Abstract
Intrinsic immunity is orchestrated by a wide range of host cellular proteins called restriction factors. They have the capacity to interfere with viral replication, and most of them are tightly regulated by interferons (IFNs). In addition, their regulation through post-translational modifications (PTMs) constitutes a major mechanism to shape their action positively or negatively. Following viral infection, restriction factor modification can be decisive. Palmitoylation of IFITM3, SUMOylation of MxA, SAMHD1 and TRIM5α or glycosylation of BST2 are some of those PTMs required for their antiviral activity. Nonetheless, for their benefit and by manipulating the PTMs machinery, viruses have evolved sophisticated mechanisms to counteract restriction factors. Indeed, many viral proteins evade restriction activity by inducing their ubiquitination and subsequent degradation. Studies on PTMs and their substrates are essential for the understanding of the antiviral defense mechanisms and provide a global vision of all possible regulations of the immune response at a given time and under specific infection conditions. Our aim was to provide an overview of current knowledge regarding the role of PTMs on restriction factors with an emphasis on their impact on viral replication.
1. Introduction
Interferons (IFNs) constitute the first line of defense against pathogens and extracellular aggression. They orchestrate immune defenses through the induction of hundreds of genes named ISGs (IFN-stimulated genes). Moreover, there is another type of immunity, referred to as intrinsic immunity. It is mediated by antiviral proteins (defined as restriction factors) that display a potency to block specific steps of the viral replication cycle (Figure 1), acting as potent intrinsic barriers against infection [1]. While most antiviral factors are IFN-induced, some of these proteins are constitutively expressed [2].
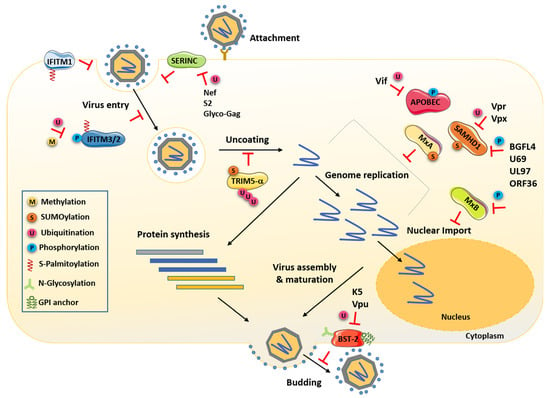
Figure 1.
Cells express several cellular antiviral factors that interfere with almost every step of the viral replication cycle. Schematic representation that highlights the known PTMs of those factors required for their antiviral activity. The viral proteins and PTMs antagonizing these factors are also indicated. Nef, Vpr, Vpx, S2 and Glyco-Gag are retroviral proteins, and BGLF4, U69, UL97 and ORF36 are herpesvirus-encoded kinases.
Mx1 was the first antiviral factor discovered in 1962 for its capacity to interfere in an inbred mouse strain with influenza A virus (IAV) infection and in A2G mice with myxoviruses [3]. However, the term restriction factor was introduced in 1970 following the discovery of the retroviral restriction factor Fv1 that protects mice against infection by murine leukemia virus (MLV) [4]. Since, a variety of additional restriction factors have been described such as TRIM5α [5,6], APOBEC3G [7,8,9], SAMHD1 [10,11], Mx2/MxB [12,13,14], Tetherin/BST2 [15,16], SERINC3/5 [17,18] and IFITMs [19].
As mentioned, the expression of most restriction factors is upregulated by type I IFNs, and this induction, therefore, constitutes the main mechanism of their regulation. However, more intricate levels of regulation have been brought to light. Indeed, post-translational modifications (PTMs) form a critical part of restriction factor regulation (Figure 1, Figure 2 and Figure 3). They can finely modulate their expression, conformation, localization, interactome, stability and therefore their functions and capacity to restrict viruses. Interestingly, these factors can be counteracted by viruses, often through hijacking PTMs for their own replication, thus forming another mechanism for their regulation.
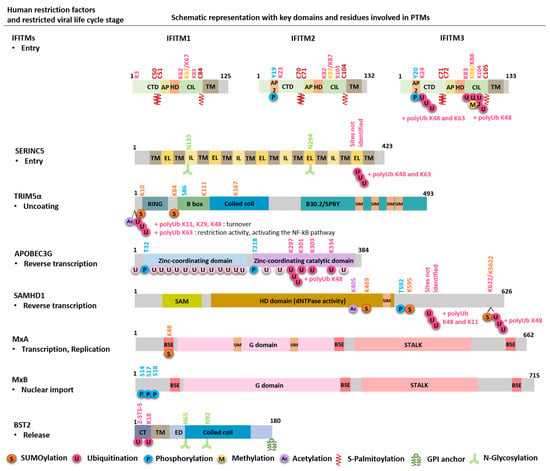
Figure 2.
Schematic representation of human restriction factors, with key domains and residues involved in PTMs. For each restriction factor, the target viral step is indicated, and the modification residue and the corresponding PTM are marked with same colors. The 20 putative lysines (K) in APOBEC are K2, 40, 42, 52, 63, 76, 79, 99, 113, 141, 150, 163, 180, 249, 270, 297, 301, 303, 334 and 344. The four residues critical for Vif-induced APOBEC proteasomal degradation are indicated, although their importance remains controversial. CTD: cytoplasmic C-terminal domain; AP2: AP2 binding domain, AP: amphipathic helix; HD: hydrophobic domain; CIL: conserved intracellular loop; TM: transmembrane domain; IL: intracellular loop; EL: extracellular loop; SIM: SUMO-interacting motif; SAM: sterile alpha motif; HD domain: histidine-aspartic-containing domain; BSE: bundle-signaling element; G domain: GTPase domain; CT: cytoplasmic tail; ED: extracellular domain.
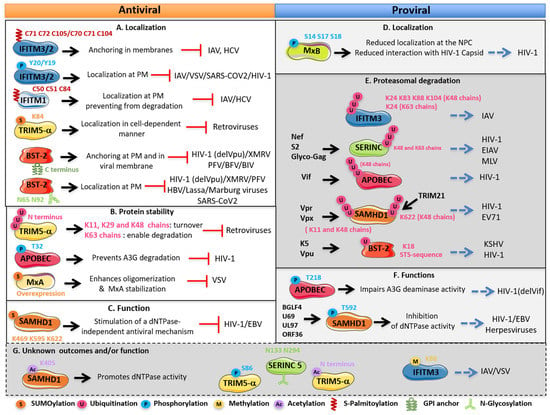
Figure 3.
Regulation of human restriction factor activities and outcomes by PTMs. (A,B,C) Antiviral effects of the PTMs, (D,E,F) proviral effects of the PTMs on the restriction factors and (G) unknown outcomes and/or function of the PTMs on the restriction factors. Restriction factors with the modified residues are illustrated. For each factor, the mechanism involved, the outcome of modified factors, the effect on viruses and the viral proteins antagonizing these factors are indicated. PM: plasma membrane; NPC: nuclear pore complex.
In eukaryotic cells, proteins can undergo a wide variety of reversible and irreversible PTMs. Four major types of PTMs are documented in literature: (i) cleavage and proteolysis of proteins, (ii) the addition of proteins or polypeptides including ubiquitination and ubiquitin-like proteins (Ublps), (iii) the addition of complex molecules such as glycosylation and palmitoylation and finally (iv) chemical changes which include phosphorylation, methylation or acetylation [20,21]. The same protein can be affected by different modifications sequentially or in response to different cellular stimuli (virus infection, stress, cell cycle, etc.). In addition, PTMs can interact with each other, modify each other and/or modify the same target in a cooperative or competitive manner. They are involved in numerous biological and cellular processes including regulation of transcription, genome integrity, cell signaling, protein degradation, IFN pathway, host–virus interactions and innate immunity [22,23,24]. Phosphorylation, methylation, acetylation, ubiquitination, SUMOylation or even glycosylation are among the most studied PTMs and are also gaining importance in the context of antiviral factor regulation.
The last years of research on PTMs have revealed the potential of these modifications in innate immunity, and the regulation of host–pathogen interactions has received considerable research interest. Emerging evidence supports that PTMs form an interface between viruses, restriction factors and cellular defense mechanisms. Thus, identifying modifications of restriction factors has become a priority in understanding the mechanisms of innate immunity, antiviral defense and IFN response (Figure 1, Figure 2 and Figure 3).
This review summarizes the recent and well-characterized regulation mechanisms of restriction factors by PTMs, highlights the importance of these regulations at the interface of virus–cell defenses and gathers many examples illustrating this diversity of the consequences of PTMs.
2. Post-Translational Modifications (PTMs)
2.1. PTMs Based on the Addition of Polypeptides: Ubiquitination and SUMOylation
Ubiquitination consists in the covalent conjugation of ubiquitin, a highly conserved protein, to lysine (K) residues of target proteins. Its main documented function is to target its substrates to the main cellular degradation machinery, the proteasome [25]. This dynamic modification implicates three main enzymatic steps involving three types of enzymes: 2 E1 ubiquitin-activating enzymes, approximately 40 E2 ubiquitin-conjugating enzymes and around 700 E3 ubiquitin ligases. It can be reversed by around 100 different deubiquitinases (DUBs) in humans [26,27].
The most common types of ubiquitination are the modification by a single ubiquitin moiety (mono-ubiquitination) or poly-ubiquitination in which several ubiquitin proteins are added in a chain at the same position on the protein substrate. Poly-ubiquitination takes place by connecting new ubiquitin proteins to either a K or Methionine (M) residue of the previous ubiquitin molecule, thus forming a chain. Indeed, ubiquitin itself contains seven K residues (K6, K11, K27, K29, K33, K48 and K63) and the N-terminal M residue to which another ubiquitin can be conjugated. The most common types of poly-ubiquitination chains are K48, K29 (which normally tags proteins for proteasomal degradation), K63, K11, K6 and M1 which, together with mono-ubiquitination, are involved in cell trafficking, signaling pathways, lysosomal degradation, activation/inactivation of enzymatic activities, translation and DNA repair [28,29,30]. Nevertheless, in addition to ensuring the turnover of cellular protein and mentioned processes, ubiquitination constitutes a main regulatory mechanism allowing viruses to evade the action of restriction factors. SERINC5 [31,32], BST2 [33,34], APOBEC [35] and SAMHD1 [36] are some of those factors highly ubiquitinated and therefore targeted to their degradation by viral proteins or under conditions of infection (Figure 2 and Figure 3).
Thus, target proteins can be subjected to a variety of ubiquitin linkage types including mono-ubiquitination, modification by multiple single ubiquitin moieties (multiubiquitination), modification of non-canonical residues serine (Ser or S), cysteine (Cys or C) or threonine (Thr or T) and, finally, modification by polyubiquitin chains on K residue or N-terminal residue of target substrates [29,37], together illustrating the diversity and complexity of regulation of the ubiquitination signal.
