Full-Genome Sequences and Phylogenetic Analysis of Archived Danish European Bat Lyssavirus 1 (EBLV-1) Emphasize a Higher Genetic Resolution and Spatial Segregation for Sublineage 1a
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Sample Preparation
2.3. Genome Assembly and Phylogenetic Analysis
3. Results
3.1. Comparison of Full-Length Sequences
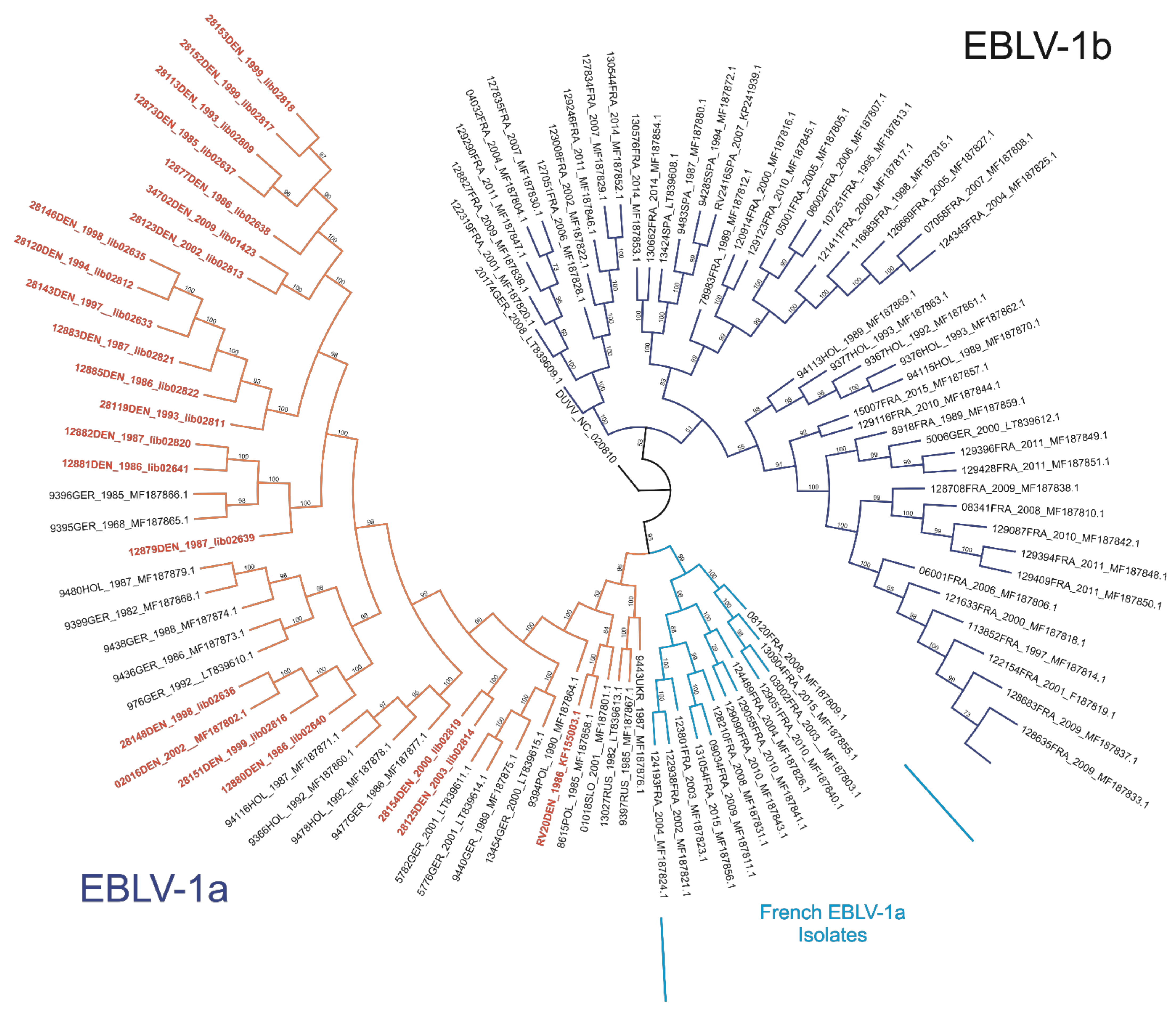
3.2. Phylogenetic Analysis Including New Danish EBLV-1 Sequences Reveal Higher Genetic Resolution for the A1 Cluster of Sublineage 1a
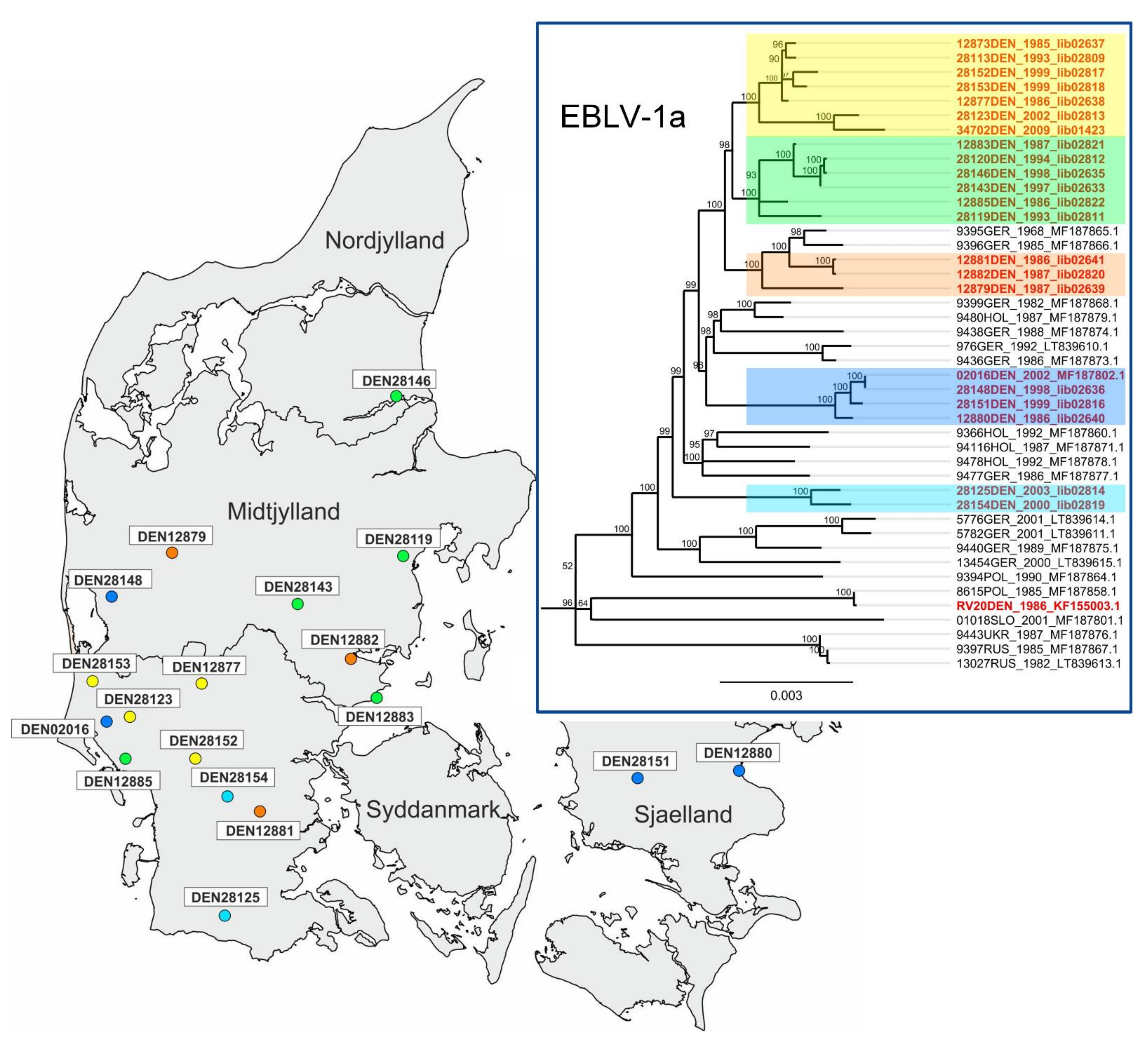
3.3. Genetic Geography Reveals Small-Scale Spatial Distribution of Danish EBLV-1a Isolates
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Banyard, A.C.; Davis, A.; Gilbert, A.T.; Markotter, W. Bat Rabies: Rabies—Scientific Basis of the Disease and Its Management, 4th ed.; Elsevier Academic Press: London, UK; San Diego, CA, USA, 2020; ISBN 9780128187050. [Google Scholar]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef]
- Amarasinghe, G.K.; Ayllón, M.A.; Bào, Y.; Basler, C.F.; Bavari, S.; Blasdell, K.R.; Briese, T.; Brown, P.A.; Bukreyev, A.; Balkema-Buschmann, A.; et al. Taxonomy of the order Mononegavirales: Update 2019. Arch. Virol. 2019, 164, 1967–1980. [Google Scholar] [CrossRef]
- International Committee on Taxonomy of Viruses. ICTV Report on Virus Classification and Taxon Nomenclatur, Genus: Lyssavirus. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/w/rhabdoviridae/795/genus-lyssavirus (accessed on 6 April 2021).
