Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus
Abstract
1. Introduction
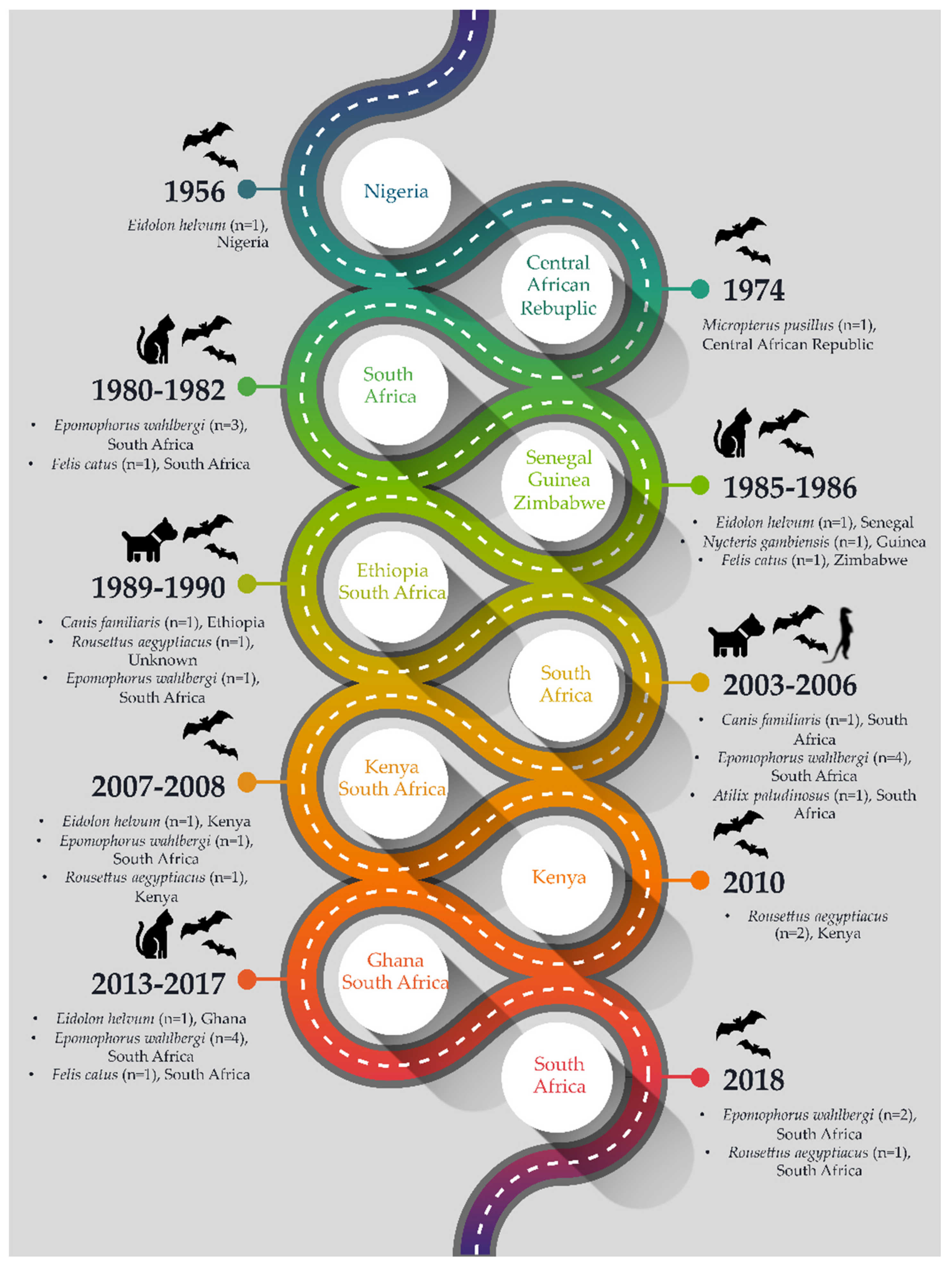
2. Lagos Bat Virus Detections
2.1. Detections in Eidolon helvum
2.2. Detections in Epomophorus wahlbergi
2.3. Detections in Rousettus sp.
2.4. Detection in Micropterus pusillus
2.5. Detection in Nycteris gambianus
2.6. Detections in Spillover Hosts