SUMOylation is orchestrated by the small ubiquitin-like modifier (SUMO) proteins that belong to the ubiquitin-like (UBL) family. In humans, five paralogs of SUMO are described [38]. However, only SUMO1, 2 and 3 are well documented. These small proteins share an important structural identity with ubiquitin despite a low percentage of sequence identity [39]. Like ubiquitination, SUMOylation is an enzymatic reaction that also involves three types of enzymes: the E1-activating enzymes SAE1/SAE2, the E2-conjugating enzyme Ubc9 and one of the several E3 ligases (PIAS1, PIAS3, PIASxα, PIASxβ, PIASy, RanBP2, ZNF451, Pc2, etc.). SUMOylation is a reversible mechanism since SUMO can be deconjugated by the SENP proteins family, thus allowing the recycling of SUMO proteins [38,40]. SUMOylation consists of the covalent conjugation of SUMO on a consensus motif on a K residue, although alternative consensus sites have been identified [38]. SUMO2 and SUMO3 comprise a consensus sequence of SUMOylation at their N-terminal and can themselves be modified at lysine in position 11, K11. Thus, modification by SUMO2 and SUMO3 is characterized by their ability to form polySUMO2/3 chains, and they are often indicated as SUMO2/3 [38]. Recently, other lysines on SUMO2/3, K7, K21 and K33 have been reported to participate in chain formation [38]. Surprisingly, SUMO1, previously thought to act as a chain terminator, can also be modified on K7 by SUMO2/3 and can form polySUMO chains [38,41,42]. In contrast to other PTMs, SUMO can also interact non-covalently with target proteins bearing motifs called SIMs (SUMO-interacting motifs) via its SIG domain (SUMO-interacting groove) [38]. Both covalent and non-covalent interactions of target proteins with SUMO are key regulators for their activities.
SUMOylation modulates protein stability, interactions, subcellular localization and the activity of SUMOylated targets in a cell- or stimuli-dependent manner. SUMOylation leads to significant structural and conformational changes of the substrate by masking or conferring additional binding surfaces for protein interactions thereby modulating several cellular processes including signaling pathways, transcriptional regulation and protein stability [38,40]. In the last years, several studies revealed that SUMOylation also helps to regulate host immunity and appears, in many cases, to contribute to an antiviral state. Indeed, we and others have shown that various antiviral factors are SUMOylated (PML, PKR, MxA, TRIM5α, SAMHD1) [43,44,45,46,47,48,49,50] or non-covalently modified by SUMO (TRIM5α, Daxx) [51,52,53]. Their modifications finely modulate their restriction activities [24,54].
2.2. PTMs Based on the Addition of Functional Groups: Glycosylation and S-Palmitoylation
Glycosylation consists in the addition of sugar on proteins or lipids. In mammalian cells, glycosylation results in a wide variety of glycosidic linkages, catalyzed by different types of enzymes—glycosidases, glycosyltransferases and nucleotide sugar transporters, which can be divided into four major types: N-linked and O-linked glycosylation, C-linked mannosylation and glypiation [55]. The most prevalent is N-glycosylation, which consists in the attachment of a carbohydrate on one or more asparagine (N) residues on an acceptor site N-X-T/S. It takes place in the endoplasmic reticulum (ER) concomitantly with translation before processing along the Golgi pathway where they acquire their mature and complex form [55,56]. Proteins can also be subjected to another type of glycosylation, the GPI (glycosylphosphatidylinositol) anchor. This modification is catalyzed by a family of GPI protein transamidases and consists in the addition of a glycolipid on the hydrophobic C-terminal end of proteins [57]. Like other types of glycosylation, a GPI precursor is synthesized in the ER where it is directly attached to the protein. Modified proteins then traffic through the Golgi where GPI undergoes maturation. This review focuses on the role of two well-documented glycosylated factors, SERINC5 and BST2 [58,59] (Figure 2 and Figure 3).
S-Palmitoylation (S-acylation) mediates the covalent attachment of fatty acids, primarily palmitic acid composed of 16 carbons, to a cysteine residue via a thioester linkage [60]. This modification enhances the hydrophobicity of proteins and contributes to membrane attachment. Palmitoylation is catalyzed, in mammalian cells, by a family of 24 transmembrane proteins named DHHC palmitoyltransferases (PATs) all of which contain a conserved catalytic Zinc domain called DHHC catalytic domain (zDHHC). Each PAT has a specific subcellular location. However, most of them are resident in the Golgi. Due to this distribution, PATs control the association of palmitoylated proteins to the plasma membrane (PM) and other intracellular membranes. Inversely, the reaction is reversed by the acyl-protein thioesterases, which induce depalmitoylation of targeted proteins leading to their translocation into the cytosol. Thus, palmitoylation allows modulating the subcellular location, membrane trafficking and therefore function of the palmitoylated proteins. The best known palmitoylated restriction factors are IFTIMs. Their palmitoylation constitutes a key regulator for their cell trafficking and antiviral function (Figure 2 and Figure 3) [61,62,63].
2.3. Protein Chemical Changes: Phosphorylation, Acetylation and Methylation
Phosphorylation consists in the addition of one or more phosphate groups (PO4) to proteins. This reversible PTM is one of the most common and important PTMs and chemical protein changes that occur in animal cells. Indeed, more than two-thirds of the proteins encoded by the human genome have been shown or predicted to be phosphorylated (for human phosphorylated proteins, see websites http://www.phosphosite.org/ (accessed on 28 September 2021) and http://www.phosphonet.ca/ (accessed on 28 September 2021)). Serine (Ser), tyrosine (Tyr or Y) and threonine (Thr) are the amino acids that are subjected to phosphorylation. However, in eukaryotic cells, about 86.4% of phosphorylation events occur on Ser, while only 11.8% and 1.8% occur on Thr and Tyr residues, respectively [64,65]. In humans, the phosphorylated state of proteins is mainly determined by the activity of approximately 568 protein kinases and 156 phosphatases on their substrates [66]. Many kinases and phosphatases are themselves phosphorylated, thereby forming mutually dependent and hierarchically regulated signaling loops and cascades [67]. Similar to other PTMs, protein phosphorylation is involved in the regulation of a broad spectrum of cellular processes and signal transduction including antiviral response [66]. As shown in Figure 2, restriction factors IFITM3/2 [62,68,69,70], APOBEC [71,72,73] and SAMHD1 [74,75,76,77] can be subjected to phosphorylation. It is notable that their phosphorylation can modulate their activity positively or negatively depending on targets and viruses (Figure 2 and Figure 3).
Acetylation refers to the addition of an acetyl group (CH3CO) in a protein. This reaction is catalyzed by various acetyltransferases [78]. Protein acetylation normally occurs in two distinct forms, which constitute the cell-wide acetylome. In humans, the first one occurs for approximately 80% of proteins that are co-translationally acetylated at the nascent polypeptide chains [78]. Nevertheless, this type of modification named N-terminal (Nt) acetylation can also occur post-translationally, and reactions are catalyzed by Nt-acetyltransferases (NATs) [78]. The second requires K residues and was first characterized on histones. These enzymatic reactions involve histone acetyltransferases (HATs). In contrast to Nt-acetylation, which is considered irreversible, the acetylation status of a lysine is reversible and tightly regulated by histone deacetylases (HDACs) [78]. However, many non-histone proteins have been identified as the substrates of HATs and HDACs, which consequently were renamed lysine (K) acetyltransferases (KATs) and deacetylases (KDACs) [78]. Thus, the antagonistic actions of these enzymes, KATs and KDACs, dynamically control the acetylation state of several proteins, their stability and their interactome and serve as an important mechanism for the epigenetic regulation of gene expression and diverse cellular processes, such as chromatin remodeling, cell division, nuclear transport and cell metabolism [79]. In contrast to other PTMs, the role of acetylation in antiviral response is not well documented. Nevertheless, some reports have recently identified some restriction factors to be acetylated, including SAMHD1 [80] and TRIM5α [81,82] (Figure 2 and Figure 3). These pioneering studies point to the fact that acetylation may play a crucial role in antiviral defenses.
Methylation, similar to phosphorylation and acetylation, refers to the transfer of one-carbon methyl groups (CH3) to lysine or arginine residues of protein substrates [83]. This process is achieved by two types of enzymes called arginine methyltransferases (PRMTs) and lysine methyltransferases (PKMTs) [83]. In stark contrast to other modifications, the global turnover of lysine methylation is low, suggesting that this modification is stable and not reversible. However, several studies have described the existence of lysine demethylases suggesting that the methylation can be reversible under tightly regulated conditions [83]. Like acetylation, methylation has been widely studied first in histones, and unlike acetylated lysine residues on histones, which are generally associated with the activation of gene expression, histone methylation can lead to gene activation or repression based on the target residue [83]. Moreover, the most studied process is DNA and histone methylation contributing to epigenetic regulation [84] including, of note, viral DNA [85]. Indeed, several studies evidenced the role of DNA and histone methylation in virus epigenetic regulation and their association with innate immune evasion by human viruses including HIV [86], other RNA viruses and DNA viruses [87,88,89]. In contrast, only IFITMs are reported to be methylated, evidencing that methylation may play a crucial role in antiviral factor regulation (Figure 2 and Figure 3). Thus, more research is required to better explore its effects on other factors [90].
4. Recent Identified Restriction Factors and Their PTMs
In recent years, an expanding number of studies emerged with screens that have identified other ISGs and antiviral host factors [190,191,192,193]. It is not surprising that these factors are also regulated by several PTMs.
Indeed, a high-throughput imaging-based screen allowed the identification of the mixed-lineage kinase 3 (MLK3) as a restriction factor against Zika virus [190]. MLK3 is a serine/threonine kinase implicated in the Jun N-terminal protein kinase (JNK) pathway that induces cytokine production. Its activation triggered by phosphorylation is induced by Zika virus infection. Recently, the lymphocyte antigen 6 complex locus E (LY6E) was also identified by an ISG screen as an antiviral factor of coronaviruses, including SARS-CoV, SARS-CoV-2 and Middle East respiratory syndrome (MERS)-CoV [191,194]. LY6E inhibits the entry by impeding with spike-protein-mediated membrane fusion [191]. It localizes on the PM thanks to a GPI anchoring probably on lipid rafts where receptors for the virus are also located. Mutating the site of the GPI anchor on the N99 residue abolishes its antiviral activity [194]. Strikingly, for other viruses including flaviviruses, Chikungunya or IAV, LY6E was described to promote infection [195,196], while concerning HIV-1, LY6E can be either disadvantageous or advantageous for the virus, depending on the level of expression of CD4 on target cells [197]. LY6E, as other factors, may be subjected to PTMs modulating positively or negatively its function. However, for the most part of those new factors, more research is required to dissect the role of PTMs in their functions.
Other studies are also evidencing new activities for known antiviral factors. Indeed, in 2015, the death-domain-associated protein 6 (Daxx) was identified as a new restriction factor inhibiting the reverse transcription of HIV-1 and endogenous retroviruses [198]. This protein contains two SIM domains and numerous SUMOylated sites [199,200]. Recently, we reported that Daxx is associated with incoming HIV-1 cores through a SIM-dependent interaction with cyclophilin A (CypA) and capsid (CA) [53]. Interestingly, we found that Daxx, by recruiting TNPO3, TRIM5α and TRIM34 and possibly other proteins onto incoming HIV-1 cores, prevents uncoating and therefore inhibits HIV-1 reverse transcription in a SIM-dependent manner. Thus, this report further suggests that non-covalent interaction with SUMO proteins can be also a critical regulation process in antiviral activity.