- WHO Expert Consultation on Rabies. WHO Expert Consultation on Rabies: Third Report, Bangkok, Thailand on 26–28 April 2017; WHO: Geneva, Switzerland, 2018; ISBN 9789241210218. [Google Scholar]
- Streicker, D.G.; Turmelle, A.S.; Vonhof, M.J.; Kuzmin, I.V.; McCracken, G.F.; Rupprecht, C.E. Host phylogeny constrains cross-species emergence and establishment of rabies virus in bats. Science 2010, 329, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Marston, D.A.; Banyard, A.C.; McElhinney, L.M.; Freuling, C.M.; Finke, S.; Lamballerie, X.; de Müller, T.; Fooks, A.R. The lyssavirus host-specificity conundrum-rabies virus-the exception not the rule. Curr. Opin. Virol. 2018, 28, 68–73. [Google Scholar] [CrossRef]
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Bourhy, H.; Kissi, B.; Lafon, M.; Sacramento, D.; Tordo, N. Antigenic and molecular characterization of bat rabies virus in Europe. J. Clin. Microbiol. 1992, 30, 2419–2426. [Google Scholar] [CrossRef] [PubMed]
- Müller, T.; Johnson, N.; Freuling, C.M.; Fooks, A.R.; Selhorst, T.; Vos, A. Epidemiology of bat rabies in Germany. Arch. Virol. 2007, 152, 273–288. [Google Scholar] [CrossRef]
- Müller, T.; Cox, J.; Peter, W.; Schäfer, R.; Johnson, N.; McElhinney, L.M.; Geue, J.L.; Tjørnehøj, K.; Fooks, A.R. Spill-over of European bat lyssavirus type 1 into a stone marten (Martes foina) in Germany. J. Vet. Med. B Infect. Dis. Vet. Public Health 2004, 51, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Tjørnehøj, K.; Fooks, A.R.; Agerholm, J.S.; Rønsholt, L. Natural and experimental infection of sheep with European bat lyssavirus type-1 of Danish bat origin. J. Comp. Pathol. 2006, 134, 190–201. [Google Scholar] [CrossRef]
- Dacheux, L.; Larrous, F.; Mailles, A.; Boisseleau, D.; Delmas, O.; Biron, C.; Bouchier, C.; Capek, I.; Muller, M.; Ilari, F.; et al. European bat Lyssavirus transmission among cats, Europe. Emerg. Infect. Dis. 2009, 15, 280–284. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Botvinkin, A.D.; Poleschuk, E.M.; Orciari, L.A.; Rupprecht, C.E. Bat rabies surveillance in the former Soviet Union. Dev. Biol. 2006, 125, 273–282. [Google Scholar]
- Troupin, C.; Picard-Meyer, E.; Dellicour, S.; Casademont, I.; Kergoat, L.; Lepelletier, A.; Dacheux, L.; Baele, G.; Monchâtre-Leroy, E.; Cliquet, F.; et al. Host Genetic Variation Does Not Determine Spatio-Temporal Patterns of European Bat 1 Lyssavirus. Genome Biol. Evol. 2017, 9, 3202–3213. [Google Scholar] [CrossRef]
- Davis, P.L.; Holmes, E.C.; Larrous, F.; van der Poel, W.H.M.; Tjørnehøj, K.; Alonso, W.J.; Bourhy, H. Phylogeography, population dynamics, and molecular evolution of European bat lyssaviruses. J. Virol. 2005, 79, 10487–10497. [Google Scholar] [CrossRef] [PubMed]
- Eggerbauer, E.; Pfaff, F.; Finke, S.; Höper, D.; Beer, M.; Mettenleiter, T.C.; Nolden, T.; Teifke, J.-P.; Müller, T.; Freuling, C.M. Comparative analysis of European bat lyssavirus 1 pathogenicity in the mouse model. PLoS Negl. Trop. Dis. 2017, 11, e0005668. [Google Scholar] [CrossRef]