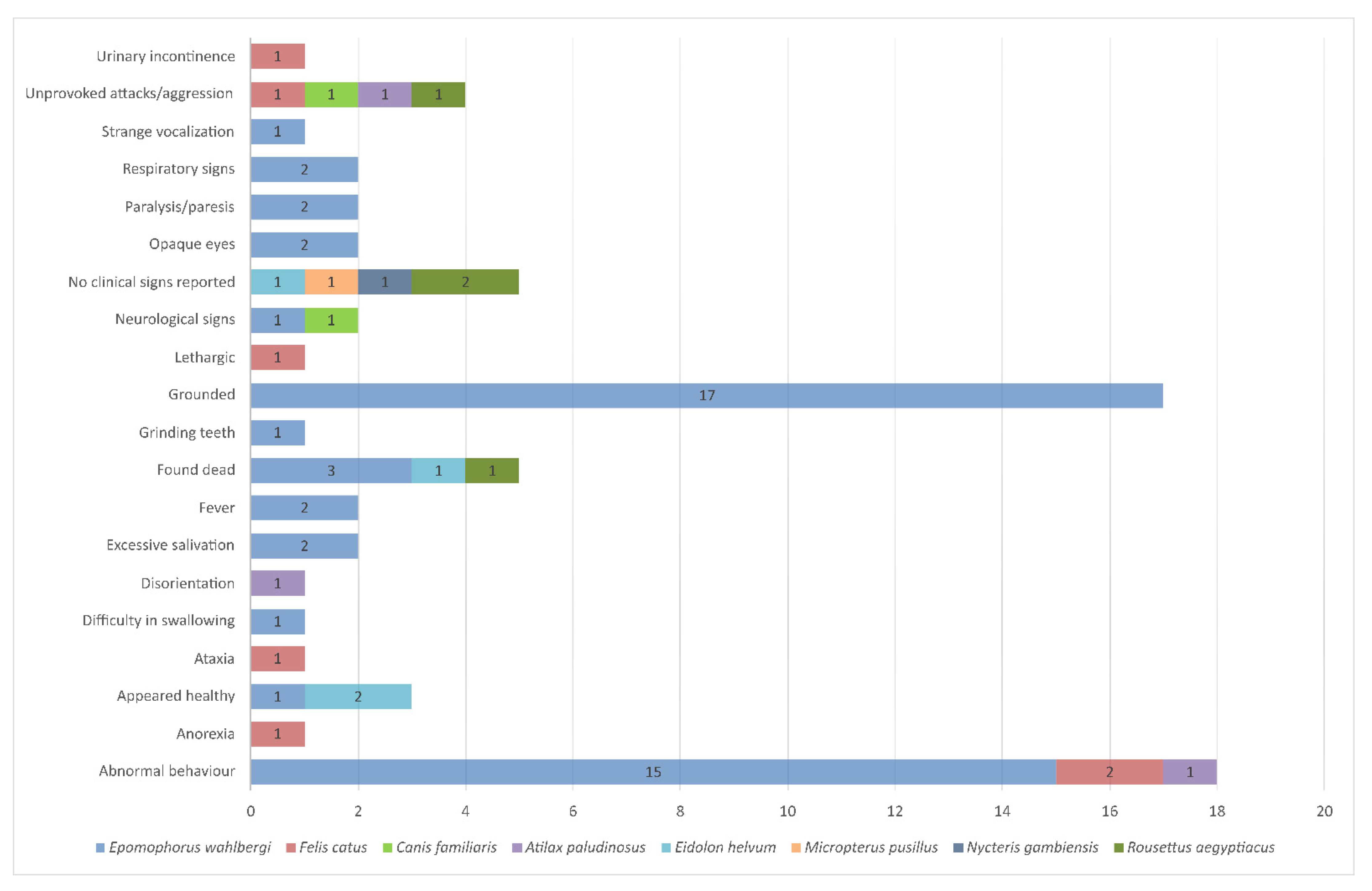
3. New Lagos Bat Virus Cases Identified in South Africa, 2013–2018
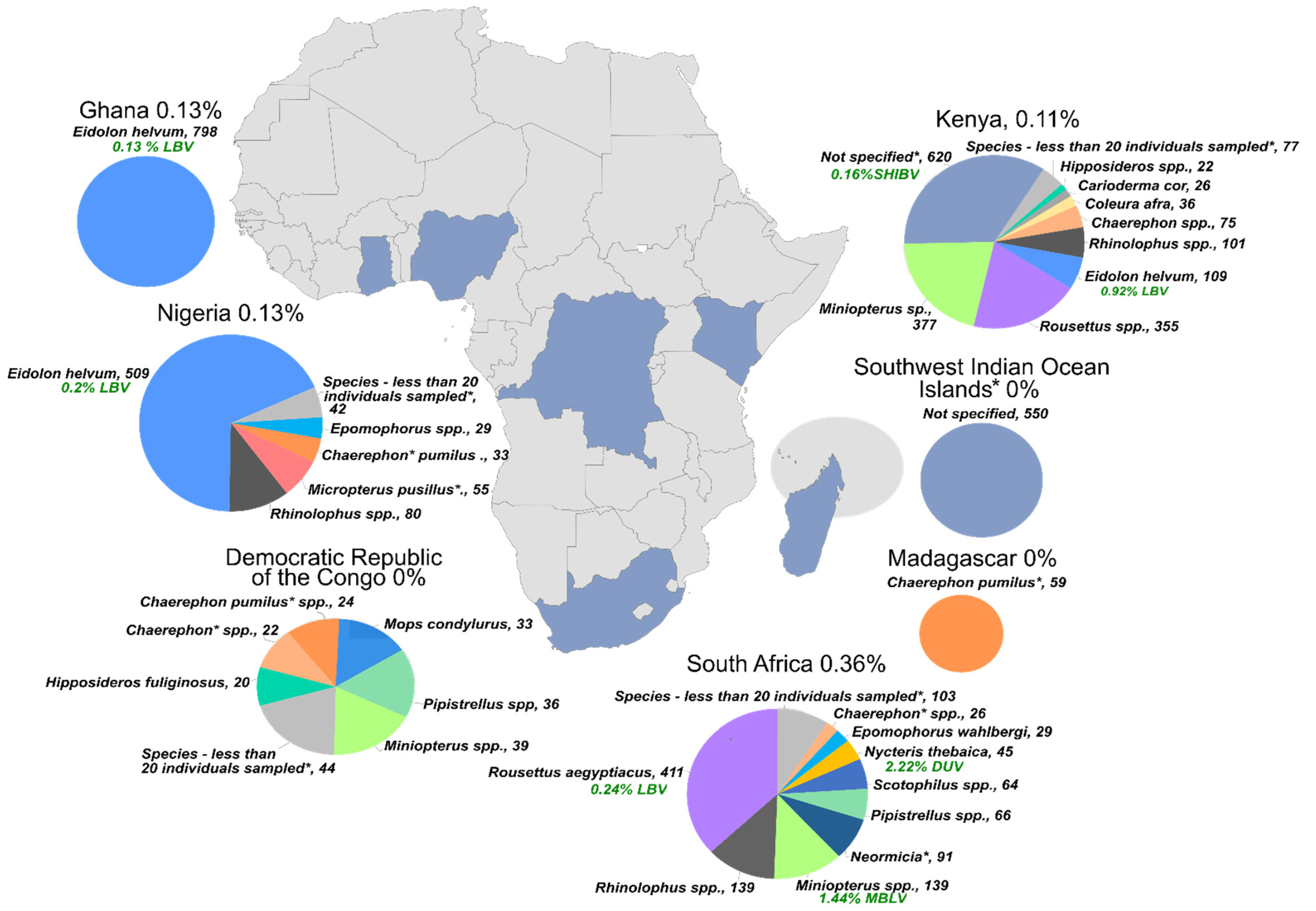
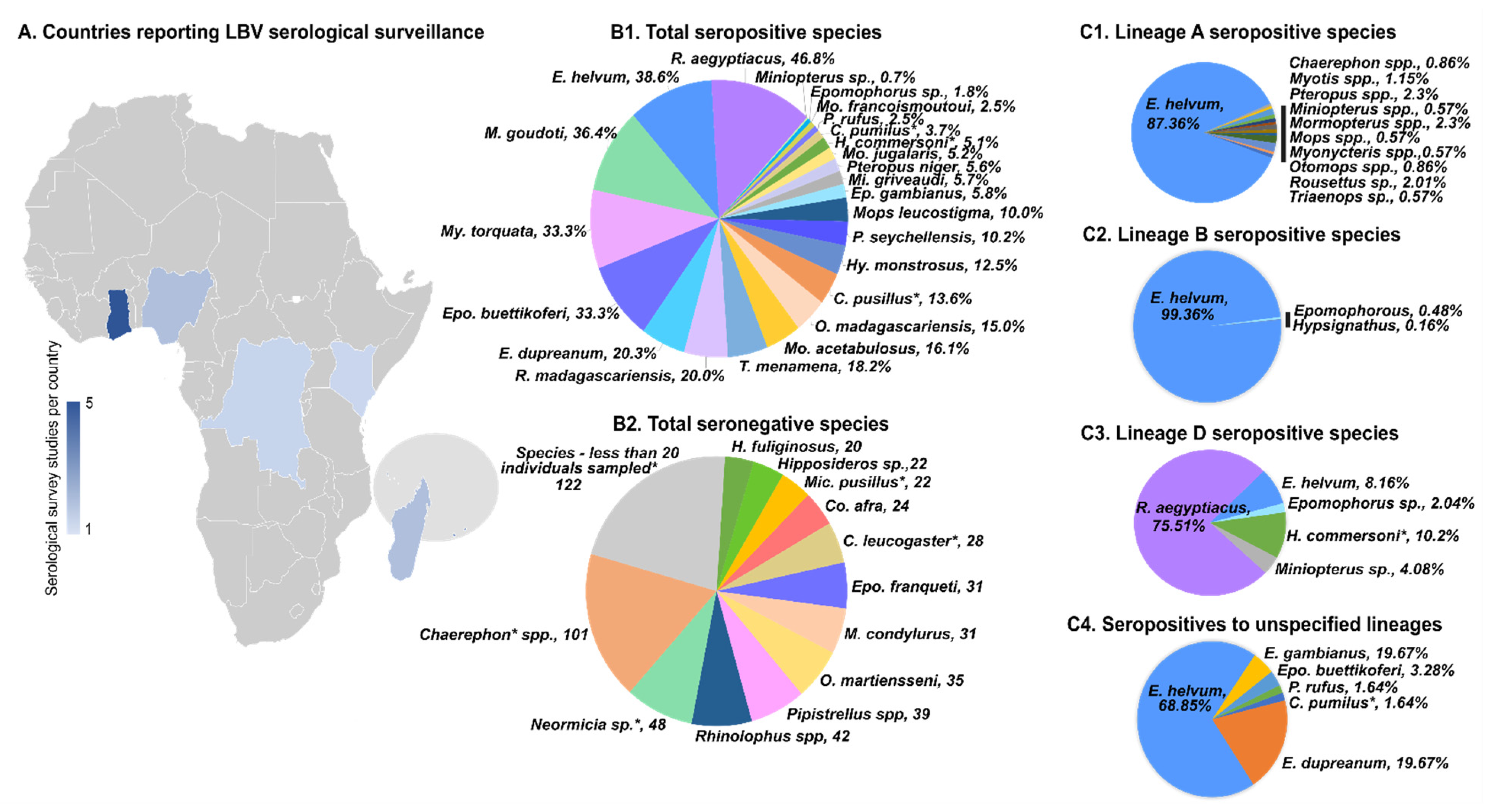
4. Surveillance for Lagos Bat Virus
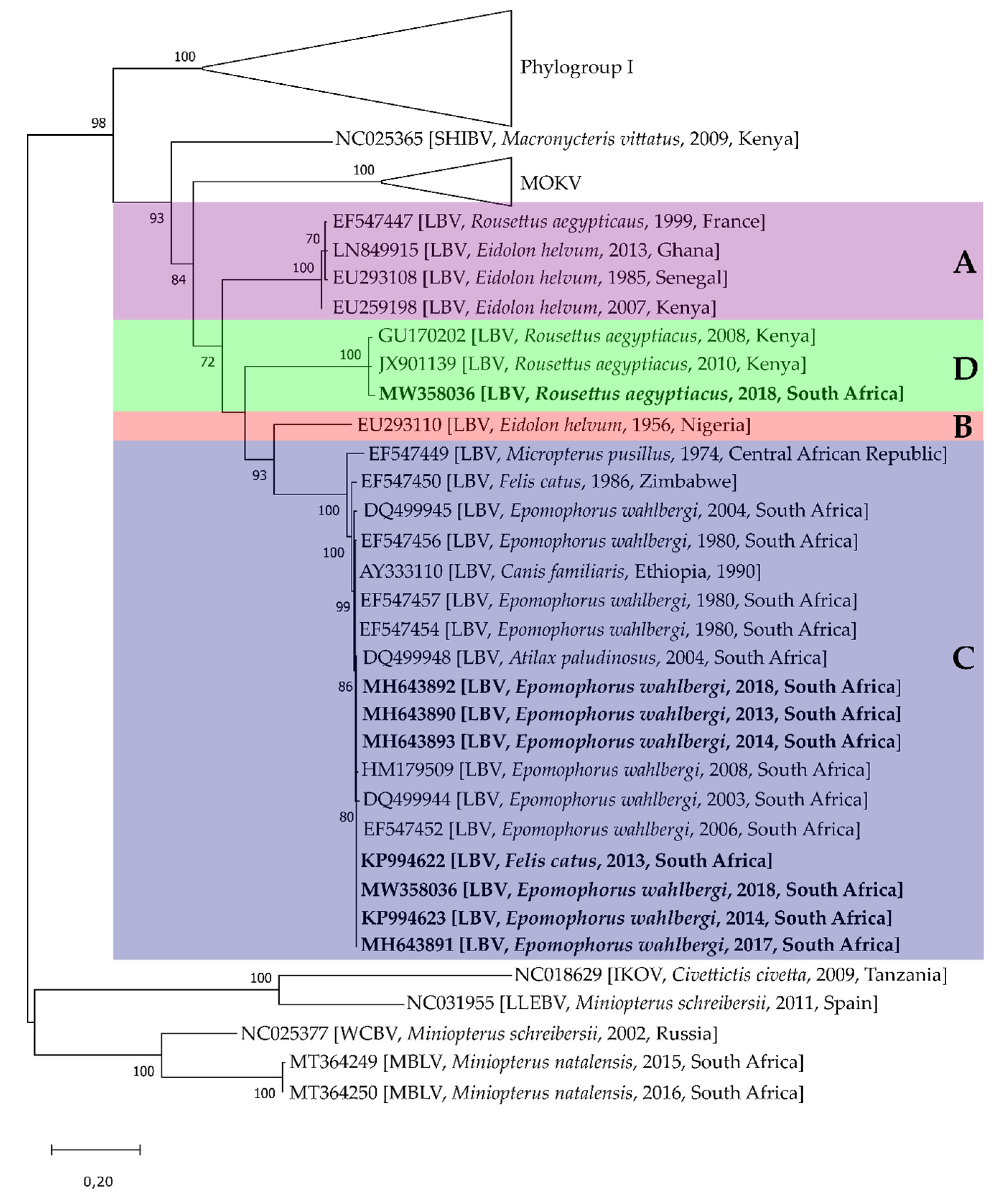
5. Lagos Bat Virus Diversity
6. Pathogenesis
7. Prevention
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boulger, L.R.; Porterfield, J.S. Isolation of a virus from Nigerian fruit bats. Trans. R. Soc. Trop. Med. Hyg. 1958, 52, 421–424. [Google Scholar] [CrossRef]
- Markotter, W.; Coertse, J. Bat lyssaviruses. Sci. Tech. Rev. 2018, 37, 385–400. [Google Scholar] [CrossRef]
- ICTV International Committee on Taxonomy of Viruses. Available online: https://talk.ictvonline.org/ (accessed on 23 February 2021).
