.; Yang, B.; Weo, J.; Wu, J.; Wang, R.; Huang, X.; Chen, J.; Yang, L. Efficient base editing in methylated regions with a human APOBEC3A-Cas9 fusion. Nat. Biotechnol. 2018, 36, 946. [Google Scholar] [CrossRef]
- Jiang, W.; Feng, S.; Huang, S.; Yu, W.; Li, G.; Yang, G.; Liu, Y.; Zhang, Y.; Zhang, L.; Hou, Y.; et al. BE-PLUS: A new base editing tool with broadened editing window and enhanced fidelity. CELL Res. 2018, 28, 855–861. [Google Scholar] [CrossRef] [Green Version]
- Hess, G.T.; Frésard, L.; Han, K.; Lee, C.H.; Li, A.; Cimprich, K.A.; Montgomery, S.B.; Bassik, M.C. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat. Methods 2016, 13, 1036. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464. [Google Scholar] [CrossRef]
- Dominguez, A.A.; Lim, W.A.; Qi, L.S. Beyond editing: Repurposing CRISPR–Cas9 for precision genome regulation and interrogation. Nat. Rev. Mol. Cell Biol. 2016, 17, 5. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Wang, G.Z.; Cingöz, O.; Goff, S.P. NP220 mediates silencing of unintegrated retroviral DNA. Nature 2018, 564, 278–282. [Google Scholar] [CrossRef]
- Chia, B.S.; Li, B.; Cui, A.; Eisenhaure, T.; Raychowdhury, R.; Lieb, D.; Hacohen, N. Loss of the nuclear protein RTF2 enhances influenza virus replication. J. Virol. 2020, 94, e00319-20. [Google Scholar] [CrossRef] [PubMed]
- OhAinle, M.; Helms, L.; Vermeire, J.; Roesch, F.; Humes, D.; Bason, R.; Delrow, J.J.; Overbaugh, J.; Emerman, M. A virus-packageable CRISPR screen identifies host factors mediating interferon inhibition of HIV. Elife 2018, 7, e39823. [Google Scholar] [CrossRef] [PubMed]
- Roesch, F.; OhAinle, M.; Emerman, M. A CRISPR screen for factors regulating SAMHD1 degradation identifies IFITMs as potent inhibitors of lentiviral particle delivery. Retrovirology 2018, 15, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, J.; Perez, J.T.; Chen, C.; Li, Y.; Benitez, A.; Kandasamy, M.; Lee, Y.; Andrade, J.; tenOever, B.; Manicassamy, B. Genome-wide CRISPR/Cas9 Screen Identifies Host Factors Essential for Influenza Virus Replication. Cell Rep. 2018, 23, 596–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orchard, R.C.; Sullender, M.E.; Dunlap, B.F.; Balce, D.R.; Doench, J.G.; Virgin, H.W. Identification of antinorovirus genes in human cells using Genome-Wide CRISPR activation screening. J. Virol. 2019, 93, e01324-18. [Google Scholar] [CrossRef] [Green Version]
- Dukhovny, A.; Lamkiewicz, K.; Chen, Q.; Fricke, M.; Jabrane-Ferrat, N.; Marz, M.; Jung, J.U.; Sklan, E.H. A CRISPR Activation Screen Identifies Genes That Protect against Zika Virus Infection. J. Virol. 2019, 93, e00211-19. [Google Scholar] [CrossRef] [Green Version]
- LaFleur, M.W.; Nguyen, T.H.; Coxe, M.A.; Yates, K.B.; Trombley, J.D.; Weiss, S.A.; Brown, F.D.; Gillis, J.E.; Coxe, D.J.; Doench, J.G.; et al. A CRISPR-Cas9 delivery system for in vivo screening of genes in the immune system. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I inteferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [Green Version]
- Lugrin, J.; Martinon, F. The AIM 2 inflammasome: Sensor of pathogens and cellular perturbations. Immunol. Rev. 2018, 281, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Paijo, J.; Döring, M.; Spanier, J.; Grabski, E.; Nooruzzaman, M.; Schmidt, T.; Witte, G.; Messerle, M.; Hornung, V.; Kaever; et al. cGAS senses human cytomegalovirus and induces type I interferon responses in human monocyte-derived cells. PLoS Pathog. 2016, 12, e1005546. [Google Scholar] [CrossRef] [PubMed]