Finally, screens for Vpx targets, which was already known to counteract SAMHD1, revealed a new restriction factor also counteracted by ubiquitination-inducing degradation [192,193]. This is the human silencing hub (HUSH) complex composed of three proteins: MPP8, TASOR (Transgene Activation SuppressOR, also named FAM208A) and periphilin that recruits a methyltransferase to mediate repression of transcription [201]. As for SAMHD1, Vpx encoded by HIV-2, but also Vpr from SIV, induces TASOR ubiquitination and degradation, thanks to the DCAF1/CUL4A/B E3 ubiquitin ligase, allowing the transcription of its integrated viral genome. This phenomenon supports again that some viruses can take advantage of the host PTM machinery and puts the antiviral factor modification at the heart of host–pathogen interactions.
5. Conclusions
Thereby, in this review we wanted to highlight the fact that regulation of the innate immune system and antiviral defenses are coordinated by a myriad of host enzymes (e.g., E3 ligases, kinases, phosphatases, acetyltransferases) that modify key innate signaling molecules and antiviral factors to fine-tune antiviral responses. These enzymes induce PTMs that act as an on/off switch to modulate protein functions and form a critical part of restriction factor regulation. These factors are highly modified by PTMs, modeling their subcellular localization, stability and activity and regulating protein–protein interaction allowing restriction factors to adapt to viral infections. Nevertheless, evolution between viral proteins and restriction factors are tightly correlated and define the ability of the virus to spread in a particular species. To this end, some viruses evolved by hijacking the PTM machinery to shut them down. Therefore, it will be important to map these factor modifications and to address what determines the specificity of these enzymes toward their target proteins or upon viral infection.
Proteomics studies based on mass spectrometry (LC-MS/MS) applied to certain PTMs make it possible to carry out large-scale studies with great specificity to define a phosphoproteome, acetylome, methylome or SUMOylome. However, given the complexity and dynamics of these interactions, mapping them remains a challenge for years to come. Indeed, it is still unknown whether these regulatory mechanisms are common or differ between different cell types and species. While ubiquitination, phosphorylation and SUMOylation are increasingly associated with antiviral responses and well documented, the roles of other PTMs, such as ISGylation, neddylation, succinylation, carbonylation, glycation, citrullination, nitration and other modifications in intrinsic and innate immunity are still poorly understood. Although signaling networks in which PTMs operate are highly complex and strongly modulated, great progress has been made in recent years. We believe that developing drugs in order to favor PTMs that enhance antiviral factor activity or in order to block viral antagonism and restore efficient restriction is a promising way to fight viral infections and identify more effective therapies.
Author Contributions
C.C., G.B., S.N., N.J.A. and G.M. all edited and provided ideas for this article; C.C. and G.M. generated the figures and wrote the initial draft of the article. All authors have read and agreed to the published version of the manuscript.
Funding
G.M. is supported by a grant from the ANRS (National Agency for Research on AIDS and Hepatitis).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Marie-France Martin (University of Montpellier, IRIM, Montpellier, France) for her critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Doyle, T.; Goujon, C.; Malim, M.H. HIV-1 and Interferons: Who’s Interfering with Whom? Nat. Rev. Microbiol. 2015, 13, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Hotter, D.; Kirchhoff, F. Interferons and beyond: Induction of Antiretroviral Restriction Factors. J. Leukoc. Biol. 2018, 103, 465–477. [Google Scholar] [CrossRef]
- Lindenmann, J. Resistance of Mice to Mouse-Adapted Influenza A Virus. Virology 1962, 16, 203–204. [Google Scholar] [CrossRef]
- Lilly, F. Fv-2: Identification and Location of a Second Gene Governing the Spleen Focus Response to Friend Leukemia Virus in Mice. J. Natl. Cancer Inst. 1970, 45, 163–169. [Google Scholar] [PubMed]
- Stremlau, M.; Owens, C.M.; Perron, M.J.; Kiessling, M.; Autissier, P.; Sodroski, J. The Cytoplasmic Body Component TRIM5a Restricts HIV-1 Infection in Old World Monkeys. Nature 2004, 427, 8. [Google Scholar] [CrossRef]
- Pertel, T.; Hausmann, S.; Morger, D.; Züger, S.; Guerra, J.; Lascano, J.; Reinhard, C.; Santoni, F.A.; Uchil, P.D.; Chatel, L.; et al. TRIM5 Is an Innate Immune Sensor for the Retrovirus Capsid Lattice. Nature 2011, 472, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a Human Gene That Inhibits HIV-1 Infection and Is Suppressed by the Viral Vif Protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Mangeat, B.; Turelli, P.; Caron, G.; Friedli, M.; Perrin, L.; Trono, D. Broad Antiretroviral Defence by Human APOBEC3G through Lethal Editing of Nascent Reverse Transcripts. Nature 2003, 424, 99–103. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, B.; Pomerantz, R.J.; Zhang, C.; Arunachalam, S.C.; Gao, L. The Cytidine Deaminase CEM15 Induces Hypermutation in Newly Synthesized HIV-1 DNA. Nature 2003, 424, 94–98. [Google Scholar] [CrossRef]
- Laguette, N.; Sobhian, B.; Casartelli, N.; Ringeard, M.; Chable-Bessia, C.; Ségéral, E.; Yatim, A.; Emiliani, S.; Schwartz, O.; Benkirane, M. SAMHD1 Is the Dendritic- and Myeloid-Cell-Specific HIV-1 Restriction Factor Counteracted by Vpx. Nature 2011, 474, 654–657. [Google Scholar] [CrossRef]
- Lahouassa, H.; Daddacha, W.; Hofmann, H.; Ayinde, D.; Logue, E.C.; Dragin, L.; Bloch, N.; Maudet, C.; Bertrand, M.; Gramberg, T.; et al. SAMHD1 Restricts the Replication of Human Immunodeficiency Virus Type 1 by Depleting the Intracellular Pool of Deoxynucleoside Triphosphates. Nat. Immunol. 2012, 13, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Goujon, C.; Moncorgé, O.; Bauby, H.; Doyle, T.; Ward, C.C.; Schaller, T.; Hué, S.; Barclay, W.S.; Schulz, R.; Malim, M.H. Human MX2 Is an Interferon-Induced Post-Entry Inhibitor of HIV-1 Infection. Nature 2013, 502, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Kane, M.; Yadav, S.S.; Bitzegeio, J.; Kutluay, S.B.; Zang, T.; Wilson, S.J.; Schoggins, J.W.; Rice, C.M.; Yamashita, M.; Hatziioannou, T.; et al. MX2 Is an Interferon-Induced Inhibitor of HIV-1 Infection. Nature 2013, 502, 563–566. [Google Scholar] [CrossRef]
- Liu, Z.; Pan, Q.; Ding, S.; Qian, J.; Xu, F.; Zhou, J.; Cen, S.; Guo, F.; Liang, C. The Interferon-Inducible MxB Protein Inhibits HIV-1 Infection. Cell Host Microbe 2013, 14, 398–410. [Google Scholar] [CrossRef]
- Neil, S.J.D.; Zang, T.; Bieniasz, P.D. Tetherin Inhibits Retrovirus Release and Is Antagonized by HIV-1 Vpu. Nature 2008, 451, 425–430. [Google Scholar] [CrossRef]
- Van Damme, N.; Goff, D.; Katsura, C.; Jorgenson, R.L.; Mitchell, R.; Johnson, M.C.; Stephens, E.B.; Guatelli, J. The Interferon-Induced Protein BST-2 Restricts HIV-1 Release and Is Downregulated from the Cell Surface by the Viral Vpu Protein. Cell Host Microbe 2008, 3, 245–252. [Google Scholar] [CrossRef]
- Usami, Y.; Wu, Y.; Göttlinger, H.G. SERINC3 and SERINC5 Restrict HIV-1 Infectivity and Are Counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef Promotes Infection by Excluding SERINC5 from Virion Incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef]
- Brass, A.L.; Huang, I.-C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM Proteins Mediate Cellular Resistance to Influenza A H1N1 Virus, West Nile Virus, and Dengue Virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Peterson, S.E.; Loring, J.F. Protein Post-Translational Modifications and Regulation of Pluripotency in Human Stem Cells. Cell Res. 2014, 24, 143–160. [Google Scholar] [CrossRef]
- Spoel, S.H. Orchestrating the Proteome with Post-Translational Modifications. J. Exp. Bot. 2018, 69, 4499–4503. [Google Scholar] [CrossRef] [PubMed]
- Orford, K.; Crockett, C.; Jensen, J.P.; Weissman, A.M.; Byers, S.W. Serine Phosphorylation-Regulated Ubiquitination and Degradation of Beta-Catenin. J. Biol. Chem. 1997, 272, 24735–24738. [Google Scholar] [CrossRef]