- Coertse, J.; Grobler, C.S.; Sabeta, C.T.; Seamark, E.C.J.; Kearney, T.; Paweska, J.T.; Markotter, W. Lyssaviruses in insectivorous bats, South Africa, 2003–2018. Emerg. Infect. Dis. 2020, 26, 3056–3060. [Google Scholar] [CrossRef]
- Badrane, H.; Tordo, N. Host switching in lyssavirus history from the Chiroptera to the Carnivora Orders. J. Virol. 2001, 75, 8096–8104. [Google Scholar] [CrossRef]
- Nel, L.H.; Rupprecht, C.E. Emergence of lyssaviruses in the Old World: The case of Africa. In Wildlife and Emerging Zoonotic Diseases: The biology, Circumstances and Consequences of Cross-Species Transmission; Springer: Berlin/Heidelberg, Germany, 2007; Volume 1, pp. 161–193. [Google Scholar]
- Shope, R.E. Rabies-related viruses. Yale J. Biol. Med. 1982, 55, 271–275. [Google Scholar]
- Shope, R.E.; Murphy, F.A.; Harrison, A.K.; Causey, O.R.; Kemp, G.E.; Simpson, D.I.H.; Moore, D.L. Two African viruses serologically and morphologically related to rabies virus. J. Virol. 1970, 6, 690–692. [Google Scholar] [CrossRef]
- Swanepoel, R.; Barnard, B.J.; Meredith, C.D.; Bishop, G.C.; Brückner, G.K.; Foggin, C.M.; Hübschle, O.J. Rabies in southern Africa. Onderstepoort J. Vet. Res. 1993, 60, 325–346. [Google Scholar]
- Kuzmin, I.V.; Niezgoda, M.; Franka, R.; Agwanda, B.; Markotter, W.; Beagley, J.C.; Urazova, O.Y.; Breiman, R.F.; Rupprecht, C.E. Lagos bat virus in Kenya. J. Clin. Microbiol. 2008, 46, 1451–1461. [Google Scholar] [CrossRef]
- Freuling, C.M.; Binger, T.; Beer, M.; Adu-Sarkodie, Y.; Schatz, J.; Fischer, M.; Hanke, D.; Hoffmann, B.; Höper, D.; Mettenleiter, T.C.; et al. Lagos bat virus transmission in an Eidolon helvum bat colony, Ghana. Virus Res. 2015, 210, 42–45. [Google Scholar] [CrossRef]
- Meredith, C.D.; Standing, E. Lagos bat virus in South Africa. Lancet 1981, 317, 832–833. [Google Scholar] [CrossRef]
- Crick, J.; Tignor, G.H.; Moreno, K. A new isolate of Lagos bat virus from the Republic of South Africa. Trans. R. Soc. Trop. Med. Hyg. 1982, 76, 211–213. [Google Scholar] [CrossRef]
- Markotter, W.; Randles, J.; Rupprecht, C.E.; Sabeta, C.T.; Taylor, P.J.; Wandeler, A.I.; Nel, L.H. Lagos bat virus, South Africa. Emerg. Infect. Dis. 2006, 12, 504–506. [Google Scholar] [CrossRef]
- Markotter, W. Molecular Epidemiology and Pathogenesis of Lagos Bat Virus, a Rabies-Related Virus Specific to Africa; University of Pretoria: Pretoria, South Africa, 2007. [Google Scholar]
- Markotter, W.; Kuzmin, I.; Rupprecht, C.E.; Nel, L.H. Phylogeny of Lagos bat virus: Challenges for lyssavirus taxonomy. Virus Res. 2008, 135, 10–21. [Google Scholar] [CrossRef]
- Kgaladi, J.; Nel, L.H.; Markotter, W. Comparison of pathogenic domains of rabies and African rabies-related lyssaviruses and pathogenicity observed in mice. Onderstepoort J. Vet. Res. 2013, 80. [Google Scholar] [CrossRef] [PubMed]
- Aubert, M.F.A. Rabies in individual countries. France. WHO Rabies Bull. Eur. 1999, Second Qua, 6. [Google Scholar]
- Aubert, M.F.A. Rabies in individual countries. France. WHO Rabies Bull. Eur. 1999, Third quar, 5. [Google Scholar]
- Kuzmin, I.V.; Markotter, W.; Agwanda, B.; Niezgoda, M.; Gilbert, A.T.; Rupprecht, C.E. Egyptian Fruit Bat (Rousettus aegyptiacus) as the Host of a Specific Lineage of Lagos Bat Virus in Kenya. Available online: https://www.researchgate.net/publication/328127074 (accessed on 25 February 2021).
- Kuzmin, I.V.; Mayer, A.E.; Niezgoda, M.; Markotter, W.; Agwanda, B.; Breiman, R.F.; Rupprecht, C.E. Shimoni bat virus, a new representative of the Lyssavirus genus. Virus Res. 2010, 149, 197–210. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Rupprecht, C. Bat lyssaviruses. In Bats and Viruses: A New Frontier of Emerging Infectious Diseases; Wang, L.-F., Cowled, C., Eds.; Wiley: Hoboken, NJ, USA, 2015; pp. 47–97. ISBN 9781118818824. [Google Scholar]
- van Cakenberghe, V.; Seamark, E.C. (Eds.) African Chiroptera Report; AfricanBats NPC: Pretoria, South Africa, 2020; ISBN 9780323609845. [Google Scholar]
- Sureau, P.; Germain, M.; Herve, J.P.; Geoffroy, B.; Cornet, J.P.; Heme, G.; Robin, Y. Isolation of the Lagos-bat virus in the Central African Republic. Bull. Soc. Pathol. Exot. Fil. 1977, 70, 467–470. [Google Scholar]
- Sureau, P.; Tignor, G.H.; Smith, A.L. Antigenic characterization of the Bangui strain (ANCB-672d) of Lagos bat virus. Ann. l’Institut Pasteur Virol. 1980, 131, 25–32. [Google Scholar] [CrossRef]
- Markotter, W.; Coertse, J.; de Vries, L.; Geldenhuys, M.; Mortlock, M. Bat-borne viruses in Africa: A critical review. J. Zool. 2020, 311, 77–98. [Google Scholar] [CrossRef]
- King, A.; Crick, J. Rabies-related viruses. In Rabies. Developments in Veterinary Virology; Campbell, J.B., Charlton, K.M., Eds.; Springer: Boston, MA, USA, 1988; pp. 177–199. [Google Scholar]
- Foggin, C.M. Rabies and Rabies-Related Viruses in Zimbabwe: Historical, Virological and Ecological Aspects; University of Zimbabwe: Harare, Zimbabwe, 1988. [Google Scholar]
- Mebatsion, T.; Cox, J.H.; Frost, J.W. Isolation and characterization of 115 street rabies virus isolates from Ethiopia by using monoclonal antibodies: Identification of 2 isolates as Mokola and Lagos bat viruses. J. Infect. Dis. 1992, 166, 972–977. [Google Scholar] [CrossRef] [PubMed]