- Marston, D.A.; McElhinney, L.M.; Johnson, N.; Muller, T.; Conzelmann, K.K.; Tordo, N.; Fooks, A.R. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ non-translated region. J. Gen. Virol. 2007, 88, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Conraths, F.J.; Freuling, C.M.; Hoffmann, B.; Selhorst, T.; Kliemt, J.; Schatz, J.; Müller, T. New insights into the genetics of EBLV-1 from Germany. Berl. Munch. Tierarztl. Wochenschr. 2012, 259–263. [Google Scholar]
- Vázquez-Moron, S. Phylogeny of European Bat Lyssavirus 1 in Eptesicus isabellinus Bats, Spain. Emerg. Infect. Dis. 2011. [Google Scholar] [CrossRef]
- Amengual, B.; Whitby, J.E.; King, A.; Cobo, J.S.; Bourhy, H. Evolution of European bat lyssaviruses. J. Gen. Virol. 1997, 78, 2319–2328. [Google Scholar] [CrossRef] [PubMed]
- McElhinney, L.M.; Marston, D.A.; Leech, S.; Freuling, C.M.; van der Poel, W.H.M.; Echevarria, J.; Vázquez-Moron, S.; Horton, D.L.; Müller, T.; Fooks, A.R. Molecular Epidemiology of Bat Lyssaviruses in Europe. Zoonoses Public Health 2013, 60, 35–45. [Google Scholar] [CrossRef]
- Grauballe, P.C.; Baagøe, H.J.; Fekadu, M.; Westergaard, J.M.; Zoffmann, H. Bat rabies in Denmark. Lancet 1987, 329, 379–380. [Google Scholar] [CrossRef]
- Schatz, J.; Fooks, A.R.; McElhinney, L.; Horton, D.; Echevarria, J.; Vázquez-Moron, S.; Kooi, E.A.; Rasmussen, T.B.; Müller, T.; Freuling, C.M. Bat rabies surveillance in Europe. Zoonoses Public Health 2013, 60, 22–34. [Google Scholar] [CrossRef]
- Freuling, C.M.; Klöss, D.; Schröder, R.; Kliemt, A.; Müller, T. The WHO Rabies Bulletin Europe: A key source of information on rabies and a pivotal tool for surveillance and epidemiology. Rev. Off. Int. Epizoot. 2012, 31, 799–807. [Google Scholar] [CrossRef]
- Wylezich, C.; Papa, A.; Beer, M.; Höper, D. A Versatile Sample Processing Workflow for Metagenomic Pathogen Detection. Sci. Rep. 2018, 8, 13108. [Google Scholar] [CrossRef] [PubMed]
- Delmas, O.; Holmes, E.C.; Talbi, C.; Larrous, F.; Dacheux, L.; Bouchier, C.; Bourhy, H. Genomic diversity and evolution of the lyssaviruses. PLoS ONE 2008, 3, e2057. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; Haeseler, A.; von Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Hoang, D.T.; Chernomor, O.; Haeseler, A.; von Minh, B.Q.; Le Vinh, S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Haeseler, A.; von Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Marston, D.A.; McElhinney, L.M.; Ellis, R.J.; Horton, D.L.; Wise, E.L.; Leech, S.L.; David, D.; Lamballerie, X.; de Fooks, A.R. Next generation sequencing of viral RNA genomes. BMC Genom. 2013, 14, 444. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; Amengual, B.; Abellán, C.; Bourhy, H. European bat lyssavirus infection in Spanish bat populations. Emerg. Infect. Dis. 2002, 8, 413–420. [Google Scholar] [CrossRef]