- Waby, J.S.; Bingle, C.D.; Corfe, B.M. Post-Translational Control of Sp-Family Transcription Factors. Curr. Genomics 2008, 9, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Hannoun, Z.; Maarifi, G.; Chelbi-Alix, M.K. The Implication of SUMO in Intrinsic and Innate Immunity. Cytokine Growth Factor Rev. 2016, 29, 3–16. [Google Scholar] [CrossRef]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Morreale, F.E.; Walden, H. Types of Ubiquitin Ligases. Cell 2016, 165, 248–248.e1. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef]
- Husnjak, K.; Dikic, I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Haglund, K.; Dikic, I. Ubiquitylation and Cell Signaling. EMBO J. 2005, 24, 3353–3359. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A.; Stanhill, A. The Complexity of Recognition of Ubiquitinated Substrates by the 26S Proteasome. Biochim. Biophys. Acta 2014, 1843, 86–96. [Google Scholar] [CrossRef]
- Li, S.; Ahmad, I.; Shi, J.; Wang, B.; Yu, C.; Zhang, L.; Zheng, Y.-H. Murine Leukemia Virus Glycosylated Gag Reduces Murine SERINC5 Protein Expression at Steady-State Levels via the Endosome/Lysosome Pathway to Counteract SERINC5 Antiretroviral Activity. J. Virol. 2018, 93, e01651-18. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, I.; Li, S.; Li, R.; Chai, Q.; Zhang, L.; Wang, B.; Yu, C.; Zheng, Y.-H. The Retroviral Accessory Proteins S2, Nef, and GlycoMA Use Similar Mechanisms for Antagonizing the Host Restriction Factor SERINC5. J. Biol. Chem. 2019, 294, 7013–7024. [Google Scholar] [CrossRef]
- Pardieu, C.; Vigan, R.; Wilson, S.J.; Calvi, A.; Zang, T.; Bieniasz, P.; Kellam, P.; Towers, G.J.; Neil, S.J.D. The RING-CH Ligase K5 Antagonizes Restriction of KSHV and HIV-1 Particle Release by Mediating Ubiquitin-Dependent Endosomal Degradation of Tetherin. PLoS Pathog. 2010, 6, e1000843. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.; Pacini, G.; Berlioz-Torrent, C.; Janvier, K. Characterization of E3 Ligases Involved in Lysosomal Sorting of the HIV-1 Restriction Factor BST2. J. Cell Sci. 2017, 130, 1596–1611. [Google Scholar] [CrossRef]
- Sheehy, A.M.; Gaddis, N.C.; Malim, M.H. The Antiretroviral Enzyme APOBEC3G Is Degraded by the Proteasome in Response to HIV-1 Vif. Nat. Med. 2003, 9, 1404–1407. [Google Scholar] [CrossRef] [PubMed]
- Hrecka, K.; Hao, C.; Gierszewska, M.; Swanson, S.K.; Kesik-Brodacka, M.; Srivastava, S.; Florens, L.; Washburn, M.P.; Skowronski, J. Vpx Relieves Inhibition of HIV-1 Infection of Macrophages Mediated by the SAMHD1 Protein. Nature 2011, 474, 658–661. [Google Scholar] [CrossRef]
- Ciechanover, A.; Ben-Saadon, R. N-Terminal Ubiquitination: More Protein Substrates Join In. Trends Cell Biol. 2004, 14, 103–106. [Google Scholar] [CrossRef]
- Celen, A.B.; Sahin, U. Sumoylation on Its 25th Anniversary: Mechanisms, Pathology, and Emerging Concepts. FEBS J. 2020, 287, 3110–3140. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, Y.; Wang, X.; Liang, Z.; He, G.; Zhang, P.; Zhu, H.; Xu, N.; Liang, S. Protein SUMOylation Modification and Its Associations with Disease. Open Biol. 2017, 7, 170167. [Google Scholar] [CrossRef]
- Boulanger, M.; Chakraborty, M.; Tempé, D.; Piechaczyk, M.; Bossis, G. SUMO and Transcriptional Regulation: The Lessons of Large-Scale Proteomic, Modifomic and Genomic Studies. Molecules 2021, 26, 828. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Vertegaal, A.C.O. A Comprehensive Compilation of SUMO Proteomics. Nat. Rev. Mol. Cell Biol. 2016, 17, 581–595. [Google Scholar] [CrossRef]
- Hendriks, I.A.; Lyon, D.; Su, D.; Skotte, N.H.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Site-Specific Characterization of Endogenous SUMOylation across Species and Organs. Nat. Commun. 2018, 9, 2456. [Google Scholar] [CrossRef]
- Maarifi, G.; Fernandez, J.; Portilho, D.M.; Boulay, A.; Dutrieux, J.; Oddos, S.; Butler-Browne, G.; Nisole, S.; Arhel, N.J. RanBP2 Regulates the Anti-Retroviral Activity of TRIM5α by SUMOylation at a Predicted Phosphorylated SUMOylation Motif. Commun. Biol. 2018, 1, 193. [Google Scholar] [CrossRef] [PubMed]
- De la Cruz-Herrera, C.F.; Campagna, M.; García, M.A.; Marcos-Villar, L.; Lang, V.; Baz-Martínez, M.; Gutiérrez, S.; Vidal, A.; Rodríguez, M.S.; Esteban, M.; et al. Activation of the Double-Stranded RNA-Dependent Protein Kinase PKR by Small Ubiquitin-like Modifier (SUMO). J. Biol. Chem. 2014, 289, 26357–26367. [Google Scholar] [CrossRef] [PubMed]
- Brantis-de-Carvalho, C.E.; Maarifi, G.; Gonçalves Boldrin, P.E.; Zanelli, C.F.; Nisole, S.; Chelbi-Alix, M.K.; Valentini, S.R. MxA Interacts with and Is Modified by the SUMOylation Machinery. Exp. Cell Res. 2015, 330, 151–163. [Google Scholar] [CrossRef] [PubMed]
- Dutrieux, J.; Portilho, D.M.; Arhel, N.J.; Hazan, U.; Nisole, S. TRIM5α Is a SUMO Substrate. Retrovirology 2015, 12, 28. [Google Scholar] [CrossRef]
- Portilho, D.M.; Fernandez, J.; Ringeard, M.; Machado, A.K.; Boulay, A.; Mayer, M.; Müller-Trutwin, M.; Beignon, A.-S.; Kirchhoff, F.; Nisole, S.; et al. Endogenous TRIM5α Function Is Regulated by SUMOylation and Nuclear Sequestration for Efficient Innate Sensing in Dendritic Cells. Cell Rep. 2016, 14, 355–369. [Google Scholar] [CrossRef]
- Maarifi, G.; Hannoun, Z.; Geoffroy, M.C.; El Asmi, F.; Zarrouk, K.; Nisole, S.; Blondel, D.; Chelbi-Alix, M.K. MxA Mediates SUMO-Induced Resistance to Vesicular Stomatitis Virus. J. Virol. 2016, 90, 6598–6610. [Google Scholar] [CrossRef]
- Maarifi, G.; El Asmi, F.; Maroui, M.A.; Dianoux, L.; Chelbi-Alix, M.K. Differential Effects of SUMO1 and SUMO3 on PKR Activation and Stability. Sci. Rep. 2018, 8, 1277. [Google Scholar] [CrossRef]
- Martinat, C.; Cormier, A.; Tobaly-Tapiero, J.; Palmic, N.; Casartelli, N.; Mahboubi, B.; Coggins, S.A.; Buchrieser, J.; Persaud, M.; Diaz-Griffero, F.; et al. SUMOylation of SAMHD1 at Lysine 595 Is Required for HIV-1 Restriction in Non-Cycling Cells. Nat. Commun. 2021, 12, 4582. [Google Scholar] [CrossRef]
- Arriagada, G.; Muntean, L.N.; Goff, S.P. SUMO-Interacting Motifs of Human TRIM5α Are Important for Antiviral Activity. PLoS Pathog. 2011, 7, e1002019. [Google Scholar] [CrossRef] [PubMed]
- Lukic, Z.; Goff, S.P.; Campbell, E.M.; Arriagada, G. Role of SUMO-1 and SUMO Interacting Motifs in Rhesus TRIM5α-Mediated Restriction. Retrovirology 2013, 10, 10. [Google Scholar] [CrossRef]
- Maillet, S.; Fernandez, J.; Decourcelle, M.; El Koulali, K.; Blanchet, F.P.; Arhel, N.J.; Maarifi, G.; Nisole, S. Daxx Inhibits HIV-1 Reverse Transcription and Uncoating in a SUMO-Dependent Manner. Viruses 2020, 12, 636. [Google Scholar] [CrossRef] [PubMed]
- Colomer-Lluch, M.; Castro-Gonzalez, S.; Serra-Moreno, R. Ubiquitination and SUMOylation in HIV Infection: Friends and Foes. Curr. Issues Mol. Biol. 2020, 35, 159–194. [Google Scholar] [CrossRef]
- Moremen, K.W.; Tiemeyer, M.; Nairn, A.V. Vertebrate Protein Glycosylation: Diversity, Synthesis and Function. Nat. Rev. Mol. Cell Biol. 2012, 13, 448–462. [Google Scholar] [CrossRef]
- Clerc, F.; Reiding, K.R.; Jansen, B.C.; Kammeijer, G.S.M.; Bondt, A.; Wuhrer, M. Human Plasma Protein N-Glycosylation. Glycoconj. J. 2016, 33, 309–343. [Google Scholar] [CrossRef]
- Kinoshita, T.; Fujita, M. Biosynthesis of GPI-Anchored Proteins: Special Emphasis on GPI Lipid Remodeling. J. Lipid Res. 2016, 57, 6–24. [Google Scholar] [CrossRef]
- Sharma, S.; Lewinski, M.K.; Guatelli, J. An N-Glycosylated Form of SERINC5 Is Specifically Incorporated into HIV-1 Virions. J. Virol. 2018, 92, e00753-18. [Google Scholar] [CrossRef] [PubMed]
- Kupzig, S.; Korolchuk, V.; Rollason, R.; Sugden, A.; Wilde, A.; Banting, G. Bst-2/HM1.24 Is a Raft-Associated Apical Membrane Protein with an Unusual Topology: A Raft Protein with an Unusual Topology. Traffic 2003, 4, 694–709. [Google Scholar] [CrossRef]
- Sobocińska, J.; Roszczenko-Jasińska, P.; Ciesielska, A.; Kwiatkowska, K. Protein Palmitoylation and Its Role in Bacterial and Viral Infections. Front. Immunol. 2018, 8, 2003. [Google Scholar] [CrossRef]
- Yount, J.S.; Karssemeijer, R.A.; Hang, H.C. S -Palmitoylation and Ubiquitination Differentially Regulate Interferon-Induced Transmembrane Protein 3 (IFITM3)-Mediated Resistance to Influenza Virus. J. Biol. Chem. 2012, 287, 19631–19641. [Google Scholar] [CrossRef]
- Narayana, S.K.; Helbig, K.J.; McCartney, E.M.; Eyre, N.S.; Bull, R.A.; Eltahla, A.; Lloyd, A.R.; Beard, M.R. The Interferon-Induced Transmembrane Proteins, IFITM1, IFITM2, and IFITM3 Inhibit Hepatitis C Virus Entry. J. Biol. Chem. 2015, 290, 25946–25959. [Google Scholar] [CrossRef]