- Markotter, W.; Kuzmin, I.; Rupprecht, C.; Randles, J.; Sabeta, C.; Wandeler, A.; Nel, L. Isolation of Lagos Bat Virus from water mongoose. Emerg. Infect. Dis. 2006, 12, 1913–1918. [Google Scholar] [CrossRef]
- Coertse, J.; Weyer, J.; Nel, L.H.; Markotter, W. Improved PCR methods for detection of African rabies and rabies-related lyssaviruses. J. Clin. Microbiol. 2010, 48, 3949–3955. [Google Scholar] [CrossRef] [PubMed]
- Coertse, J.; Markotter, W.; le Roux, K.; Stewart, D.; Sabeta, C.T.; Nel, L.H. New isolations of the rabies-related Mokola virus from South Africa. BMC Vet. Res. 2016, 13, 37. [Google Scholar] [CrossRef] [PubMed]
- Coertse, J. Is Mokola Virus the Most Recent Known Lyssavirus Ancestor of an Insect Virus Progenitor? In Insights from Cell Culture Studies and Molecular Surveillance; University of Pretoria: Pretoria, South Africa, 2015. [Google Scholar]
- Coertse, J.; Weyer, J.; Nel, L.H.; Markotter, W. Reverse transcription recombinase polymerase amplification assay for rapid detection of canine associated rabies virus in Africa. PLoS ONE 2019, 14, e0219292. [Google Scholar] [CrossRef] [PubMed]
- Monadjem, A.; Demos, T.C.; Dalton, D.L.; Webala, P.W.; Musila, S.; Kerbis Peterhans, J.C.; Patterson, B.D. A revision of pipistrelle-like bats (Mammalia: Chiroptera: Vespertilionidae) in East Africa with the description of new genera and species. Zool. J. Linn. Soc. 2020. [Google Scholar] [CrossRef]
- Smith, T.; Gilbert, A. Comparison of a micro-neutralization test with the rapid fluorescent focus inhibition test for measuring rabies virus neutralizing antibodies. Trop. Med. Infect. Dis. 2017, 2, 24. [Google Scholar] [CrossRef]
- Gilbert, A.T.; Fooks, A.R.; Hayman, D.T.S.; Horton, D.L.; Müller, T.; Plowright, R.; Peel, A.J.; Bowen, R.; Wood, J.L.N.; Mills, J.; et al. Deciphering serology to understand the ecology of infectious diseases in wildlife. Ecohealth 2013, 10, 298–313. [Google Scholar] [CrossRef]
- Mélade, J.; McCulloch, S.; Ramasindrazana, B.; Lagadec, E.; Turpin, M.; Pascalis, H.; Goodman, S.M.; Markotter, W.; Dellagi, K. Serological evidence of lyssaviruses among bats on Southwestern Indian Ocean Islands. PLoS ONE 2016, 11, e0160553. [Google Scholar] [CrossRef]
- Badrane, H.; Bahloul, C.; Perrin, P.; Tordo, N. Evidence of two lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 2001, 75, 3268–3276. [Google Scholar] [CrossRef]
- Horton, D.L.; McElhinney, L.M.; Marston, D.A.; Wood, J.L.N.; Russell, C.A.; Lewis, N.; Kuzmin, I.V.; Fouchier, R.A.M.; Osterhaus, A.D.M.E.; Fooks, A.R.; et al. Quantifying antigenic relationships among the lyssaviruses. J. Virol. 2010, 84, 11841–11848. [Google Scholar] [CrossRef] [PubMed]
- Hanlon, C.A.; Kuzmin, I.V.; Blanton, J.D.; Weldon, W.C.; Manangan, J.S.; Rupprecht, C.E. Efficacy of rabies biologics against new lyssaviruses from Eurasia. Virus Res. 2005, 111, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Hayman, D.T.S.; Fooks, A.R.; Horton, D.; Suu-Ire, R.; Breed, A.C.; Cunningham, A.A.; Wood, J.L.N. Antibodies against Lagos Bat Virus in Megachiroptera from West Africa. Emerg. Infect. Dis. 2008, 14, 926–928. [Google Scholar] [CrossRef]
- Wright, E.; Hayman, D.T.S.; Vaughan, A.; Temperton, N.J.; Wood, J.L.N.; Cunningham, A.A.; Suu-Ire, R.; Weiss, R.A.; Fooks, A.R. Virus neutralising activity of African fruit bat (Eidolon helvum) sera against emerging lyssaviruses. Virology 2010, 408, 183–189. [Google Scholar] [CrossRef]
- Dietzschold, B.; Rupprecht, C.E.; Tollis, M.; Lafon, M.; Mattei, J.; Wiktor, T.J.; Koprowski, H. Antigenic diversity of the glycoprotein and nucleocapsid proteins of rabies and rabies-related viruses: Implications for epidemiology and control of rabies. Clin. Infect. Dis. 1988, 10, S785–S798. [Google Scholar] [CrossRef]
- Bourhy, H.; Kissi, B.; Tordo, N. Molecular diversity of the Lyssavirus genus. Virology 1993, 194, 70–81. [Google Scholar] [CrossRef]
- Kissi, B.; Tordo, N.; Bourhy, H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology 1995, 209, 526–537. [Google Scholar] [CrossRef]
- Delmas, O.; Holmes, E.C.; Talbi, C.; Larrous, F.; Dacheux, L.; Bouchier, C.; Bourhy, H. Genomic diversity and evolution of the lyssaviruses. PLoS ONE 2008, 3, e2057. [Google Scholar] [CrossRef]
- Tignor, G.H.; Shope, R.E.; Bhatt, P.N.; Percy, D.H. Experimental infection of dogs and monkeys with two rabies serogroup viruses, Lagos bat and Mokola (IbAn 27377): Clinical, serologic, virologic, and fluorescent-antibody studies. J. Infect. Dis. 1973, 128, 471–478. [Google Scholar] [CrossRef] [PubMed]
- Markotter, W.; Kuzmin, I.V.; Rupprecht, C.E.; Nel, L.H. Lagos bat virus virulence in mice inoculated by the peripheral route. Epidemiol. Infect. 2009, 137, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Masatani, T.; Ito, N.; Shimizu, K.; Ito, Y.; Nakagawa, K.; Abe, M.; Yamaoka, S.; Sugiyama, M. Amino acids at positions 273 and 394 in rabies virus nucleoprotein are important for both evasion of host RIG-I-mediated antiviral response and pathogenicity. Virus Res. 2011, 155, 168–174. [Google Scholar] [CrossRef]
- Poisson, N.; Real, E.; Gaudin, Y.; Vaney, M.-C.; King, S.; Jacob, Y.; Tordo, N.; Blondel, D. Molecular basis for the interaction between rabies virus phosphoprotein P and the dynein light chain LC8: Dissociation of dynein-binding properties and transcriptional functionality of P. J. Gen. Virol. 2001, 82, 2691–2696. [Google Scholar] [CrossRef]
