Study of a SARS-CoV-2 Outbreak in a Belgian Military Education and Training Center in Maradi, Niger
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples for COVID-19 Diagnosis
2.2. SARS-CoV-2 RT-qPCR
2.3. SARS-CoV-2 Immunoassays
2.4. SARS-CoV-2 Genome Sequencing
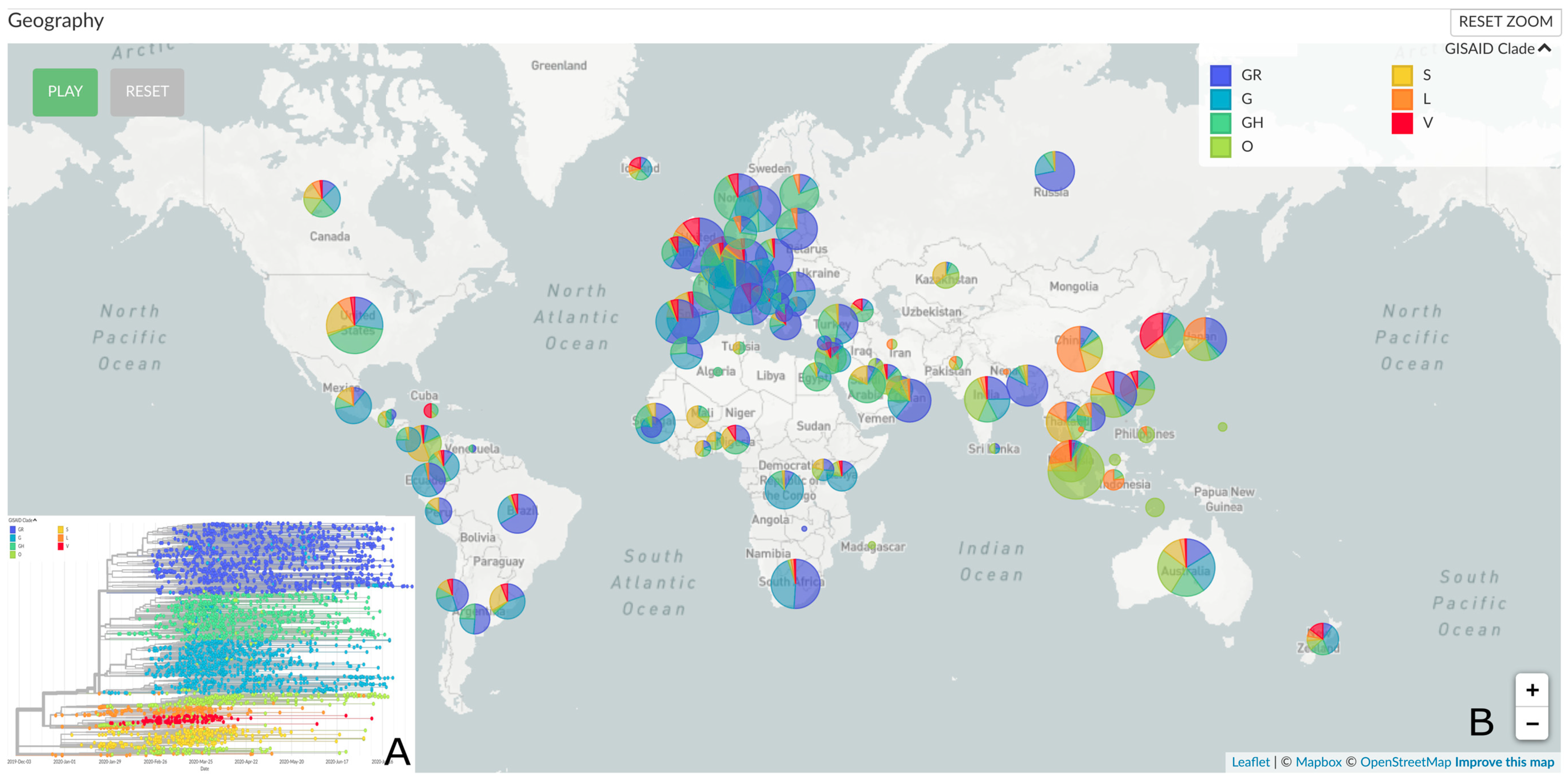
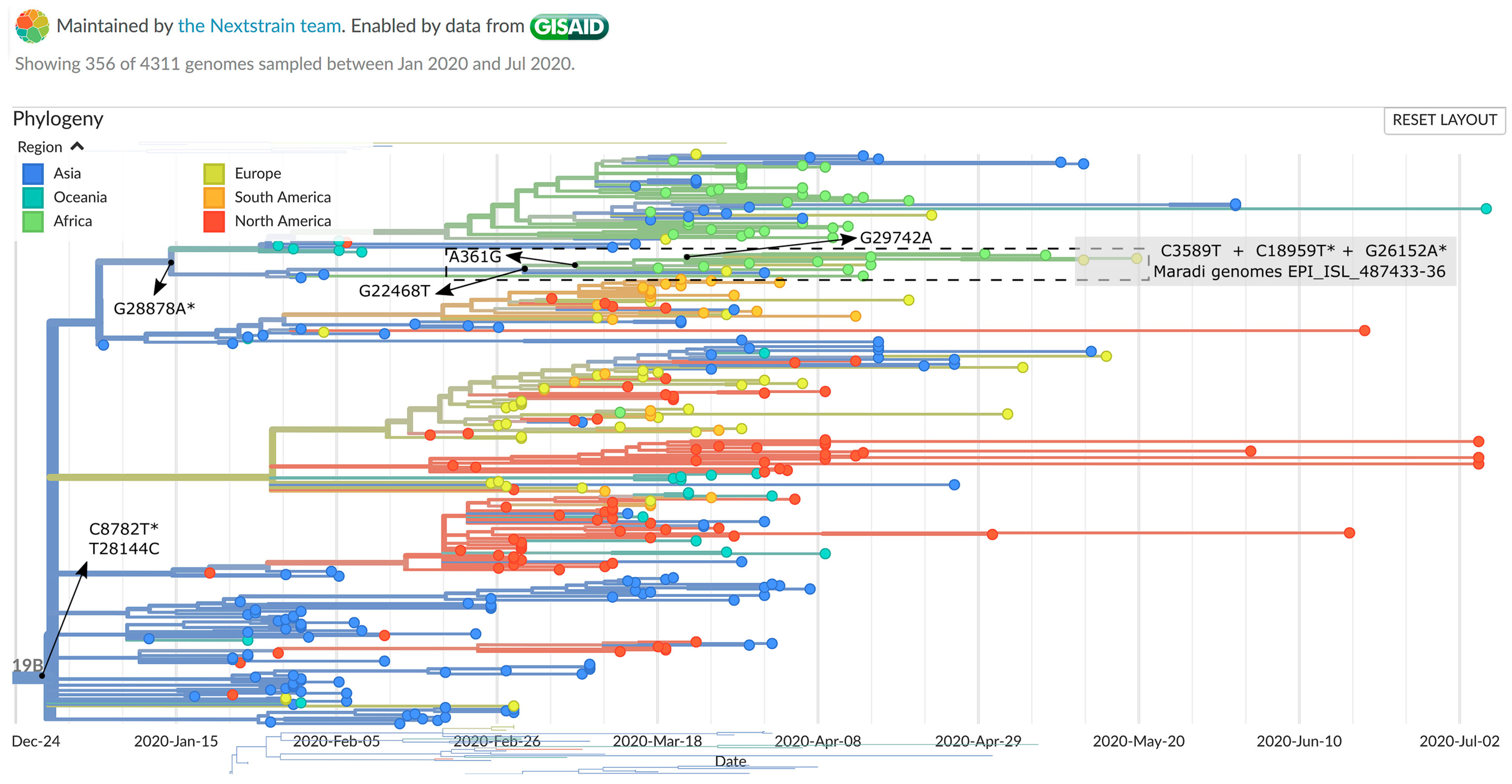
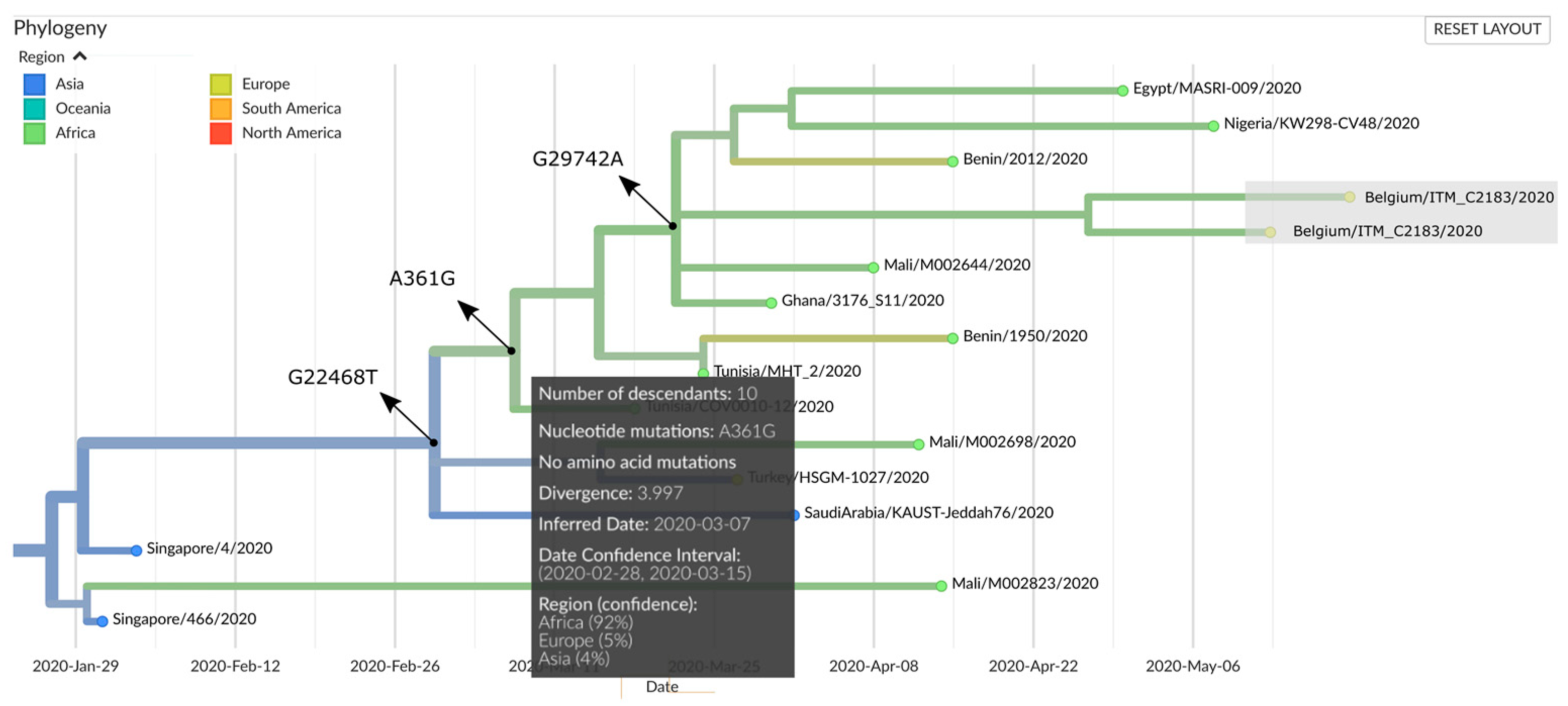