- Picard-Meyer, E.; Robardet, E.; Arthur, L.; Larcher, G.; Harbusch, C.; Servat, A.; Cliquet, F. Bat rabies in France: A 24-year retrospective epidemiological study. PLoS ONE 2014, 9, e98622. [Google Scholar] [CrossRef] [PubMed]
- Schatz, J.; Freuling, C.M.; Auer, E.; Goharriz, H.; Harbusch, C.; Johnson, N.; Kaipf, I.; Mettenleiter, T.C.; Mühldorfer, K.; Mühle, R.-U.; et al. Enhanced passive bat rabies surveillance in indigenous bat species from Germany-a retrospective study. PLoS Negl. Trop. Dis. 2014, 8, e2835. [Google Scholar] [CrossRef] [PubMed]
- Bogdanowicz, W.; Lesiński, G.; Sadkowska-Todys, M.; Gajewska, M.; Rutkowski, R. Population Genetics and Bat Rabies: A Case Study of Eptesicus serotinus in Poland. Acta Chiropt. 2013, 15, 35–56. [Google Scholar] [CrossRef]
- Hutterer, R.; Ivanova, T.; Meyer-Cords, C.H.; Rodrigues, L. Bat Migration in Europe: A Review of Banding Data and Literature; Federal Agency for Nature Conservation: Bonn, Germany, 2005; ISBN 378433928X. [Google Scholar]
- McElhinney, L.M.; Marston, D.A.; Wise, E.L.; Freuling, C.M.; Bourhy, H.; Zanoni, R.; Moldal, T.; Kooi, E.A.; Neubauer-Juric, A.; Nokireki, T.; et al. Molecular Epidemiology and Evolution of European Bat Lyssavirus 2. Int. J. Mol. Sci. 2018, 19, 156. [Google Scholar] [CrossRef] [PubMed]
| Sample ID | Library No. | Species | Sample Matrix | Year of Collection | Location | Sequence Length |
|---|---|---|---|---|---|---|
| 12873DEN | 2637 | Bat (E. serotinus) | Brain | 1985 | n.d. | 11,963 |
| 12877DEN | 2638 | Bat (E. serotinus) | Brain | 1986 | Grindsted | 11,925 |
| 12879DEN | 2639 | Bat (E. serotinus) | Brain | 1987 | Vildbjerg | 11,966 |
| 12880DEN | 2640 | Bat (E. serotinus) | Brain | 1986 | Køge | 11,945 |
| 12881DEN | 2641 | Bat (E. serotinus) | Brain | 1986 | Sommersted | 11,953 |
| 12882DEN | 2820 | Bat (E. serotinus) | Brain | 1987 | Horsens | 11,953 |
| 12883DEN | 2821 | Bat (E. serotinus) | Brain | 1987 | Juelsminde | 11,934 |
| 12885DEN | 2822 | Bat (E. serotinus) | Brain | 1986 | Esbjerg | 11,955 |
| 28113DEN | 2809 | Bat (E. serotinus) | Brain | 1993 | n.d. | 11,967 |
| 28119DEN | 2811 | Bat (E. serotinus) | CCS | 1993 | Arhus | 11,965 |
| 28120DEN | 2812 | Bat (E. serotinus) | CCS | 1994 | Arhus | 11,958 |
| 28123DEN | 2813 | Bat (E. serotinus) | CCS | 2002 | Varde | 11,966 |
| 28125DEN | 2814 | Bat (E. serotinus) | CCS | 2003 | Bylderup | 11,965 |
| 28143DEN | 2633 | Bat (E. serotinus) | CCS | 1997 | Bryrup | 11,966 |
| 28146DEN | 2635 | Bat (P. Pygmaeus) | CCS | 1998 | Hadsund | 11,966 |
| 28148DEN | 2636 | Sheep (Ovis aries) | CCS | 1998 | Lem | 11,966 |
| 28151DEN | 2816 | Bat (E. serotinus) | CCS | 1999 | Zealand Soro | 11,965 |
| 28152DEN | 2817 | Bat (E. serotinus) | CCS | 1999 | Holsted | 11,966 |
| 28153DEN | 2818 | Bat (E. serotinus) | CCS | 1999 | Nebel | 11,966 |
| 28154DEN | 2819 | Bat (E. serotinus) | CCS | 2000 | Rodding | 11,964 |