- Irie, T.; Carnero, E.; Okumura, A.; García-Sastre, A.; Harty, R.N. Modifications of the PSAP region of the matrix protein lead to attenuation of vesicular stomatitis virus in vitro and in vivo. J. Gen. Virol. 2007, 88, 2559–2567. [Google Scholar] [CrossRef]
- Wirblich, C.; Tan, G.S.; Papaneri, A.; Godlewski, P.J.; Orenstein, J.M.; Harty, R.N.; Schnell, M.J. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J. Virol. 2008, 82, 9730–9738. [Google Scholar] [CrossRef]
- Gholami, A.; Kassis, R.; Real, E.; Delmas, O.; Guadagnini, S.; Larrous, F.; Obach, D.; Prevost, M.-C.; Jacob, Y.; Bourhy, H. Mitochondrial dysfunction in lyssavirus-induced apoptosis. J. Virol. 2008, 82, 4774–4784. [Google Scholar] [CrossRef]
- Mita, T.; Shimizu, K.; Ito, N.; Yamada, K.; Ito, Y.; Sugiyama, M.; Minamoto, N. Amino acid at position 95 of the matrix protein is a cytopathic determinant of rabies virus. Virus Res. 2008, 137, 33–39. [Google Scholar] [CrossRef]
- Prehaud, C.; Diallo, A.; Martient-Edelist, C.; Coulon, P.; Flamand, A. Characterization of a new temperature-sensitive and avirulent mutant of the rabies virus. Virus Res. 1988, 11, 88. [Google Scholar] [CrossRef]
- Faber, M.; Pulmanausahakul, R.; Nagao, K.; Prosniak, M.; Rice, A.B.; Koprowski, H.; Schnell, M.J.; Dietzschold, B. Identification of viral genomic elements responsible for rabies virus neuroinvasiveness. Proc. Natl. Acad. Sci. USA 2004, 101, 16328–16332. [Google Scholar] [CrossRef]
- Takayama-Ito, M.; Ito, N.; Yamada, K.; Sugiyama, M.; Minamoto, N. Multiple amino acids in the glycoprotein of rabies virus are responsible for pathogenicity in adult mice. Virus Res. 2006, 115, 169–175. [Google Scholar] [CrossRef]
- Langevin, C.; Jaaro, H.; Bressanelli, S.; Fainzilber, M.; Tuffereau, C. Rabies virus glycoprotein (RVG) is a trimeric ligand for the N-terminal cysteine-rich domain of the mammalian p75 neurotrophin receptor. J. Biol. Chem. 2002, 277, 37655–37662. [Google Scholar] [CrossRef] [PubMed]
- Coulon, P.; Ternaux, J.-P.; Flamand, A.; Tuffereau, C. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J. Virol. 1998, 72, 273–278. [Google Scholar] [CrossRef]
- Dietzschold, B.; Wunner, W.H.; Wiktor, T.J.; Lopes, A.D.; Lafon, M.; Smith, C.L.; Koprowski, H. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. USA 1983, 80, 70–74. [Google Scholar] [CrossRef]
- Tuffereau, C.; Benejean, J.; Blondel, D.; Kieffer, B.; Flamand, A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998, 17, 7250–7259. [Google Scholar] [CrossRef]
- Begeman, L.; Suu-Ire, R.; Banyard, A.C.; Drosten, C.; Eggerbauer, E.; Freuling, C.M.; Gibson, L.; Goharriz, H.; Horton, D.L.; Jennings, D.; et al. Experimental Lagos bat virus infection in straw-colored fruit bats: A suitable model for bat rabies in a natural reservoir species. PLoS Negl. Trop. Dis. 2020, 14, e0008898. [Google Scholar] [CrossRef]
- Suu-Ire, R.; Fooks, A.; Banyard, A.; Selden, D.; Amponsah-Mensah, K.; Riesle, S.; Ziekah, M.; Ntiamoa-Baidu, Y.; Wood, J.; Cunningham, A. Lagos bat virus infection dynamics in free-ranging straw-colored fruit bats (Eidolon helvum). Trop. Med. Infect. Dis. 2017, 2, 25. [Google Scholar] [CrossRef]
- World Health Organization. Rabies Vaccines: WHO Position Paper, April 2018—Recommendations. Vaccine 2018, 36, 5500–5503. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.C.; Morimoto, K.; Bette, M.; Weihe, E.; Koprowski, H.; Dietzschold, B. Collaboration of antibody and inflammation in clearance of rabies virus from the central nervous system. J. Virol. 1998, 72, 3711–3719. [Google Scholar] [CrossRef]
- Malerczyk, C.; Freuling, C.; Gniel, D.; Giesen, A.; Selhorst, T.; Müller, T. Cross-neutralization of antibodies induced by vaccination with Purified Chick Embryo Cell Vaccine (PCECV) against different Lyssavirus species. Hum. Vaccin. Immunother. 2014, 10, 2799–2804. [Google Scholar] [CrossRef] [PubMed]
- Fekadu, M.; Shaddock, J.; Sanderlin, D.; Smith, J. Efficacy of rabies vaccines against Duvenhage virus isolated from European house bats (Eptesicus serotinus), classic rabies and rabies-related viruses. Vaccine 1988, 6, 533–539. [Google Scholar] [CrossRef]
- Overduin, L.A.; van Dongen, J.J.; Visser, L.G.; Dongen, V. The cellular immune response to rabies vaccination: A systematic review. Vaccines 2019, 7, 110. [Google Scholar] [CrossRef]
- Mallewa, M.; Fooks, A.R.; Banda, D.; Chikungwa, P.; Mankhambo, L.; Molyneux, E.; Molyneux, M.E.; Solomon, T. Rabies encephalitis in malaria-endemic area, Malawi, Africa. Emerg. Infect. Dis. 2007, 13, 136–139. [Google Scholar] [CrossRef] [PubMed]
- Weyer, J.; Dermaux-Msimang, V.; Grobbelaar, A.; le Roux, C.; Moolla, N.; Paweska, J.; Blumberg, L. Epidemiology of human rabies in South Africa, 2008–2018. S. Afr. Med. J. 2020, 110, 877. [Google Scholar] [CrossRef]
- Hampson, K.; Coudeville, L.; Lembo, T.; Sambo, M.; Kieffer, A.; Attlan, M.; Barrat, J.; Blanton, J.D.; Briggs, D.J.; Cleaveland, S.; et al. Estimating the global burden of endemic canine rabies. PLoS Negl. Trop. Dis. 2015, 9, e0003709. [Google Scholar] [CrossRef]
- Rupprecht, C.E.; Kuzmin, I.V.; Yale, G.; Nagarajan, T.; Meslin, F.-X. Priorities in applied research to ensure programmatic success in the global elimination of canine rabies. Vaccine 2019, 37, A77–A84. [Google Scholar] [CrossRef]