2.5. Genomic Epidemiology
2.6. Conventional Epidemiology
2.7. Ethics Statement
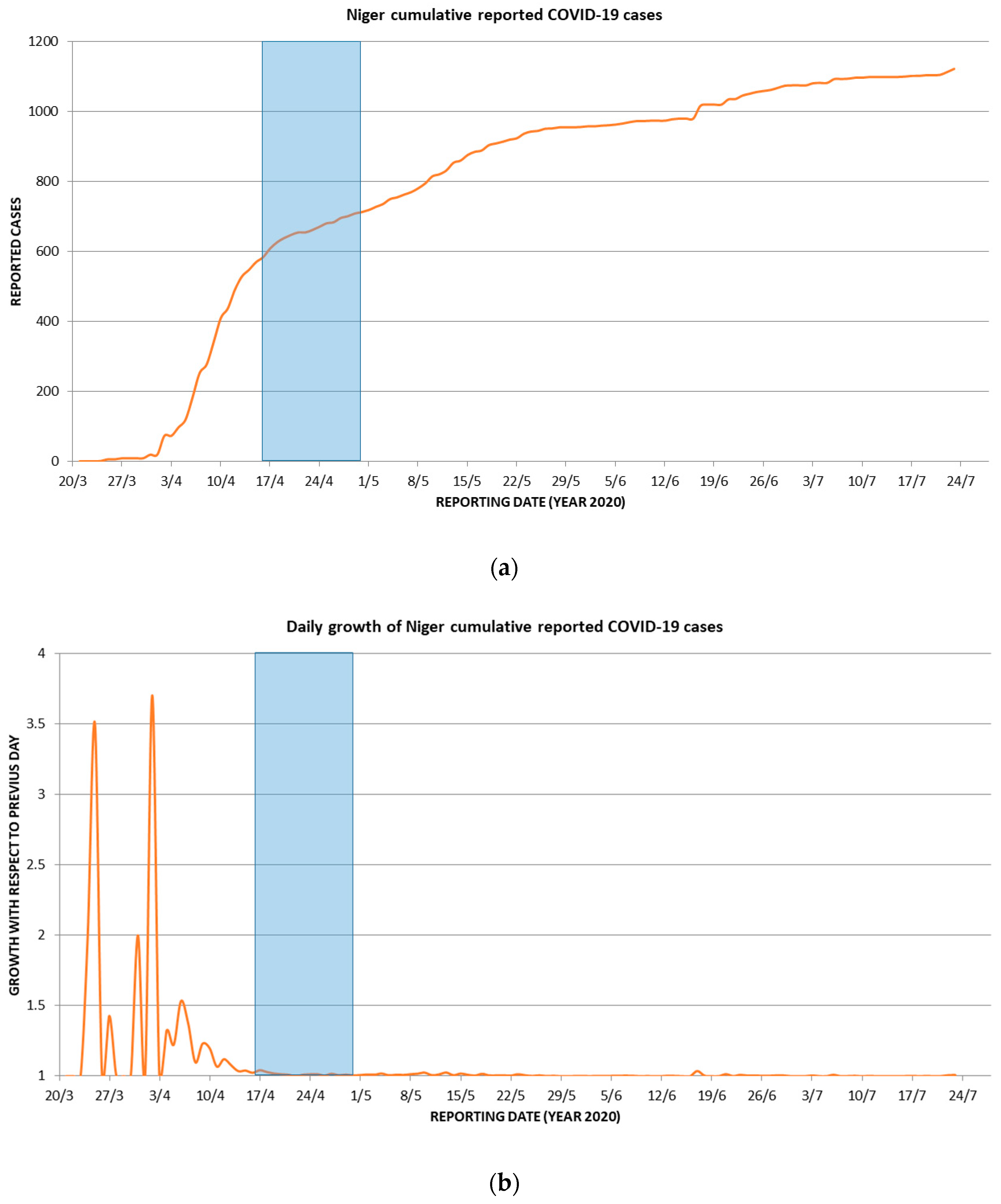
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Schlein, L. UNHCR: Attacks in NW Nigeria Send Thousands Fleeing to Niger. News 24. 27 September 2019. Available online: https://www.voanews.com/africa/unhcr-attacks-nw-nigeria-send-thousands-fleeing-niger (accessed on 6 August 2020).