- Lio, C.-W. J.; McDonald, B.; Takahashi, M.; Dhanwani, R.; Sharma, N.; Huang, J.; Pham, E.; Benedict, C.A.; Sharma, S. cGAS-STING signaling regulates initial innate control of cytomegalovirus infection. J. Virol. 2016, 90, 7789–7797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sui, H.; Zhou, M.; Imamichi, H.; Jiao, X.; Sherman, B.T.; Lane, H.C.; Imamichi, T. STING is an essential mediator of the Ku70-mediated production of IFN-lambda 1 in response to exogenous DNA. Sci. Signal. 2017, 10, eaah5054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gray, E.E.; Winship, D.; Snyder, J.M.; Chuld, S.J.; Geballe, A.P.; Stetson, D.B. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity 2016, 45, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diner, B.A.; Lum, K.K.; Toettcher, J.E.; Cristea, I.M. Viral DNA sensors IFI16 and cyclic GMP-AMP synthase possess distinct functions in regulating viral gene expression, immune defenses, and apoptotic responses during herpesvirus infection. MBio 2016, 7, e01553-16. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Banerjee, S.; Wang, Y.; Goldstein, S.A.; Dong, B.; Gaughan, C.; Silverman, R.H.; Weiss, S.R. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci. USA 2016, 113, 2241–2246. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhang, X.; Cao, W.; Yang, F.; Du, X.; Shi, Z.; Zhang, M.; Liu, X.; Zhu, Z.; Zheng, H. RIG-I is responsible for activation of type I interferon pathway in Seneca Valley virus-infected porcine cells to suppress viral replication. Virol. J. 2018, 15, 162. [Google Scholar] [CrossRef] [Green Version]
- Schilling, M.; Bridgeman, A.; Gray, N.; Hertzog, J.; Hubiltz, P.; Kohl, A.; Rehwinkel, J. RIG-I plays a dominant role in the induction of transcriptional changes in Zika virus-infected cells, which protect from virus-induced cell death. Cells 2020, 9, 1476. [Google Scholar] [CrossRef]
- Zhang, Q.; Zeng, L.-P.; Zhou, P.; Irving, A.T.; Li, S.; Shi, Z-L.; Wang, L-F. IFNAR2-dependent gene expression profile induced by IFN-α in Pteropus alecto bat cells and impact of IFNAR2 knockout on virus infection. PLoS ONE 2017, 12, e0182866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urin, V.; Shemesh, M.; Schreiber, G. CRISPR/Cas9-based Knockout Strategy Elucidates Components Essential for Type 1 Interferon Signaling in Human HeLa Cells. J. Mol. Biol. 2019, 431, 3324–3338. [Google Scholar] [CrossRef]
- Yamauchi, S.; Takeuchi, K.; Chihara, K.; Honjoh, C.; Kato, Y.; Yoshiki, H.; Hotta, H.; Sada, K. STAT1 is essential for the inhibition of hepatitis C virus replication by interferon-λ but not by interferon-α. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.S.; Shin, M.J.; Kim, K.H. Increase of viral hemorrhagic septicemia virus growth by knockout of IRF9 gene in Epithelioma papulosum cyprini cells. Fish Shellfish Immunol. 2018, 83, 443–448. [Google Scholar] [CrossRef] [PubMed]
- Ashley, C.L.; Abendroth, A.; McSharry, B.P.; Slobedman, B. Interferon-independent upregulation of interferon-stimulated genes during human cytomegalovirus infection is dependent on IRF3 expression. Viruses 2019, 11, 246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, W.M.; Chevillotte, M.D.; Rice, C.M. Interferon-stimulated genes: A complex web of host defenses. Annu. Rev. Immunol. 2014, 32, 513–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Wu, Y.; Qin, Y.; Hu, J.; Xie, W.; Qin, F.X.-F.; Cui, J. Broad and diverse mechanisms used by deubiquitinase family members in regulating the type I interferon signaling pathway during antiviral responses. Sci. Adv. 2018, 4, eaar2824. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Wu, R.; Guo, W.; Xie, L.; Qiao, Z.; Chen, S.; Zhu, J.; Huang, C.; Huang, J.; Chen, B.; et al. STING-Mediated IFI16 Degradation Negatively Controls Type I Interferon Production. Cell Rep. 2019, 29, 1249–1260. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.-S.; Zhang, Z.-Y.; Cai, H.; Zhao, M.; Mao, J.; Dai, J.; Xia, T.; Zhang, X-M.; Li, T. RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci. 2018, 8, 35. [Google Scholar] [CrossRef] [Green Version]