- Sällman Almén, M.; Bringeland, N.; Fredriksson, R.; Schiöth, H.B. The Dispanins: A Novel Gene Family of Ancient Origin That Contains 14 Human Members. PLoS ONE 2012, 7, e31961. [Google Scholar] [CrossRef]
- Roskoski, R. ERK1/2 MAP Kinases: Structure, Function, and Regulation. Pharmacol. Res. 2012, 66, 105–143. [Google Scholar] [CrossRef]
- Li, X.; Wilmanns, M.; Thornton, J.; Köhn, M. Elucidating Human Phosphatase-Substrate Networks. Sci. Signal. 2013, 6, rs10. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The Crucial Role of Protein Phosphorylation in Cell Signaling and Its Use as Targeted Therapy. Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef]
- Bononi, A.; Agnoletto, C.; De Marchi, E.; Marchi, S.; Patergnani, S.; Bonora, M.; Giorgi, C.; Missiroli, S.; Poletti, F.; Rimessi, A.; et al. Protein Kinases and Phosphatases in the Control of Cell Fate. Enzyme Res. 2011, 2011, 329098. [Google Scholar] [CrossRef]
- Jia, R.; Pan, Q.; Ding, S.; Rong, L.; Liu, S.-L.; Geng, Y.; Qiao, W.; Liang, C. The N-Terminal Region of IFITM3 Modulates Its Antiviral Activity by Regulating IFITM3 Cellular Localization. J. Virol. 2012, 86, 13697–13707. [Google Scholar] [CrossRef]
- Foster, T.L.; Wilson, H.; Iyer, S.S.; Coss, K.; Doores, K.; Smith, S.; Kellam, P.; Finzi, A.; Borrow, P.; Hahn, B.H.; et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell Host Microbe 2016, 20, 429–442. [Google Scholar] [CrossRef]
- Winstone, H.; Lista, M.J.; Reid, A.C.; Bouton, C.; Pickering, S.; Galao, R.P.; Kerridge, C.; Doores, K.J.; Swanson, C.M.; Neil, S.J.D. The Polybasic Cleavage Site in SARS-CoV-2 Spike Modulates Viral Sensitivity to Type I Interferon and IFITM2. J. Virol. 2021, 95, 9. [Google Scholar] [CrossRef]
- Shirakawa, K.; Takaori-Kondo, A.; Yokoyama, M.; Izumi, T.; Matsui, M.; Io, K.; Sato, T.; Sato, H.; Uchiyama, T. Phosphorylation of APOBEC3G by Protein Kinase A Regulates Its Interaction with HIV-1 Vif. Nat. Struct. Mol. Biol. 2008, 15, 1184–1191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Demorest, Z.L.; Li, M.; Harris, R.S. Phosphorylation Directly Regulates the Intrinsic DNA Cytidine Deaminase Activity of Activation-Induced Deaminase and APOBEC3G Protein. J. Biol. Chem. 2011, 286, 26568–26575. [Google Scholar] [CrossRef]
- Matsumoto, T.; Shirakawa, K.; Yokoyama, M.; Fukuda, H.; Sarca, A.D.; Koyabu, S.; Yamazaki, H.; Kazuma, Y.; Matsui, H.; Maruyama, W.; et al. Protein Kinase A Inhibits Tumor Mutator APOBEC3B through Phosphorylation. Sci. Rep. 2019, 9, 8307. [Google Scholar] [CrossRef]
- Cribier, A.; Descours, B.; Valadão, A.L.C.; Laguette, N.; Benkirane, M. Phosphorylation of SAMHD1 by Cyclin A2/CDK1 Regulates Its Restriction Activity toward HIV-1. Cell Rep. 2013, 3, 1036–1043. [Google Scholar] [CrossRef]
- Welbourn, S.; Dutta, S.M.; Semmes, O.J.; Strebel, K. Restriction of Virus Infection but Not Catalytic DNTPase Activity Is Regulated by Phosphorylation of SAMHD1. J. Virol. 2013, 87, 11516–11524. [Google Scholar] [CrossRef]
- White, T.E.; Brandariz-Nuñez, A.; Valle-Casuso, J.C.; Amie, S.; Nguyen, L.A.; Kim, B.; Tuzova, M.; Diaz-Griffero, F. The Retroviral Restriction Ability of SAMHD1, but Not Its Deoxynucleotide Triphosphohydrolase Activity, Is Regulated by Phosphorylation. Cell Host Microbe 2013, 13, 441–451. [Google Scholar] [CrossRef] [PubMed]
- St. Gelais, C.; de Silva, S.; Hach, J.C.; White, T.E.; Diaz-Griffero, F.; Yount, J.S.; Wu, L. Identification of Cellular Proteins Interacting with the Retroviral Restriction Factor SAMHD1. J. Virol. 2014, 88, 5834–5844. [Google Scholar] [CrossRef]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The World of Protein Acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef]
- Choudhary, C.; Kumar, C.; Gnad, F.; Nielsen, M.L.; Rehman, M.; Walther, T.C.; Olsen, J.V.; Mann, M. Lysine Acetylation Targets Protein Complexes and Co-Regulates Major Cellular Functions. Science 2009, 325, 834–840. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Seo, J.H.; Park, J.-H.; Vo, T.T.L.; An, S.; Bae, S.-J.; Le, H.; Lee, H.S.; Wee, H.-J.; Lee, D.; et al. SAMHD1 Acetylation Enhances Its Deoxynucleotide Triphosphohydrolase Activity and Promotes Cancer Cell Proliferation. Oncotarget 2017, 8, 68517–68529. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Christensen, D.E.; Nelson, C.; Tan, C.P.; Schaller, T.; Lehner, P.J.; Sundquist, W.I.; Towers, G.J. TRIM 5α Requires Ube2W to Anchor Lys63-linked Ubiquitin Chains and Restrict Reverse Transcription. EMBO J. 2015, 34, 2078–2095. [Google Scholar] [CrossRef]
- Fletcher, A.J.; Vaysburd, M.; Maslen, S.; Zeng, J.; Skehel, J.M.; Towers, G.J.; James, L.C. Trivalent RING Assembly on Retroviral Capsids Activates TRIM5 Ubiquitination and Innate Immune Signaling. Cell Host Microbe 2018, 24, 761–775. [Google Scholar] [CrossRef]
- Murn, J.; Shi, Y. The Winding Path of Protein Methylation Research: Milestones and New Frontiers. Nat. Rev. Mol. Cell Biol. 2017, 18, 517–527. [Google Scholar] [CrossRef]
- Ravichandran, M.; Jurkowska, R.Z.; Jurkowski, T.P. Target Specificity of Mammalian DNA Methylation and Demethylation Machinery. Org. Biomol. Chem. 2018, 16, 1419–1435. [Google Scholar] [CrossRef]
- Weber, M.; Schübeler, D. Genomic Patterns of DNA Methylation: Targets and Function of an Epigenetic Mark. Curr. Opin. Cell Biol. 2007, 19, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Ringeard, M.; Marchand, V.; Decroly, E.; Motorin, Y.; Bennasser, Y. FTSJ3 Is an RNA 2’-O-Methyltransferase Recruited by HIV to Avoid Innate Immune Sensing. Nature 2019, 565, 500–504. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immun. 2020, 12, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Milavetz, B.I.; Balakrishnan, L. Viral Epigenetics. Methods Mol. Biol. 2015, 1238, 569–596. [Google Scholar] [CrossRef]
- Chen, Y.; Guo, D. Molecular Mechanisms of Coronavirus RNA Capping and Methylation. Virol. Sin. 2016, 31, 3–11. [Google Scholar] [CrossRef]
- Shan, Z.; Han, Q.; Nie, J.; Cao, X.; Chen, Z.; Yin, S.; Gao, Y.; Lin, F.; Zhou, X.; Xu, K.; et al. Negative Regulation of Interferon-Induced Transmembrane Protein 3 by SET7-Mediated Lysine Monomethylation. J. Biol. Chem. 2013, 288, 35093–35103. [Google Scholar] [CrossRef]
- Chesarino, N.M.; Compton, A.A.; McMichael, T.M.; Kenney, A.D.; Zhang, L.; Soewarna, V.; Davis, M.; Schwartz, O.; Yount, J.S. IFITM 3 Requires an Amphipathic Helix for Antiviral Activity. EMBO Rep. 2017, 18, 1740–1751. [Google Scholar] [CrossRef]
- Rahman, K.; Coomer, C.A.; Majdoul, S.; Ding, S.Y.; Padilla-Parra, S.; Compton, A.A. Homology-Guided Identification of a Conserved Motif Linking the Antiviral Functions of IFITM3 to Its Oligomeric State. eLife 2020, 9, e58537. [Google Scholar] [CrossRef]
- Spence, J.S.; He, R.; Hoffmann, H.-H.; Das, T.; Thinon, E.; Rice, C.M.; Peng, T.; Chandran, K.; Hang, H.C. IFITM3 Directly Engages and Shuttles Incoming Virus Particles to Lysosomes. Nat. Chem. Biol. 2019, 15, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Bailey, C.C.; Zhong, G.; Huang, I.-C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.-F.; Nisole, S. West Nile Virus Restriction in Mosquito and Human Cells: A Virus under Confinement. Vaccines 2020, 8, 256. [Google Scholar] [CrossRef]
- Marziali, F.; Cimarelli, A. Membrane Interference Against HIV-1 by Intrinsic Antiviral Factors: The Case of IFITMs. Cells 2021, 10, 1171. [Google Scholar] [CrossRef]
- Shi, G.; Kenney, A.D.; Kudryashova, E.; Zani, A.; Zhang, L.; Lai, K.K.; Hall-Stoodley, L.; Robinson, R.T.; Kudryashov, D.S.; Compton, A.A.; et al. Opposing Activities of IFITM Proteins in SARS-CoV-2 Infection. EMBO J. 2020, 40, e106501. [Google Scholar] [PubMed]
- Zhao, X.; Guo, F.; Liu, F.; Cuconati, A.; Chang, J.; Block, T.M.; Guo, J.-T. Interferon Induction of IFITM Proteins Promotes Infection by Human Coronavirus OC43. Proc. Natl. Acad. Sci. USA 2014, 111, 6756–6761. [Google Scholar] [CrossRef] [PubMed]
- Prelli Bozzo, C.; Nchioua, R.; Volcic, M.; Koepke, L.; Krüger, J.; Schütz, D.; Heller, S.; Stürzel, C.M.; Kmiec, D.; Conzelmann, C.; et al. IFITM Proteins Promote SARS-CoV-2 Infection and Are Targets for Virus Inhibition in Vitro. Nat. Commun. 2021, 12, 4584. [Google Scholar] [CrossRef]
- Gea-Mallorquí, E. Does a Host Restriction Factor Facilitate Entry of SARS-CoV-2? Nat. Rev. Immunol. 2020, 20, 648. [Google Scholar] [CrossRef]