- Weemaes, M.; Martens, S.; Cuypers, L.; van Elslande, J.; Hoet, K.; Welkenhuysen, J.; Goossens, R.; Wouters, S.; Houben, E.; Jeuris, K.; et al. Laboratory information system requirements to manage the COVID-19 pandemic: A report from the Belgian national reference testing center. J. Am. Med. Inform. Assoc. 2020. [Google Scholar] [CrossRef] [PubMed]
- COVID 19: Un huitième cas annoncé au Niger. 2020. Available online: http://www.faapa.info/blog/covid-19-un-huitieme-cas-annonce-au-niger/ (accessed on 6 August 2020).
- Segal, D.; Rotschield, J.; Ankory, R.; Kutikov, S.; Moaddi, B.; Verhovsky, G.; Benov, A.; Twig, G.; Glassberg, E.; Fink, N.; et al. Measures to Limit COVID-19 Outbreak Effects Among Military Personnel: Preliminary Data. Mil. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Quick, J.; Grubaugh, N.D.; Pullan, S.T.; Claro, I.M.; Smith, A.D.; Gangavarapu, K.; Oliveira, G.; Robles-Sikisaka, R.; Rogers, T.F.; Beutler, N.A.; et al. Multiplex PCR method for MinION and Illumina sequencing of zika and other virus genomes directly from clinical samples. Nat. Protoc. 2017, 12, 1261–1266. [Google Scholar] [CrossRef] [PubMed]
- Nanopore Pipeline Wrapper. Available online: https://github.com/ColinAnthony/nanopore_pipeline_wrapper (accessed on 11 June 2020).
- nCoV-2019 Novel Coronavirus Bioinformatics Protocol. Available online: https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html (accessed on 11 June 2020).
- Nanopolish. Available online: https://github.com/jts/nanopolish (accessed on 11 June 2020).
- Elbe, S.; Buckland-Merrett, G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017, 1, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [PubMed]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [PubMed]
- Nextstrain/ncov. Available online: https://github.com/nextstrain/ncov (accessed on 11 August 2020).
- Long, Q.X.; Liu, B.Z.; Deng, H.J.; Wu, G.C.; Deng, K.; Chen, Y.K.; Liao, P.; Qiu, J.F.; Lin, Y.; Cai, X.F.; et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, L.; Kou, G.; Zheng, Y.; Ding, Y.; Ni, W.; Wang, Q.; Tan, L.; Wu, W.; Tang, S.; et al. Evaluation of Nucleocapsid and Spike Protein-Based Enzyme-Linked Immunosorbent Assays for Detecting Antibodies against SARS-CoV-2. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.C.; Smith-Jeffcoat, S.E.; Nowak, G.; Chukwuma, U.; Geibe, J.R.; Hawkins, R.J.; Johnson, J.A.; Thornburg, N.J.; Schiffer, J.; Weiner, Z.; et al. SARS-CoV-2 Infections and Serologic Responses from a Sample of U.S. Navy Service Members—USS Theodore Roosevelt, April 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 714–721. [Google Scholar] [CrossRef]
- Reinhard, A.; Ikonomidis, C.; Broome, M.; Gorostidi, F. Anosmia and COVID-19. Rev. Med. Suisse 2020, 16, 849–851. [Google Scholar]
- Rockett, R.J.; Arnott, A.; Lam, C.; Sadsad, R.; Timms, V.; Gray, K.A.; Eden, J.S.; Chang, S.; Gall, M.; Draper, J.; et al. Revealing COVID-19 transmission in Australia by SARS-CoV-2 genome sequencing and agent-based modeling. Nat. Med. 2020. [Google Scholar] [CrossRef] [PubMed]
- Mercatelli, D.; Giorgi, F.M. Geographic and Genomic Distribution of SARS-CoV-2 Mutations. Front. Microbiol. 2020, 11, 1800. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Platt, D.; Parida, L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020, 98, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Coppée, F.; Lechien, J.R.; Declèves, A.E.; Tafforeau, L.; Saussez, S. Severe acute respiratory syndrome coronavirus 2: Virus mutations in specific European populations. New Microbes New Infect. 2020, 36, 100696. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control (ECDC). Q & A on Novel Coronavirus. Available online: https://www.ecdc.europa.eu/en/covid-19/questions-answers (accessed on 10 August 2020).