- Weyer, J.; Kuzmin, I.V.; Rupprecht, C.E.; Nel, L.H. Cross-protective and cross-reactive immune responses to recombinant vaccinia viruses expressing full-length lyssavirus glycoprotein genes. Epidemiol. Infect. 2008, 136, 670–678. [Google Scholar] [CrossRef]
- Fisher, C.R.; Lowe, D.E.; Smith, T.G.; Yang, Y.; Hutson, C.L.; Wirblich, C.; Cingolani, G.; Schnell, M.J. Lyssavirus vaccine with a chimeric glycoprotein protects across phylogroups. Cell Rep. 2020, 32, 107920. [Google Scholar] [CrossRef]
- Evans, J.S.; Horton, D.L.; Easton, A.J.; Fooks, A.R.; Banyard, A.C. Rabies virus vaccines: Is there a need for a pan-lyssavirus vaccine? Vaccine 2012, 30, 7447–7454. [Google Scholar] [CrossRef]
- Jallet, C.; Jacob, Y.; Bahloul, C.; Drings, A.; Desmezieres, E.; Tordo, N.; Perrin, P. Chimeric lyssavirus glycoproteins with increased immunological potential. J. Virol. 1999, 73, 225–233. [Google Scholar] [CrossRef]
- Kgaladi, J.; Faber, M.; Dietzschold, B.; Nel, L.; Markotter, W. Pathogenicity and immunogenicity of recombinant rabies viruses expressing the Lagos bat virus matrix and glycoprotein: Perspectives for a pan-lyssavirus vaccine. Trop. Med. Infect. Dis. 2017, 2, 37. [Google Scholar] [CrossRef]
- Nel, L.; Niezgoda, M.; Hanlon, C.; Morril, P.; Yager, P.; Rupprecht, C. A comparison of DNA vaccines for the rabies-related virus, Mokola. Vaccine 2003, 21, 2598–2606. [Google Scholar] [CrossRef]
- Olival, K.J. To cull, or not to cull, bat is the question. Ecohealth 2016, 13, 6–8. [Google Scholar] [CrossRef] [PubMed]
- Banyard, A.; Evans, J.; Luo, T.; Fooks, A. Lyssaviruses and bats: Emergence and zoonotic threat. Viruses 2014, 6, 2974–2990. [Google Scholar] [CrossRef]
- Rupprecht, C.; Kuzmin, I.; Meslin, F. Lyssaviruses and rabies: Current conundrums, concerns, contradictions and controversies. F1000Research 2017, 6, 184. [Google Scholar] [CrossRef] [PubMed]
- Streicker, D.G.; Turmelle, A.S.; Vonhof, M.J.; Kuzmin, I.V.; McCracken, G.F.; Rupprecht, C.E. Host Phylogeny Constrains Cross-Species Emergence and Establishment of Rabies Virus in Bats. Science 2010, 329, 676–679. [Google Scholar] [CrossRef]
- Mollentze, N.; Streicker, D.G.; Murcia, P.R.; Hampson, K.; Biek, R. Virulence mismatches in index hosts shape the outcomes of cross-species transmission. Proc. Natl. Acad. Sci. USA 2020, 117, 28859–28866. [Google Scholar] [CrossRef]
| Lyssavirus Species | Phylogroup | Geographical Distribution | Host(s) 1 |
|---|---|---|---|
| Aravan lyssavirus | I | Eurasia | Myotis blythi |
| Australian bat lyssavirus | I | Australasia | Pteropus alecto Saccolaimus flaviventris |
| Bokeloh bat lyssavirus | I | Europe | Myotis nattereri |
| Duvenhage lyssavirus | I | Africa | Nycteris thebaica |
| European bat 1 lyssavirus | I | Europe | Eptesicus serotinus |
| European bat 2 lyssavirus | I | Europe | Myotis daubentonii |
| Gannoruwa bat lyssavirus | I | Asia | Pteropus medius |
| Ikoma lyssavirus | Africa | Civettictis civetta2 | |
| Irkut lyssavirus | I | Eurasia | Murina leucogaster |
| Khujand lyssavirus | I | Eurasia | Myotis mystacinus |
| Kotalathi bat lyssavirus3 | I | Europe | Myotis brandtii |
| Lagos bat lyssavirus | II | Africa | Eidolon helvum Rousettus aegyptiacus Epomophorus wahlbergi |
| Lleida bat lyssavirus | Europe | Miniopterus schreibersii | |
| Matlo bat lyssavirus4 | Africa | Miniopterus natalensis | |
| Mokola lyssavirus | II | Africa | Felis catus2 |
| Rabies lyssavirus | I | Almost worldwide | Most mammalian species |
| Shimoni bat lyssavirus | II | Africa | Macronycteris vittatus |
| Taiwan bat lyssavirus | I | Asia | Pipistrellus abramus |
| West Caucasian bat lyssavirus | Eurasia | Miniopterus schreibersii |
| Host Family | Genera | Species 1 | Genera (Tested Species 2) | Tested Individuals | Positive Individuals | Virus |
|---|---|---|---|---|---|---|
| PTEROPODIDAE | 13 | 44 | 5 (8) | 2350 | 4 | Lagos bat virus |
| HIPPOSIDERIDAE | 4 | 21 | 2 (3) | 62 | 1 | Shimoni bat virus |
| MEGADERMATIDAE | 2 | 2 | 2 (2) | 29 | 0 | |
| RHINOLOPHIDAE | 1 | 38 | 1 (11) | 321 | 0 | |
| RHINONYCTERIDAE | 3 | 6 | 2 (2) | 21 | 0 | |
| RHINOPOMATIDAE | 1 | 3 | 1 (1) | 4 | 0 | |
| MYZOPODIDAE | 1 | 2 | 0 (0) | 0 | 0 | |
| EMBALLONURIDAE | 4 | 12 | 2 (3) | 44 | 0 | |
| NYCTERIDAE | 1 | 15 | 1 (3) | 69 | 1 | Duvenhage virus |
| MOLOSSIDAE | 7 | 44 | 4 (7) | 333 | 0 | |
| CISTUGONIDAE | 1 | 2 | 0 (0) | 0 | 0 | |
| MINIOPTERIDAE | 1 | 26 | 1 (5) | 555 | 2 | Matlo bat lyssavirus |
| VESPERTILIONIDAE | 19 | 124 | 11 (19) | 303 | 0 | |
| Not determined | - | - | - | 1193 | 0 | |
| TOTALS | 58 | 339 | 32 (64) | 5284 | 8 | 0.15% |
| Host Family | Genera | Species 1 | Genera (Tested Species 2) | Tested Individuals | Positive Individuals | Percentage Positive |
|---|---|---|---|---|---|---|
| PTEROPODIDAE | 13 | 44 | 8 (14) | 3064 | 1048 | 34.2 |
| HIPPOSIDERIDAE | 4 | 21 | 2 (3) | 148 | 5 | 3.4 |
| MEGADERMATIDAE | 2 | 2 | 1 (1) | 3 | 0 | 0 |
| RHINOLOPHIDAE | 1 | 38 | 1 (1) | 44 | 0 | 0 |
| RHINONYCTERIDAE | 3 | 6 | 1 (2) | 19 | 2 | 17.8 |
| RHINOPOMATIDAE | 1 | 3 | 0 (0) | 0 | 0 | 0 |
| MYZOPODIDAE | 1 | 2 | 0 (0) | 0 | 0 | 0 |
| EMBALLONURIDAE | 4 | 12 | 1 (1) | 24 | 0 | 0 |
| NYCTERIDAE | 1 | 15 | 1 (1) | 14 | 0 | 0 |