- Ju, L.-G.; Zhu, Y.; Lei, P.-J.; Yan, D.; Zhu, K.; Wang, X.; Li, Q.-L.; Li, X.-J.; Chen, J.-W.; Li, L.-Y.; et al. TTLL12 inhibits the activation of cellular antiviral signaling through interaction with VISA/MAVS. J. Immunol. 2017, 198, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Carvajal, L.; Singh, N.; de los Santos, T.; Rodríguez, L.L.; Long, C.R. Depletion of elongation initiation factor 4E binding proteins by CRISPR/Cas9 enhances the antiviral response in porcine cells. Antiviral Res. 2016, 125, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Atianand, M.K.; Fitzgerald, K.A. Molecular basis of DNA recognition in the immune system. J. Immunol. 2013, 190, 1911–1918. [Google Scholar] [CrossRef]
- Ma, H.; Qian, W.; Bamouskova, M.; Collins, P.L.; Porter, S.I.; Byrum, A.K.; Zhang, R.; Artyomov, M.; Oltz, E.M.; Mosammaparast, N.; et al. Barrier-to-Autointegration Factor 1 Protects against a Basal cGAS-STING Response. MBio 2020, 11, e00136-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, S.; Diep, J.; Feng, N.; Ren, L.; Li, B.; Ooi, Y.S.; Wang, X.; Brulois, K.F.; Yasukawa, L.L.; Li, X.; et al. STAG2 deficiency induces interferon responses via cGAS-STING pathway and restricts virus infection. Nat. Commun. 2018, 9, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Morrison, J.H.; Dingli, D.; Poeschla, E. HIV-1 activation of innate immunity depends strongly on the intracellular level of TREX1 and sensing of incomplete reverse transcription products. J. Virol. 2018, 92, e00001-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whelan, J.N.; Li, Y.; Silverman, R.H.; Weiss, S.R. Zika virus production is resistant to RNase L antiviral activity. J. Virol. 2019, 93, e00313-19. [Google Scholar] [CrossRef] [Green Version]
- Schafer, A.R.M.; Smith, J.L.; Pryke, K.M.; DeFilippis, V.R.; Hirsch, A.J. The E3 Ubiquitin Ligase SIAH1 Targets MyD88 for Proteasomal Degradation During Dengue Virus Infection. Front. Microbiol. 2020, 11, 24. [Google Scholar] [CrossRef] [Green Version]
- Taylor, J.P.; Cash, M.N.; Santostefano, K.E.; Nakanishi, M.; Terada, N.; Wallet, M.A. CRISPR/Cas9 knockout of USP18 enhances type I IFN responsiveness and restricts HIV-1 infection in macrophages. J. Leukoc. Biol. 2018, 103, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Kuffour, E.O.; Schott, K.; Vasudevan, A.A.J.; Holler, J.; Schulz, W.A.; Lang, P.A.; Lang, K.S.; Kim, B.; Häussinger, D.; König, R.; et al. USP18 (UBP43) abrogates p21-mediated inhibition of HIV-1. J. Virol. 2018, 92, e00592-18. [Google Scholar] [CrossRef] [Green Version]
- Goodwin, C.M.; Schafer, X.; Munger, J. UL26 Attenuates IKKβ-Mediated Induction of Interferon-Stimulated Gene (ISG) Expression and Enhanced Protein ISGylation during Human Cytomegalovirus Infection. J. Virol. 2019, 93, e01052-19. [Google Scholar] [CrossRef]