- Yount, J.S.; Moltedo, B.; Yang, Y.-Y.; Charron, G.; Moran, T.M.; López, C.B.; Hang, H.C. Palmitoylome Profiling Reveals S-Palmitoylation–Dependent Antiviral Activity of IFITM3. Nat. Chem. Biol. 2010, 6, 610–614. [Google Scholar] [CrossRef] [PubMed]
- Percher, A.; Ramakrishnan, S.; Thinon, E.; Yuan, X.; Yount, J.S.; Hang, H.C. Mass-Tag Labeling Reveals Site-Specific and Endogenous Levels of Protein S-Fatty Acylation. Proc. Natl. Acad. Sci. USA 2016, 113, 4302–4307. [Google Scholar] [CrossRef] [PubMed]
- John, S.P.; Chin, C.R.; Perreira, J.M.; Feeley, E.M.; Aker, A.M.; Savidis, G.; Smith, S.E.; Elia, A.E.H.; Everitt, A.R.; Vora, M.; et al. The CD225 Domain of IFITM3 Is Required for Both IFITM Protein Association and Inhibition of Influenza A Virus and Dengue Virus Replication. J. Virol. 2013, 87, 7837–7852. [Google Scholar] [CrossRef]
- McMichael, T.M.; Zhang, L.; Chemudupati, M.; Hach, J.C.; Kenney, A.D.; Hang, H.C.; Yount, J.S. The Palmitoyltransferase ZDHHC20 Enhances Interferon-Induced Transmembrane Protein 3 (IFITM3) Palmitoylation and Antiviral Activity. J. Biol. Chem. 2017, 292, 21517–21526. [Google Scholar] [CrossRef]
- Hach, J.C.; McMichael, T.; Chesarino, N.M.; Yount, J.S. Palmitoylation on Conserved and Nonconserved Cysteines of Murine IFITM1 Regulates Its Stability and Anti-Influenza A Virus Activity. J. Virol. 2013, 87, 9923–9927. [Google Scholar] [CrossRef] [PubMed]
- Jia, R.; Xu, F.; Qian, J.; Yao, Y.; Miao, C.; Zheng, Y.-M.; Liu, S.-L.; Guo, F.; Geng, Y.; Qiao, W.; et al. Identification of an Endocytic Signal Essential for the Antiviral Action of IFITM3: Endocytosis of IFITM3 and Its Antiviral Activity. Cell. Microbiol. 2014, 16, 1080–1093. [Google Scholar] [CrossRef]
- Chesarino, N.M.; McMichael, T.M.; Hach, J.C.; Yount, J.S. Phosphorylation of the Antiviral Protein Interferon-Inducible Transmembrane Protein 3 (IFITM3) Dually Regulates Its Endocytosis and Ubiquitination. J. Biol. Chem. 2014, 289, 11986–11992. [Google Scholar] [CrossRef]
- Perreira, J.M.; Chin, C.R.; Feeley, E.M.; Brass, A.L. IFITMs Restrict the Replication of Multiple Pathogenic Viruses. J. Mol. Biol. 2013, 425, 4937–4955. [Google Scholar] [CrossRef]
- Chesarino, N.M.; McMichael, T.M.; Yount, J.S. E3 Ubiquitin Ligase NEDD4 Promotes Influenza Virus Infection by Decreasing Levels of the Antiviral Protein IFITM3. PLoS Pathog. 2015, 11, e1005095. [Google Scholar] [CrossRef]
- Malakhova, O.A.; Zhang, D.-E. ISG15 Inhibits Nedd4 Ubiquitin E3 Activity and Enhances the Innate Antiviral Response. J. Biol. Chem. 2008, 283, 8783–8787. [Google Scholar] [CrossRef]
- Shan, J.; Zhao, B.; Shan, Z.; Nie, J.; Deng, R.; Xiong, R.; Tsun, A.; Pan, W.; Zhao, H.; Chen, L.; et al. Histone Demethylase LSD1 Restricts Influenza A Virus Infection by Erasing IFITM3-K88 Monomethylation. PLoS Pathog. 2017, 13, e1006773. [Google Scholar] [CrossRef]
- Matheson, N.J.; Sumner, J.; Wals, K.; Rapiteanu, R.; Weekes, M.P.; Vigan, R.; Weinelt, J.; Schindler, M.; Antrobus, R.; Costa, A.S.H.; et al. Cell Surface Proteomic Map of HIV Infection Reveals Antagonism of Amino Acid Metabolism by Vpu and Nef. Cell Host Microbe 2015, 18, 409–423. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, H.; Zhang, J.; Yang, J.; Bai, L.; Zheng, B.; Zheng, T.; Wang, Y.; Li, J.; Zhang, W. SERINC5 Inhibits the Secretion of Complete and Genome-Free Hepatitis B Virions Through Interfering With the Glycosylation of the HBV Envelope. Front. Microbiol. 2020, 11, 697. [Google Scholar] [CrossRef]
- Qiu, X.; Eke, I.E.; Johnson, S.F.; Ding, C.; Zheng, Y.-H. Proteasomal Degradation of Human SERINC4: A Potent Host Anti-HIV-1 Factor That Is Antagonized by Nef. Curr. Res. Virol. Sci. 2020, 1, 100002. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ji, C.; Wang, L.; Cao, Y.; Dai, J.; Ye, X.; Zeng, L.; Dai, J.; Wu, Q.; Xie, Y.; et al. Cloning and Expression of a Novel Human C5orf12 Gene*, a Member of the TMS_TDE Family. Mol. Biol. Rep. 2003, 30, 47–52. [Google Scholar] [CrossRef]
- Sood, C.; Marin, M.; Chande, A.; Pizzato, M.; Melikyan, G.B. SERINC5 Protein Inhibits HIV-1 Fusion Pore Formation by Promoting Functional Inactivation of Envelope Glycoproteins. J. Biol. Chem. 2017, 292, 6014–6026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhou, T.; Yang, J.; Lin, Y.; Shi, J.; Zhang, X.; Frabutt, D.A.; Zeng, X.; Li, S.; Venta, P.J.; et al. Identification of SERINC5-001 as the Predominant Spliced Isoform for HIV-1 Restriction. J. Virol. 2017, 91, e00137-17. [Google Scholar] [CrossRef]
- Chande, A.; Cuccurullo, E.C.; Rosa, A.; Ziglio, S.; Carpenter, S.; Pizzato, M. S2 from Equine Infectious Anemia Virus Is an Infectivity Factor Which Counteracts the Retroviral Inhibitors SERINC5 and SERINC3. Proc. Natl. Acad. Sci. USA 2016, 113, 13197–13202. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Xiong, R.; Zhou, T.; Su, P.; Zhang, X.; Qiu, X.; Li, H.; Li, S.; Yu, C.; Wang, B.; et al. HIV-1 Nef Antagonizes SERINC5 Restriction by Downregulation of SERINC5 via the Endosome/Lysosome System. J. Virol. 2018, 92, e00196-18. [Google Scholar] [CrossRef]
- Ganser-Pornillos, B.K.; Pornillos, O. Restriction of HIV-1 and Other Retroviruses by TRIM5. Nat. Rev. Microbiol. 2019, 17, 546–556. [Google Scholar] [CrossRef] [PubMed]
- Perron, M.J.; Stremlau, M.; Song, B.; Ulm, W.; Mulligan, R.C.; Sodroski, J. TRIM5 Mediates the Postentry Block to N-Tropic Murine Leukemia Viruses in Human Cells. Proc. Natl. Acad. Sci. USA 2004, 101, 11827–11832. [Google Scholar] [CrossRef]
- Yap, M.W.; Nisole, S.; Lynch, C.; Stoye, J.P. Trim5 Protein Restricts Both HIV-1 and Murine Leukemia Virus. Proc. Natl. Acad. Sci. USA 2004, 101, 10786–10791. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Griffero, F.; Kar, A.; Lee, M.; Stremlau, M.; Poeschla, E.; Sodroski, J. Comparative Requirements for the Restriction of Retrovirus Infection by TRIM5α and TRIMCyp. Virology 2007, 369, 400–410. [Google Scholar] [CrossRef]
- Stremlau, M.; Perron, M.; Lee, M.; Li, Y.; Song, B.; Javanbakht, H.; Diaz-Griffero, F.; Anderson, D.J.; Sundquist, W.I.; Sodroski, J. Specific Recognition and Accelerated Uncoating of Retroviral Capsids by the TRIM5 Restriction Factor. Proc. Natl. Acad. Sci. USA 2006, 103, 5514–5519. [Google Scholar] [CrossRef]
- Yu, A.; Skorupka, K.A.; Pak, A.J.; Ganser-Pornillos, B.K.; Pornillos, O.; Voth, G.A. TRIM5α Self-Assembly and Compartmentalization of the HIV-1 Viral Capsid. Nat. Commun. 2020, 11, 1307. [Google Scholar] [CrossRef]
- Roa, A.; Hayashi, F.; Yang, Y.; Lienlaf, M.; Zhou, J.; Shi, J.; Watanabe, S.; Kigawa, T.; Yokoyama, S.; Aiken, C.; et al. RING Domain Mutations Uncouple TRIM5 Restriction of HIV-1 from Inhibition of Reverse Transcription and Acceleration of Uncoating. J. Virol. 2012, 86, 1717–1727. [Google Scholar] [CrossRef] [PubMed]
- Yudina, Z.; Roa, A.; Johnson, R.; Biris, N.; de Souza Aranha Vieira, D.A.; Tsiperson, V.; Reszka, N.; Taylor, A.B.; Hart, P.J.; Demeler, B.; et al. RING Dimerization Links Higher-Order Assembly of TRIM5α to Synthesis of K63-Linked Polyubiquitin. Cell Rep. 2015, 12, 788–797. [Google Scholar] [CrossRef]
- Yamauchi, K.; Wada, K.; Tanji, K.; Tanaka, M.; Kamitani, T. Ubiquitination of E3 Ubiquitin Ligase TRIM5α and Its Potential Role: Ubiquitination of TRIM5α and Its Role. FEBS J. 2008, 275, 1540–1555. [Google Scholar] [CrossRef]
- Danielson, C.M.; Cianci, G.C.; Hope, T.J. Recruitment and Dynamics of Proteasome Association with RhTRIM5α Cytoplasmic Complexes During HIV-1 Infection: Proteasomes Associate with RhTRIM5α and HIV-1. Traffic 2012, 13, 1206–1217. [Google Scholar] [CrossRef]
- Campbell, E.M.; Weingart, J.; Sette, P.; Opp, S.; Sastri, J.; O’Connor, S.K.; Talley, S.; Diaz-Griffero, F.; Hirsch, V.; Bouamr, F. TRIM5α-Mediated Ubiquitin Chain Conjugation Is Required for Inhibition of HIV-1 Reverse Transcription and Capsid Destabilization. J. Virol. 2016, 90, 1849–1857. [Google Scholar] [CrossRef]
- Forshey, B.M.; von Schwedler, U.; Sundquist, W.I.; Aiken, C. Formation of a Human Immunodeficiency Virus Type 1 Core of Optimal Stability Is Crucial for Viral Replication. JVI 2002, 76, 5667–5677. [Google Scholar] [CrossRef]
- Imam, S.; Kömürlü, S.; Mattick, J.; Selyutina, A.; Talley, S.; Eddins, A.; Diaz-Griffero, F.; Campbell, E.M. K63-Linked Ubiquitin Is Required for Restriction of HIV-1 Reverse Transcription and Capsid Destabilization by Rhesus TRIM5α. J. Virol. 2019, 93, e00558-19. [Google Scholar] [CrossRef] [PubMed]