| 34702DEN | 1423 | Bat (E. serotinus) | CCS | 2009 | n.d. | 11,967 * |
| Group (Color) | Sample ID | Host Species | Region | Year |
|---|---|---|---|---|
| 1 (yellow) | 12873DEN | Serotine Bat | - | 1985 |
| 28113DEN | Serotine Bat | - | 1993 | |
| 28152DEN | Serotine Bat | Syddanmark | 1999 | |
| 28153DEN | Serotine Bat | Syddanmark | 1999 | |
| 12877DEN | Serotine Bat | Syddanmark | 1986 | |
| 28123DEN | Serotine Bat | Syddanmark | 2002 | |
| 34702DEN | Serotine Bat | - | 2009 | |
| 2 (green) | 12883DEN | Serotine Bat | Midtjylland | 1987 |
| 12885DEN | Serotine Bat | Syddanmark | 1986 | |
| 28119DEN | Serotine Bat | Midtjylland | 1993 | |
| 28120DEN | Serotine Bat | Midtjylland | 1994 | |
| 28143DEN | Serotine Bat | Midtjylland | 1997 | |
| 28146DEN | Soprano Pipistrelle | Nordjylland | 1998 | |
| 3 (orange) | 12879DEN | Serotine Bat | Midtjylland | 1987 |
| 12881DEN | Serotine Bat | Syddanmark | 1986 | |
| 12882DEN | Serotine Bat | Midtjylland | 1987 | |
| 4 (blue) | 02016DEN * | Serotine Bat | Syddanmark | 2002 |
| 12880DEN | Serotine Bat | Sjælland | 1986 | |
| 28148DEN | Sheep | Midtjylland | 1998 | |
| 28151DEN | Serotine Bat | Sjælland | 1999 | |
| 5 (cyan) | 28125DEN | Serotine Bat | Syddanmark | 2003 |
| 28154DEN | Serotine Bat | Syddanmark | 2000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Calvelage, S.; Freuling, C.M.; Fooks, A.R.; Höper, D.; Marston, D.A.; McElhinney, L.; Rasmussen, T.B.; Finke, S.; Beer, M.; Müller, T. Full-Genome Sequences and Phylogenetic Analysis of Archived Danish European Bat Lyssavirus 1 (EBLV-1) Emphasize a Higher Genetic Resolution and Spatial Segregation for Sublineage 1a. Viruses 2021, 13, 634. https://doi.org/10.3390/v13040634
Calvelage S, Freuling CM, Fooks AR, Höper D, Marston DA, McElhinney L, Rasmussen TB, Finke S, Beer M, Müller T. Full-Genome Sequences and Phylogenetic Analysis of Archived Danish European Bat Lyssavirus 1 (EBLV-1) Emphasize a Higher Genetic Resolution and Spatial Segregation for Sublineage 1a. Viruses. 2021; 13(4):634. https://doi.org/10.3390/v13040634
Chicago/Turabian StyleCalvelage, Sten, Conrad M. Freuling, Anthony R. Fooks, Dirk Höper, Denise A. Marston, Lorraine McElhinney, Thomas Bruun Rasmussen, Stefan Finke, Martin Beer, and Thomas Müller. 2021. "Full-Genome Sequences and Phylogenetic Analysis of Archived Danish European Bat Lyssavirus 1 (EBLV-1) Emphasize a Higher Genetic Resolution and Spatial Segregation for Sublineage 1a" Viruses 13, no. 4: 634. https://doi.org/10.3390/v13040634
APA StyleCalvelage, S., Freuling, C. M., Fooks, A. R., Höper, D., Marston, D. A., McElhinney, L., Rasmussen, T. B., Finke, S., Beer, M., & Müller, T. (2021). Full-Genome Sequences and Phylogenetic Analysis of Archived Danish European Bat Lyssavirus 1 (EBLV-1) Emphasize a Higher Genetic Resolution and Spatial Segregation for Sublineage 1a. Viruses, 13(4), 634. https://doi.org/10.3390/v13040634