- Teng, Y.; Luo, M.; Yu, T.; Chen, L.; Huang, Q.; Chen, S.; Xie, L.; Zeng, Y.; Luo, F.; Xiong, H.; et al. CRISPR/Cas9-mediated deletion of miR-146a enhances antiviral response in HIV-1 infected cells. Genes Immun. 2019, 20, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Dufrasne, F.E.; Lombard, C.; Goubau, P.; Ruelle, J. Single amino acid substitution N659D in HIV-2 envelope glycoprotein (Env) impairs viral release and hampers BST-2 antagonism. Viruses 2016, 8, 285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hahn, A.S.; Großkopf, A.K.; Jungnickl, D.; Scholz, B.; Ensser, A. Viral FGARAT homolog ORF75 of rhesus monkey rhadinovirus effects proteasomal degradation of the ND10 components SP100 and PML. J. Virol. 2016, 90, 8013–8028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botto, S.; Abraham, J.; Mizuno, N.; Pryke, K.; Gall, B.; Landais, I.; Streblow, D.N.; Fruh, K.J.; DeFilippis, V.R. Human cytomegalovirus immediate early 86-kDa protein blocks transcription and induces degradation of the immature interleukin-1β protein during virion-mediated activation of the AIM2 inflammasome. MBio 2019, 10, e02510-18. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Pan, Q.; Liang, C. Role of MxB in alpha interferon-mediated inhibition of HIV-1 Infection. J. Virol. 2018, 92, e00422-18. [Google Scholar] [CrossRef] [Green Version]
- Bonifati, S.; Daly, M.B.; Gelais, C.St.; Kim, S.H.; Hollenbaugh, J.A.; Shepard, C.; Kennedy, E.M.; Kim, D.-H.; Schinazi, R.F.; Kim, B.; et al. SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 2016, 495, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Feng, C.; Fang, Y.; Zhou, X.; Xu, L.; Wang, W.; Kong, X.; Peppelenbosch, M.P.; Pan, Q.; Yin, Y. The eukaryotic translation initiation factor 4F complex restricts rotavirus infection via regulating the expression of IRF1 and IRF7. Int. J. Mol. Sci. 2019, 20, 1580. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wei, Z.; Zhang, Y.; Ma, X.; Chen, Y.; Yu, M.; Ma, C.; Li, X.; Cao, Y.; Liu, J.; et al. Activation of liver X receptor plays a central role in antiviral actions of 25-hydroxycholesterol. J. Lipid Res. 2018, 59, 2287–2296. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.S.; Kim, K.H. Effect of CRISPR/Cas9-mediated knockout of either Mx1 or ISG15 gene in EPC cells on resistance against VHSV infection. Fish Shellfish Immunol. 2019, 93, 1041–1046. [Google Scholar] [CrossRef]
- Brezgin, S.; Kostyusheva, A.; Bayurova, E.; Gordeychuk, I.; Isaguliants, M.; Goptar, I.; Nikiforova, A.; Smirnov, V.; Volchkova, E.; Glebe, D.; et al. Replenishment of Hepatitis B Virus cccDNA Pool Is Restricted by Baseline Expression of Host Restriction Factors In Vitro. Microorganisms 2019, 7, 533. [Google Scholar] [CrossRef] [Green Version]
- Zang, R.; Case, J.B.; Yutuc, E.; Ma, X.; Shen, S.; Castro, M.F.G.; Liu, Z.; Zeng, Q.; Zhao, H.; Son, J.; et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. Proc. Natl. Acad. Sci. USA 2020, 117, 32105–32113. [Google Scholar] [CrossRef] [PubMed]
- Passos, V.; Zillinger, T.; Casartelli, N.; Wachs, A.S.; Xu, S.; Malassa, A.; Steppich, K.; Schilling, H.; Franz, S.; Todt, D.; et al. Characterization of endogenous SERINC5 protein as anti-HIV-1 factor. J. Virol. 2019, 93, e01221-19. [Google Scholar] [CrossRef] [Green Version]
- Wąchalska, M.; Graul, M.; Praest, P.; Luteijn, R.D.; Babnis, A.W.; Wiertz, E.J.H.J.; Bieńkowska-Szewczyk, K.; Lipińska, A.D. Fluorescent TAP as a Platform for Virus-Induced Degradation of the Antigenic Peptide Transporter. Cells 2019, 8, 1590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Haan, J.M.M.; Lehar, S.M.; Bevan, M.J. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 2000, 192, 1685–1696. [Google Scholar] [CrossRef] [PubMed]
- Sali, T.M.; Pryke, K.M.; Abraham, J.; Liu, A.; Archer, I.; Broeckel, R.; Staverosky, J.A.; Smith, J.L.; Al-Shammari, A.; Amsler, L.; et al. Characterization of a novel human-specific STING agonist that elicits antiviral activity against emerging alphaviruses. PLoS Pathog. 2015, 11, e1005324. [Google Scholar] [CrossRef]