- Nepveu-Traversy, M.-É.; Demogines, A.; Fricke, T.; Plourde, M.B.; Riopel, K.; Veillette, M.; Diaz-Griffero, F.; Sawyer, S.L.; Berthoux, L. A Putative SUMO Interacting Motif in the B30.2/SPRY Domain of Rhesus Macaque TRIM5α Important for NF-ΚB/AP-1 Signaling and HIV-1 Restriction. Heliyon 2016, 2, e00056. [Google Scholar] [CrossRef] [PubMed]
- Nepveu-Traversy, M.-É.; Berthoux, L. The Conserved Sumoylation Consensus Site in TRIM5α Modulates Its Immune Activation Functions. Virus Res. 2014, 184, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and Virus Restriction. Virology 2015, 479–480, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Stopak, K.; de Noronha, C.; Yonemoto, W.; Greene, W.C. HIV-1 Vif Blocks the Antiviral Activity of APOBEC3G by Impairing Both Its Translation and Intracellular Stability. Mol. Cell 2003, 12, 591–601. [Google Scholar] [CrossRef]
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA Deamination Mediates Innate Immunity to Retroviral Infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef]
- Seissler, T.; Marquet, R.; Paillart, J.-C. Hijacking of the Ubiquitin/Proteasome Pathway by the HIV Auxiliary Proteins. Viruses 2017, 9, 322. [Google Scholar] [CrossRef]
- Zhang, W.; Du, J.; Evans, S.L.; Yu, Y.; Yu, X.-F. T-Cell Differentiation Factor CBF-β Regulates HIV-1 Vif-Mediated Evasion of Host Restriction. Nature 2012, 481, 376–379. [Google Scholar] [CrossRef]
- Jäger, S.; Kim, D.Y.; Hultquist, J.F.; Shindo, K.; LaRue, R.S.; Kwon, E.; Li, M.; Anderson, B.D.; Yen, L.; Stanley, D.; et al. Vif Hijacks CBF-β to Degrade APOBEC3G and Promote HIV-1 Infection. Nature 2012, 481, 371–375. [Google Scholar] [CrossRef]
- Anderson, B.D.; Harris, R.S. Transcriptional Regulation of APOBEC3 Antiviral Immunity through the CBF-β/RUNX Axis. Sci. Adv. 2015, 1, e1500296. [Google Scholar] [CrossRef]
- Turner, T.; Shao, Q.; Wang, W.; Wang, Y.; Wang, C.; Kinlock, B.; Liu, B. Differential Contributions of Ubiquitin-Modified APOBEC3G Lysine Residues to HIV-1 Vif-Induced Degradation. J. Mol. Biol. 2016, 428, 3529–3539. [Google Scholar] [CrossRef] [PubMed]
- Albin, J.S.; Anderson, J.S.; Johnson, J.R.; Harjes, E.; Matsuo, H.; Krogan, N.J.; Harris, R.S. Dispersed Sites of HIV Vif-Dependent Polyubiquitination in the DNA Deaminase APOBEC3F. J. Mol. Biol. 2013, 425, 1172–1182. [Google Scholar] [CrossRef]
- Iwatani, Y.; Chan, D.S.B.; Liu, L.; Yoshii, H.; Shibata, J.; Yamamoto, N.; Levin, J.G.; Gronenborn, A.M.; Sugiura, W. HIV-1 Vif-Mediated Ubiquitination/Degradation of APOBEC3G Involves Four Critical Lysine Residues in Its C-Terminal Domain. Proc. Natl. Acad. Sci. USA 2009, 106, 19539–19544. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Song, Z.; Wu, L.; Liu, G.; Ma, X.; Peng, Z.; Zhou, M.; Liang, L.; Liu, B.; Liu, J.; et al. USP49 Potently Stabilizes APOBEC3G Protein by Removing Ubiquitin and Inhibits HIV-1 Replication. eLife 2019, 8, e48318. [Google Scholar] [CrossRef] [PubMed]
- Chesarino, N.M.; Emerman, M. Polymorphisms in Human APOBEC3H Differentially Regulate Ubiquitination and Antiviral Activity. Viruses 2020, 12, 378. [Google Scholar] [CrossRef]
- Refsland, E.W.; Hultquist, J.F.; Luengas, E.M.; Ikeda, T.; Shaban, N.M.; Law, E.K.; Brown, W.L.; Reilly, C.; Emerman, M.; Harris, R.S. Natural Polymorphisms in Human APOBEC3H and HIV-1 Vif Combine in Primary T Lymphocytes to Affect Viral G-to-A Mutation Levels and Infectivity. PLoS Genet. 2014, 10, e1004761. [Google Scholar] [CrossRef]
- Jiang, Z.-Q.; Yao, X.-R.; Yu, H.; Lu, Y.-E.; Liu, B.-L.; Liu, F.-L.; Jin, Y.-B.; Zhuo, M.; Zheng, Y.-T.; Ling, F. Polymorphisms in the APOBEC3G Gene of Chinese Rhesus Macaques Affect Resistance to Ubiquitination and Degradation Mediated by HIV-2 Vif. Arch. Virol. 2019, 164, 1353–1360. [Google Scholar] [CrossRef]
- Cartier, C.; Hemonnot, B.; Gay, B.; Bardy, M.; Sanchiz, C.; Devaux, C.; Briant, L. Active CAMP-Dependent Protein Kinase Incorporated within Highly Purified HIV-1 Particles Is Required for Viral Infectivity and Interacts with Viral Capsid Protein. J. Biol. Chem. 2003, 278, 35211–35219. [Google Scholar] [CrossRef]
- Coggins, S.A.; Mahboubi, B.; Schinazi, R.F.; Kim, B. SAMHD1 Functions and Human Diseases. Viruses 2020, 12, 382. [Google Scholar] [CrossRef]
- Majer, C.; Schüssler, J.M.; König, R. Intertwined: SAMHD1 Cellular Functions, Restriction, and Viral Evasion Strategies. Med. Microbiol. Immunol. 2019, 208, 513–529. [Google Scholar] [CrossRef] [PubMed]
- Goldstone, D.C.; Ennis-Adeniran, V.; Hedden, J.J.; Groom, H.C.T.; Rice, G.I.; Christodoulou, E.; Walker, P.A.; Kelly, G.; Haire, L.F.; Yap, M.W.; et al. HIV-1 Restriction Factor SAMHD1 Is a Deoxynucleoside Triphosphate Triphosphohydrolase. Nature 2011, 480, 379–382. [Google Scholar] [CrossRef]
- Baldauf, H.-M.; Pan, X.; Erikson, E.; Schmidt, S.; Daddacha, W.; Burggraf, M.; Schenkova, K.; Ambiel, I.; Wabnitz, G.; Gramberg, T.; et al. SAMHD1 Restricts HIV-1 Infection in Resting CD4+ T Cells. Nat. Med. 2012, 18, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, S.; Behrendt, R.; Eissmann, K.; Volkmann, B.; Thomas, D.; Ebert, T.; Cribier, A.; Benkirane, M.; Hornung, V.; Bouzas, N.F.; et al. Phosphorylation of Murine SAMHD1 Regulates Its Antiretroviral Activity. Retrovirology 2015, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Hao, C.; DeLucia, M.; Swanson, S.; Florens, L.; Washburn, M.P.; Ahn, J.; Skowronski, J. CyclinA2-Cyclin-Dependent Kinase Regulates SAMHD1 Protein Phosphohydrolase Domain. J. Biol. Chem. 2015, 290, 13279–13292. [Google Scholar] [CrossRef]
- Schott, K.; Fuchs, N.V.; Derua, R.; Mahboubi, B.; Schnellbächer, E.; Seifried, J.; Tondera, C.; Schmitz, H.; Shepard, C.; Brandariz-Nuñez, A.; et al. Dephosphorylation of the HIV-1 Restriction Factor SAMHD1 Is Mediated by PP2A-B55α Holoenzymes during Mitotic Exit. Nat. Commun. 2018, 9, 2227. [Google Scholar] [CrossRef] [PubMed]
- Szaniawski, M.A.; Spivak, A.M.; Cox, J.E.; Catrow, J.L.; Hanley, T.; Williams, E.S.C.P.; Tremblay, M.J.; Bosque, A.; Planelles, V. SAMHD1 Phosphorylation Coordinates the Anti-HIV-1 Response by Diverse Interferons and Tyrosine Kinase Inhibition. mBio 2018, 9, e00819-18. [Google Scholar] [CrossRef]
- Tramentozzi, E.; Ferraro, P.; Hossain, M.; Stillman, B.; Bianchi, V.; Pontarin, G. The DNTP Triphosphohydrolase Activity of SAMHD1 Persists during S-Phase When the Enzyme Is Phosphorylated at T592. Cell Cycle 2018, 17, 1102–1114. [Google Scholar] [CrossRef]
- Zhang, K.; Lv, D.-W.; Li, R. Conserved Herpesvirus Protein Kinases Target SAMHD1 to Facilitate Virus Replication. Cell Rep. 2019, 28, 449–459. [Google Scholar] [CrossRef]
- Bermejo, M.; López-Huertas, M.R.; García-Pérez, J.; Climent, N.; Descours, B.; Ambrosioni, J.; Mateos, E.; Rodríguez-Mora, S.; Rus-Bercial, L.; Benkirane, M.; et al. Dasatinib Inhibits HIV-1 Replication through the Interference of SAMHD1 Phosphorylation in CD4+ T Cells. Biochem. Pharmacol. 2016, 106, 30–45. [Google Scholar] [CrossRef]
- Saiada, F.; Zhang, K.; Li, R. PIAS1 Potentiates the Anti-EBV Activity of SAMHD1 through SUMOylation. Cell Biosci. 2021, 11, 127. [Google Scholar] [CrossRef]
- Srivastava, S.; Swanson, S.K.; Manel, N.; Florens, L.; Washburn, M.P.; Skowronski, J. Lentiviral Vpx Accessory Factor Targets VprBP/DCAF1 Substrate Adaptor for Cullin 4 E3 Ubiquitin Ligase to Enable Macrophage Infection. PLoS Pathog. 2008, 4, e1000059. [Google Scholar] [CrossRef]
- Fregoso, O.I.; Ahn, J.; Wang, C.; Mehrens, J.; Skowronski, J.; Emerman, M. Evolutionary Toggling of Vpx/Vpr Specificity Results in Divergent Recognition of the Restriction Factor SAMHD1. PLoS Pathog. 2013, 9, e1003496. [Google Scholar] [CrossRef]
- Schaller, T.; Pollpeter, D.; Apolonia, L.; Goujon, C.; Malim, M.H. Nuclear Import of SAMHD1 Is Mediated by a Classical Karyopherin α/Β1 Dependent Pathway and Confers Sensitivity to VpxMAC Induced Ubiquitination and Proteasomal Degradation. Retrovirology 2014, 11, 29. [Google Scholar] [CrossRef]
- Petroski, M.D.; Deshaies, R.J. Function and Regulation of Cullin-RING Ubiquitin Ligases. Nat. Rev. Mol. Cell Biol. 2005, 6, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.; Norton, T.D.; Schultz, M.L.; Polsky, S.B.; Sunseri, N.; Landau, N.R. Inhibition of CUL4A Neddylation Causes a Reversible Block to SAMHD1-Mediated Restriction of HIV-1. J. Virol. 2013, 87, 11741–11750. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Z.; Huan, C.; Wang, H.; Liu, Y.; Liu, X.; Su, X.; Yu, J.; Zhao, Z.; Yu, X.; Zheng, B.; et al. TRIM 21-mediated Proteasomal Degradation of SAMHD 1 Regulates Its Antiviral Activity. EMBO Rep. 2020, 21, e47528. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Staeheli, P.; Schwemmle, M.; Kochs, G. Mx GTPases: Dynamin-like Antiviral Machines of Innate Immunity. Trends Microbiol. 2015, 23, 154–163. [Google Scholar] [CrossRef]