| MOLOSSIDAE | 7 | 44 | 3 (13) | 517 | 20 | 3.9 |
| CISTUGONIDAE | 1 | 2 | 0 (0) | 0 | 0 | 0 |
| MINIOPTERIDAE | 1 | 26 | 1 (5) | 343 | 4 | 1.2 |
| VESPERTILIONIDAE | 19 | 124 | 8 (8) | 120 | 4 | 3.3 |
| TOTALS | 58 | 339 | 27 (49) | 4296 | 1083 | 25.2 |
| Lineage A | Lineage B | Lineage C | Lineage D | ||||
|---|---|---|---|---|---|---|---|
| LN849915 1 | NC020807 2 | EU259198 3 | EU293110 4 | MH643893 5 | GU170202 6 | JX901139 7 | |
| LN849915 | 98.7 | 98.8 | 76.3 | 76 | 76.3 | 76.4 | |
| NC020807 | 99.7 | 98.8 | 76.2 | 76 | 76.2 | 76.4 | |
| EU259198 | 99.6 | 99.6 | 76.2 | 76 | 76.3 | 76.5 | |
| EU293110 | 87.9 | 87.9 | 87.8 | 79.6 | 76.7 | 76.9 | |
| MH643893 | 88.9 | 88.9 | 88.8 | 92.2 | 77.1 | 77.2 | |
| GU170202 | 89.5 | 89.6 | 89.5 | 88.4 | 89.2 | 98.3 | |
| JX901139 | 89.5 | 89.6 | 89.5 | 88.3 | 89.3 | 99.4 | |
| Protein 2 | Region | Ref 3 | Lineage A | Lineage B | Lineage C | Lineage D | |||
|---|---|---|---|---|---|---|---|---|---|
| EU 259198 | NC 020807 | LN 849915 | EU 293110 | MH 643893 | JX 901139 | GU 170202 | |||
| N | 273 | [50] | F | F | F | F | F | F | F |
| 394 | F | F | F | F | F | F | F | ||
| P | 144–148 | [51] | RQTQT | RQTQT | RQTQT | KQTQT | KQTQT | KNTQT | KNTQT |
| M | 22–25 | [52] | ASAP | ASAP | ASAP | PSAP | PSAP | ASAP | ASAP |
| 35–38 | [53] | PPEY | PPEY | PPEY | PPEY | PPEY | PPEY | PPEY | |
| 77 | [54] | K | K | K | K | K | K | K | |
| 81 | N | N | N | N | S | N | N | ||
| 95 | [55] | I | I | I | M | V | I | I | |
| G | 132 | [56] | L | L | L | L | L | L | L |
| 194 | [57] | T | T | T | T | T | T | T | |
| 198 | [56] | R | R | R | K | K | R | R | |
| 242 | [58] | S | S | S | S | S | S | S | |
| 255 | D | D | D | N | N | N | N | ||
| 268 | I | I | I | V | I | L | L | ||
| 318 | [59] | L | L | L | L | I | L | L | |
| 352 | M | M | M | V | V | L | L | ||
| 330–333 | [60,61,62] | KRVD | KRVD | KRVD | LKVD | LRVD | RRVD | RRVD | |
| Host | Route1 | Dose | Isolate | Clinical Signs | Mortality | Reference |
|---|---|---|---|---|---|---|
| Macaca mulatta | IC | 6.2 log ICDL50/mL | 8619NGA (EU293110) | Agitation | 100% (n = 2) | [48] |
| IM | Paresis | 20% (n = 5) | ||||
| Canine | IC | Depression, incoordination | 100% (n = 2) | |||
| IM | No clinical signs | 0% (n = 2) | ||||
| Mice (BALB/3H) | IM | 3 × 105 LD50 | 8619NGA (EU293110) | N/A | 0% | [39] |
| Mice (ICR) | IC | 102 MICLD50 | 0406SEN (EU93108) | Not specified | 100% (n = 5) | [49] |
| IM | 103 MICLD50 | 60% (n = 5) | ||||
| IM | 106 MICLD50 | 100% (n = 5) | ||||
| IC | 102 MICLD50 | Afr1999 (EF547447) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 20% (n = 5) | ||||
| IM | 106 MICLD50 | 100% (n = 5) | ||||
| IC | 102 MICLD50 | Zim1986 (EF547450) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 20% (n = 5) | ||||
| IM | 106 MICLD50 | 20% (n = 5) | ||||
| IM | 102 MICLD50 | CAR1974 (EF547449) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 0% (n = 5) | ||||
| IM | 106 MICLD50 | 20% (n = 5) | ||||
| IC | 102 MICLD50 | Mong2004 (DQ499948) | 100% (n = 5) | |||
| IM | 103 MICDL50 | 20% (n = 5) | ||||
| IM | 106 MICLD50 | 20% (n = 5) | ||||
| IC | 102 MICLD50 | SA2004 (EF547458) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 40% (n = 5) | ||||
| IM | 106 MICLD50 | 60% (n = 5) | ||||
| IC | 102 MICLD50 | SA2003 (EF547421) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 20% (n = 5) | ||||
| IM | 106 MICLD50 | 20% (n = 5) | ||||
| IC | 102 MICLD50 | SA2006 (EF547422) | 100% (n = 5) | |||
| IM | 103 MICLD50 | 0% (n = 5) | ||||
| IM | 106 MICLD50 | 20% (n = 5) | ||||
| Mice (BALB/c) | IM | 103 TCID50 | SA2008 (HM179509) | Hind limb paralysis, ruffled fur, weight loss, walking in circles | 0% (n = 4) | [17] |
| 105 TCID50 | SA2008 (HM179509) | 50% (n = 4) | ||||
| 8619NGA (EU293110) | 0% (n = 4) | |||||
| 107 TCID50 | SA2004 (EF547458) | 60% (n = 5) | ||||
| 108 TCID50 | Mong2004 (DQ499948) | 40% (n = 5) | ||||
| 108 TCID50 | Afr1999 (EF547447) | 80% (n = 5) | ||||
| Eidolon helvum | IC | 103.5 TCID50 | 0406SEN (EU293108) | Hindleg paresis, muscle spasms, hyperaesthesia, foam around mouth, anorexia | 100% (n = 3) | [64] |
| 8619NGA (EU293110) | 100% (n = 3) | |||||
| 31225 (LN849915) | 100% (n = 3) | |||||
| Eidolon helvum | IM | 100.1 TCID50 | 31225 (LN849915) | No clinical signs | 25% (n = 4) | [63] |
| 101.1 TCID50 | Vocalization, muscle spasms, salivation, aggression | 50% (n = 4) | ||||
| 102.1 TCID50 | 100% (n = 4) | |||||
| 103.1 TCID50 | 50% (n = 4) | |||||
| 104.1 TCID50 | 50% (n = 4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coertse, J.; Geldenhuys, M.; le Roux, K.; Markotter, W. Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus. Viruses 2021, 13, 576. https://doi.org/10.3390/v13040576
Coertse J, Geldenhuys M, le Roux K, Markotter W. Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus. Viruses. 2021; 13(4):576. https://doi.org/10.3390/v13040576
Chicago/Turabian StyleCoertse, Jessica, Marike Geldenhuys, Kevin le Roux, and Wanda Markotter. 2021. "Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus" Viruses 13, no. 4: 576. https://doi.org/10.3390/v13040576
APA StyleCoertse, J., Geldenhuys, M., le Roux, K., & Markotter, W. (2021). Lagos Bat Virus, an Under-Reported Rabies-Related Lyssavirus. Viruses, 13(4), 576. https://doi.org/10.3390/v13040576