- Vanwalscappel, B.; Tada, T.; Landau, N.R. Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 2018, 522, 199–208. [Google Scholar] [CrossRef]
- Wang, B.; Lam, T.H.; Soh, M.K.; Ye, Z.; Chen, J.; Ren, E.C. Influenza A virus facilitates its infectivity by activating p53 to inhibit the expression of interferon-induced transmembrane proteins. Front. Immunol. 2018, 9, 1193. [Google Scholar] [CrossRef] [PubMed]
- Puray-Chavez, M.N.; Farghali, M.H.; Yapo, V.; Huber, A.D.; Liu, D.; Ndongwe, T.P.; Casey, M.C.; Laughlin, T.G.; Hannik, M.; Tedbury, P.R.; et al. Effects of Moloney Leukemia Virus 10 Protein on Hepatitis B Virus Infection and Viral Replication. Viruses 2019, 11, 651. [Google Scholar] [CrossRef] [Green Version]
- James, C.D.; Prabhakar, A.T.; Otoa, R.; Evans, M.R.; Wang, X.; Bristol, M.L.; Zhang, K.; Li, R.; Morgan, I.M. SAMHD1 Regulates Human Papillomavirus 16-Induced Cell Proliferation and Viral Replication during Differentiation of Keratinocytes. mSphere 2019, 4, e00448-19. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; McLaughlin, R.N., Jr.; Basom, R.S.; Kikawa, C.; OhAinle, M.; Yount, J.S.; Emerman, M.; Overbaugh, J. Macaque interferon-induced transmembrane proteins limit replication of SHIV strains in an Envelope-dependent manner. PLoS Pathog. 2019, 15, e1007925. [Google Scholar] [CrossRef]
- Spence, J.S.; He, R.; Hoffmann, H.-H.; Das, T.; Thinon, E.; Rice, C.M.; Peng, T.; Chandran, K.; Hang, H.C. IFITM3 directly engages and shuttles incoming virus particles to lysosomes. Nat. Chem. Biol. 2019, 15, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Sivan, G.; Ormanoglu, P.; Buehler, E.C.; Martin, S.E.; Moss, B. Identification of restriction factors by human genome-wide RNA interference screening of viral host range mutants exemplified by discovery of SAMD9 and WDR6 as inhibitors of the vaccinia virus K1L− C7L−mutant. MBio 2015, 6, e01122-15. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Zhang, F.; Yan, B.; Si, C.; Honda, H.; Nagamachi, A.; Sun, L.Z.; Xian, Y. A paralogous pair of mammalian host restriction factors form a critical host barrier against poxvirus infection. PLoS Pathog. 2018, 14, e1006884. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.E.; Bottero, V.; Flaherty, S.; Dutta, S.; Singh, V.V.; Chandran, B. IFI16 restricts HSV-1 replication by accumulating on the hsv-1 genome, repressing HSV-1 gene expression, and directly or indirectly modulating histone modifications. PLoS Pathog. 2014, 10, e1004503. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoek, K.H.; Eyre, N.S.; Shue, B.; Khantisitthiporn, O.; Glab-Ampi, K.; Carr, J.M.; Gartner, M.J.; Jolly, L.A.; Thomas, P.Q.; Adikusuma, F.; et al. Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 2017, 7, 1–14. [Google Scholar]
- Cacciarelli, T.V.; Martinez, O.M.; Gish, R.G.; Villanueva, J.C.; Krams, S.M. Immunoregulatory cytokines in chronic hepatitis C virus infection: Pre-and posttreatment with interferon alfa. Hepatology 1996, 24, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Sheen, I.; Chien, R.; Chu, C.; Liaw, Y. Long-term beneficial effect of interferon therapy in patients with chronic hepatitis B virus infection. Hepatology 1999, 29, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Kühl, U.; Pauschinger, M.; Schwimmbeck, P.L.; Seeberg, B.; Lober, C.; Noutsias, M.; Poller, W.; Schultheiss, H.P. Interferon-β treatment eliminates cardiotropic viruses and improves left ventricular function in patients with myocardial persistence of viral genomes and left ventricular dysfunction. Circulation 2003, 107, 2793–2798. [Google Scholar] [CrossRef] [Green Version]