- Betancor, G.; Jimenez-Guardeño, J.M.; Lynham, S.; Antrobus, R.; Khan, H.; Sobala, A.; Dicks, M.D.J.; Malim, M.H. MX2-Mediated Innate Immunity against HIV-1 Is Regulated by Serine Phosphorylation. Nat. Microbiol. 2021, 6, 1031–1042. [Google Scholar] [CrossRef]
- Jouvenet, N.; Neil, S.J.D.; Zhadina, M.; Zang, T.; Kratovac, Z.; Lee, Y.; McNatt, M.; Hatziioannou, T.; Bieniasz, P.D. Broad-Spectrum Inhibition of Retroviral and Filoviral Particle Release by Tetherin. JVI 2009, 83, 1837–1844. [Google Scholar] [CrossRef]
- Sakuma, T.; Noda, T.; Urata, S.; Kawaoka, Y.; Yasuda, J. Inhibition of Lassa and Marburg Virus Production by Tetherin. JVI 2009, 83, 2382–2385. [Google Scholar] [CrossRef] [PubMed]
- Perez-Caballero, D.; Zang, T.; Ebrahimi, A.; McNatt, M.W.; Gregory, D.A.; Johnson, M.C.; Bieniasz, P.D. Tetherin Inhibits HIV-1 Release by Directly Tethering Virions to Cells. Cell 2009, 139, 499–511. [Google Scholar] [CrossRef]
- Venkatesh, S.; Bieniasz, P.D. Mechanism of HIV-1 Virion Entrapment by Tetherin. PLoS Pathog. 2013, 9, e1003483. [Google Scholar] [CrossRef] [PubMed]
- Iwabu, Y.; Fujita, H.; Kinomoto, M.; Kaneko, K.; Ishizaka, Y.; Tanaka, Y.; Sata, T.; Tokunaga, K. HIV-1 Accessory Protein Vpu Internalizes Cell-Surface BST-2/Tetherin through Transmembrane Interactions Leading to Lysosomes. J. Biol. Chem. 2009, 284, 35060–35072. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Pang, X.; Li, J.; Cen, S.; Jin, Q.; Guo, F. The Role of the Structural Domains of Human BST-2 in Inhibiting the Release of Xenotropic Murine Leukemia Virus-Related Virus. Biochem. Biophys. Res. Commun. 2012, 428, 17–23. [Google Scholar] [CrossRef]
- Liang, Z.; Zhang, Y.; Song, J.; Zhang, H.; Zhang, S.; Li, Y.; Tan, J.; Qiao, W. The Effect of Bovine BST2A1 on the Release and Cell-to-Cell Transmission of Retroviruses. Virol. J. 2017, 14, 173. [Google Scholar] [CrossRef]
- Andrew, A.J.; Miyagi, E.; Kao, S.; Strebel, K. The Formation of Cysteine-Linked Dimers of BST-2/Tetherin Is Important for Inhibition of HIV-1 Virus Release but Not for Sensitivity to Vpu. Retrovirology 2009, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.; Gitzen, A.; Swiderski, M.; Freed, E. High-Mannose But Not Complex-Type Glycosylation of Tetherin Is Required for Restriction of HIV-1 Release. Viruses 2018, 10, 26. [Google Scholar] [CrossRef]
- Fukuma, A.; Abe, M.; Morikawa, Y.; Miyazawa, T.; Yasuda, J. Cloning and Characterization of the Antiviral Activity of Feline Tetherin/BST-2. PLoS ONE 2011, 6, e18247. [Google Scholar] [CrossRef]
- Wang, W.; Wang, J.; Qu, M.; Li, X.; Zhang, J.; Zhang, H.; Wu, J.; Yu, B.; Wu, H.; Kong, W.; et al. Viral Restriction Activity of Feline BST2 Is Independent of Its N-Glycosylation and Induction of NF-ΚB Activation. PLoS ONE 2015, 10, e0138190. [Google Scholar] [CrossRef]
- Bai, B.; Wang, X.-F.; Zhang, M.; Na, L.; Zhang, X.; Zhang, H.; Yang, Z.; Wang, X. The N-Glycosylation of Equine Tetherin Affects Antiviral Activity by Regulating Its Subcellular Localization. Viruses 2020, 12, 220. [Google Scholar] [CrossRef]
- Han, Z.; Lv, M.; Shi, Y.; Yu, J.; Niu, J.; Yu, X.-F.; Zhang, W. Mutation of Glycosylation Sites in BST-2 Leads to Its Accumulation at Intracellular CD63-Positive Vesicles without Affecting Its Antiviral Activity against Multivesicular Body-Targeted HIV-1 and Hepatitis B Virus. Viruses 2016, 8, 62. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Tan, J.; Liu, R.; Xu, D.; Li, Y.; Geng, Y.; Liang, C.; Qiao, W. Tetherin Inhibits Prototypic Foamy Virus Release. Virol. J. 2011, 8, 198. [Google Scholar] [CrossRef]
- Taylor, J.K.; Coleman, C.M.; Postel, S.; Sisk, J.M.; Bernbaum, J.G.; Venkataraman, T.; Sundberg, E.J.; Frieman, M.B. Severe Acute Respiratory Syndrome Coronavirus ORF7a Inhibits Bone Marrow Stromal Antigen 2 Virion Tethering through a Novel Mechanism of Glycosylation Interference. J. Virol. 2015, 89, 11820–11833. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Wang, J.-J.; Qi, M.; Yoon, J.-J.; Chen, X.; Wen, X.; Hammonds, J.; Ding, L.; Spearman, P. Tetherin/BST-2 Is Essential for the Formation of the Intracellular Virus-Containing Compartment in HIV-Infected Macrophages. Cell Host Microbe 2012, 12, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, M.; Viswanathan, K.; Douglas, J.L.; Hines, J.; Gustin, J.; Moses, A.V.; Früh, K. Molecular Mechanism of BST2/Tetherin Downregulation by K5/MIR2 of Kaposi’s Sarcoma-Associated Herpesvirus. JVI 2009, 83, 9672–9681. [Google Scholar] [CrossRef] [PubMed]
- Agromayor, M.; Soler, N.; Caballe, A.; Kueck, T.; Freund, S.M.; Allen, M.D.; Bycroft, M.; Perisic, O.; Ye, Y.; McDonald, B.; et al. The UBAP1 Subunit of ESCRT-I Interacts with Ubiquitin via a SOUBA Domain. Structure 2012, 20, 414–428. [Google Scholar] [CrossRef]
- Mitchell, R.S.; Katsura, C.; Skasko, M.A.; Fitzpatrick, K.; Lau, D.; Ruiz, A.; Stephens, E.B.; Margottin-Goguet, F.; Benarous, R.; Guatelli, J.C. Vpu Antagonizes BST-2–Mediated Restriction of HIV-1 Release via β-TrCP and Endo-Lysosomal Trafficking. PLoS Pathog. 2009, 5, e1000450. [Google Scholar] [CrossRef]
- Janvier, K.; Pelchen–Matthews, A.; Renaud, J.-B.; Caillet, M.; Marsh, M.; Berlioz-Torrent, C. The ESCRT-0 Component HRS Is Required for HIV-1 Vpu-Mediated BST-2/Tetherin Down-Regulation. PLoS Pathog. 2011, 7, e1001265. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Cheng, M.; Chi, X.; Liu, X.; Zhou, J.; Lin, T.; Yang, W. High-Throughput Screening Identifies Mixed-Lineage Kinase 3 as a Key Host Regulatory Factor in Zika Virus Infection. J. Virol. 2019, 93, e00758-19. [Google Scholar] [CrossRef]
- Pfaender, S.; Mar, K.B.; Michailidis, E.; Kratzel, A.; Boys, I.N.; V’kovski, P.; Fan, W.; Kelly, J.N.; Hirt, D.; Ebert, N.; et al. LY6E Impairs Coronavirus Fusion and Confers Immune Control of Viral Disease. Nat. Microbiol. 2020, 5, 1330–1339. [Google Scholar] [CrossRef]
- Chougui, G.; Munir-Matloob, S.; Matkovic, R.; Martin, M.M.; Morel, M.; Lahouassa, H.; Leduc, M.; Ramirez, B.C.; Etienne, L.; Margottin-Goguet, F. HIV-2/SIV Viral Protein X Counteracts HUSH Repressor Complex. Nat. Microbiol. 2018, 3, 891–897. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Guney, M.H.; Kim, K.; Goh, S.L.; McCauley, S.; Dauphin, A.; Diehl, W.E.; Luban, J. Primate Immunodeficiency Virus Proteins Vpx and Vpr Counteract Transcriptional Repression of Proviruses by the HUSH Complex. Nat. Microbiol. 2018, 3, 1354–1361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zheng, S.; Chen, D.; Zheng, M.; Li, X.; Li, G.; Lin, H.; Chang, J.; Zeng, H.; Guo, J.-T. LY6E Restricts Entry of Human Coronaviruses, Including Currently Pandemic SARS-CoV-2. J. Virol. 2020, 94, e00562-20. [Google Scholar] [CrossRef] [PubMed]
- Hackett, B.A.; Cherry, S. Flavivirus Internalization Is Regulated by a Size-Dependent Endocytic Pathway. Proc. Natl. Acad. Sci. USA 2018, 115, 4246–4251. [Google Scholar] [CrossRef] [PubMed]
- Mar, K.B.; Rinkenberger, N.R.; Boys, I.N.; Eitson, J.L.; McDougal, M.B.; Richardson, R.B.; Schoggins, J.W. LY6E Mediates an Evolutionarily Conserved Enhancement of Virus Infection by Targeting a Late Entry Step. Nat. Commun. 2018, 9, 3603. [Google Scholar] [CrossRef]
- Yu, J.; Liang, C.; Liu, S.-L. CD4-Dependent Modulation of HIV-1 Entry by LY6E. J. Virol. 2019, 93, e01866-18. [Google Scholar] [CrossRef] [PubMed]
- Dutrieux, J.; Maarifi, G.; Portilho, D.M.; Arhel, N.J.; Chelbi-Alix, M.K.; Nisole, S. PML/TRIM19-Dependent Inhibition of Retroviral Reverse-Transcription by Daxx. PLoS Pathog. 2015, 11, e1005280. [Google Scholar] [CrossRef] [PubMed]
- Lin, D.-Y.; Huang, Y.-S.; Jeng, J.-C.; Kuo, H.-Y.; Chang, C.-C.; Chao, T.-T.; Ho, C.-C.; Chen, Y.-C.; Lin, T.-P.; Fang, H.-I.; et al. Role of SUMO-Interacting Motif in Daxx SUMO Modification, Subnuclear Localization, and Repression of Sumoylated Transcription Factors. Mol. Cell 2006, 24, 341–354. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.-S.; Ryu, S.-W.; Kim, E. Modification of Daxx by Small Ubiquitin-Related Modifier-1. Biochem. Biophys. Res. Commun. 2002, 295, 495–500. [Google Scholar] [CrossRef]
- Chougui, G.; Margottin-Goguet, F. HUSH, a Link Between Intrinsic Immunity and HIV Latency. Front. Microbiol. 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).