- Bogerd, H.P.; Kornepati, A.V.R.; Marshall, J.B.; Kennedy, E.M.; Cullen, B.R. Specific induction of endogenous viral restriction factors using CRISPR/Cas-derived transcriptional activators. Proc. Natl. Acad. Sci. USA 2015, 112, E7249–E7256. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ozono, S.; Yao, W.; Tobiume, M.; Yamaoka, S.; Kishigami, S.; Fujita, H.; Tokunaga, K. CRISPR-mediated activation of endogenous BST-2/tetherin expression inhibits wild-type HIV-1 production. Sci. Rep. 2019, 9, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Liu, G.; Kitamura, K.; Wang, Z.; Chowdhury, S.; Monjurul, A.M.; Wakae, K.; Koura, M.; Shimadu, M.; Kinoshita, K.; et al. TGF-β suppression of HBV RNA through AID-dependent recruitment of an RNA exosome complex. PLoS Pathog 2015, 11, e1004780. [Google Scholar] [CrossRef]
- Brezgin, S.; Kostyusheva, A.; Ponomareva, N.; Volia, V.; Goptar, I.; Nikiforova, A.; Shilovskiy, I.; Smirnov, V.; Kostyushev, D.; Chulanov, V. Clearing of Foreign Episomal DNA from Human Cells by CRISPRa-Mediated Activation of Cytidine Deaminases. Int. J. Mol. Sci. 2020, 21, 6865. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 1–8. [Google Scholar] [CrossRef]
- Schröfelbauer, B.; Chen, D.; Landau, N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc. Natl. Acad. Sci. USA 2004, 101, 3927–3932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pham, Q.T.; Bouchard, A.; Grütter, M.G.; Berthoux, L. Generation of human TRIM5 α mutants with high HIV-1 restriction activity. Gene Ther. 2010, 17, 859–871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dufour, C.; Claudel, A.; Joubarne, N.; Merindol, N.; Maisonnet, T.; Masroori, N.; Plourde, M.B.; Berthoux, L. Editing of the human TRIM5 gene to introduce mutations with the potential to inhibit HIV-1. PLoS ONE 2018, 13, e0191709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saayman, S.M.; Lazar, D.C.; Scott, T.A.; Hart, J.R.; Takahashi, M.; Burnett, J.C.; Planelles, V.; Morris, K.V.; Weinberg, M.S. Potent and Targeted Activation of Latent HIV-1 Using the CRISPR/dCas9 Activator Complex. Mol. Ther. 2016, 24, 488–498. [Google Scholar] [CrossRef] [Green Version]
- Limsirichai, P.; Gaj, T.; Schaffer, D.V. CRISPR-mediated Activation of Latent HIV-1 Expression. Mol. Ther. 2016, 24, 499–507. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yin, C.; Zhang, T.; Li, F.; Yang, W.; Kaminski, R.; Fagan, P.R.; Putatunda, R.; Young, W.-B.; Khalili, K.; et al. CRISPR/gRNA-directed synergistic activation mediator (SAM) induces specific, persistent and robust reactivation of the HIV-1 latent reservoirs. Sci. Rep. 2015, 5, 1–14. [Google Scholar] [CrossRef]
- Ji, H.; Jiang, Z.; Lu, P.; Ma, L.; Li, C.; Pan, H.; Fu, Z.; Qu, X.; Wang, P.; Deng, J.; et al. Specific Reactivation of Latent HIV-1 by dCas9-SunTag-VP64-mediated Guide RNA Targeting the HIV-1 Promoter. Mol. Ther. 2016, 24, 508–521. [Google Scholar] [CrossRef] [Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Choe, J.; Park, O.H.; Kim, Y.K. Molecular mechanisms driving mRNA degradation by m6A modification. Trends Genet. 2020, 36, 177–188. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scarrow, M.; Chen, N.; Sun, G. Insights into the N6-methyladenosine mechanism and its functionality: Progress and questions. Crit. Rev. Biotechnol. 2020, 40, 639–652. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Li, H.; Wu, A.; Peng, Y.; Shu, G.; Yin, G. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 2019, 18, 176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, F.; et al. Association of N6-methyladenosine with viruses and related diseases. Virol. J. 2019, 16, 133. [Google Scholar] [CrossRef] [Green Version]
- Winkler, R.; Gillis, E.; Lasman, L.; Safra, M.; Geula, S.; Soyris, C.; Nachshon, A.; Tai-Schmeidel, J.; Freidman, N.; Le-Trilling, V.T.K.; et al. m 6 A modification controls the innate immune response to infection by targeting type I interferons. Nat. Immunol. 2019, 20, 173–182. [Google Scholar] [CrossRef]
- Wang, L.; Wen, M.; Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019, 365, eaav0758. [Google Scholar] [CrossRef]
- Lu, M.; Zhang, Z.; Xue, M.; Zhao, B.S.; Harder, O.; Li, A.; Liang, X.; Gao, T.Z.; Xu, Y.; Zhou, J.; et al. N 6-methyladenosine modification enables viral RNA to escape recognition by RNA sensor RIG-I. Nat. Microbiol. 2020, 5, 584–598. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 2019, 76, 96–109. [Google Scholar] [CrossRef]
- Lewis, C.J.T.; Pan, T.; Kalsotra, A. RNA modifications and structures cooperate to guide RNA–protein interactions. Nat. Rev. Mol. Cell Biol. 2017, 18, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.M.; Zhou, J.; Mao, Y.H.; Ji, Q.Q.; Qian, S.B. Programmable RNA N-6-methyladenosine editing by CRISPR-Cas9 conjugates. Nat. Chem. Biol. 2019, 15, 865. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Chen, P.J.; Miao, Z.; Liu, D.R. Programmable m 6 A modification of cellular RNAs with a Cas13-directed methyltransferase. Nat. Biotechnol. 2020, 38, 1431–1440. [Google Scholar] [CrossRef] [PubMed]
- Tycko, J.; Myer, V.E.; Hsu, P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell 2016, 63, 355–370. [Google Scholar] [CrossRef] [Green Version]
- Kocak, D.D.; Josephs, E.A.; Bhandarkar, V.; Adkar, S.S.; Kwon, J.B.; Gersbach, C.A. Increasing the specificity of CRISPR systems with engineered RNA secondary structures. Nat. Biotechnol. 2019, 37, 657. [Google Scholar] [CrossRef]
- Slaymaker, I.M.; Gao, L.; Zetscje, N.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Pattanayal, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [Green Version]
- Agudelo, D.; Carter, S.; Velimirovic, M.; Duringer, A.; Rivest, J.-F.; Levesque, S.; Loehr, J.; Mouchiroud, M.; Cyr, D.; Waters, P.J.; et al. Versatile and robust genome editing with Streptococcus thermophilus CRISPR1-Cas9. Genome Res. 2020, 30, 107–117. [Google Scholar] [CrossRef]
- Kostyushev, D.; Brezgin, S.; Kostyusheva, A.; Zarifyan, D.; Goptar, I.; Chulanov, V. Orthologous CRISPR/Cas9 systems for specific and efficient degradation of covalently closed circular DNA of hepatitis B virus. Cell. Mol. LIFE Sci. 2019, 76, 1779–1794. [Google Scholar] [CrossRef]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249. [Google Scholar] [CrossRef]
- Ferdosi, S.R.; Ewaisha, R.; Moghadam, F.; Krishna, S.; Park, J.G.; Ebrahimkhani, M.R.; Kiani, S.; Anderson, K.S. Multifunctional CRISPR-Cas9 with engineered immunosilenced human T cell epitopes. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostyushev, D.; Kostyusheva, A.; Brezgin, S.; Smirnov, V.; Volchkova, E.; Lukashev, A.; Chulanov, V. Gene Editing by Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 7362. [Google Scholar] [CrossRef]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connel, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wang, J.; Liu, Y.; Xie, L.; Su, B.; Mou, D.; Wang, L.; Liu, T.; Wang, X.; Zhao, L.; et al. CRISPR-edited stem cells in a patient with HIV and acute lymphocytic leukemia. N. Engl. J. Med. 2019, 381, 1240–1247. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).