| Symptoms | COVID-19 Diagnosis | |
|---|---|---|
| At Least One Positive Test (n = 5) | No Positive Tests (n = 17) | |
| Anosmia/ageusia | 4 | 0 |
| Diarrhea | 1 | 6 |
| Sore throat/blocked nose | 2 | 8 |
| Cough | 0 | 4 |
| Myalgia | 2 | 5 |
| Post exercise fatigue | 2 | 5 |
| Fever | 3 | 1 |
| Headache | 2 | 6 |
| ID | Demographics | Symptomatology | COVID-19 Diagnostic Test Results | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gender (M/F) | Age (Years) | Function | Stay in the Maradi Training Center | Symptoms (Y/N) | Date Onset of Symptoms | Symptoms 1 | Viral Load (Ct) | Antibodies | |||||||||
| D0 | D14 | D0 | D14 | ||||||||||||||
| N gene | ORF 1ab | N gene | ORF 1ab | IgG 2 | IgM 2 | Elecsys 3 (Signal) | IgG 2 | IgM 2 | Elecsys 3 (Signal) | ||||||||
| M1 | M | 32 | Trainer | Feb 1–May 19 | Y | 30 April | AA, D, F | Neg | Neg | Neg | Neg | POS | Neg | Neg (0.405) | POS | Neg | POS (5.25) |
| M2 | M | 23 | Trainer | Feb 1–May 19 | Y | 3 May | STBN, M, PEF, F | Neg | Neg | Neg | Neg | Neg | POS | Neg (0.135) | POS | Neg | POS (3.42) |
| M3 | M | 26 | Trainer | Feb 1–May 19 | Y | 6 May | AA, M, F, H | Neg | Neg | Neg | Neg | POS | POS | Neg (0.271) | POS | Neg | POS (2.67) |
| M4 | M | 29 | Trainer | Feb 1–May 19 | Y | 15 May | AA, STBN, PEF, H | POS (28.9) | POS (29.8) | Neg | Neg | Neg | Neg | Neg (0.091) | POS | Neg | POS (2.33) |
| M5 | M | 28 | JSD | Feb 1–May 19 | Y | 16 May | AA | POS (24.5) | POS (25.0) | POS (36.9) | Neg | Neg | Neg | Neg (0.94) | POS | POS | POS (1.57) |
| M6 | M | 45 | JSD | Dec 12–May 13 | N | NA | NA | POS (26.1) | POS (26.9) | POS (36.6) | POS (36.8) | Neg | Neg | Neg (0.089) | POS | POS | POS (8.14) |
| M7 | M | 23 | Trainer | Feb 1–May 19 | N | NA | NA | POS (34.1) | POS (34.5) | POS (35.6) | POS (36.2) | Neg | Neg | Neg (0.106) | POS | Neg | POS (1.23) |
| M8 | M | 44 | Medic | Feb 1–May 19 | N | NA | NA | Neg | Neg | Neg | Neg | POS | POS | Neg (0.09) | POS | Neg | Neg (0.086) |
| M9 | M | 25 | FP | Dec 12–May 13 | N | NA | NA | Neg | Neg | Neg | Neg | POS | Neg | Neg (0.087) | Neg | Neg | Neg (0.086) |
| Genome Name | Patient ID | Nucleotide Variations (Positions) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 361 | 3589 | 8095 | 8782 | 18,959 | 19,164 | 20,477 | 22,468 | 26,152 | 28,144 | 28,878 | 29,742 | ||
| Reference (NC_045512.2) | A | C | A | C | C | C | G | G | G | T | G | G | |
| ITM C2181 (EPI_ISL_487433) | M4 | G | T | A | T | T | C | G | T | A | C | A | A |
| ITM C2185 (EPI_ISL_487436) | M7 | G | T | A | T | T | C | G | T | A | C | A | A |
| ITM C2183 (EPI_ISL_487435) | M6 | G | T | A | T | T | C | T | T | A | C | A | A |
| ITM C2182 (EPI_ISL_487434) | M5 | G | T | C | T | T | T | G | T | A | C | A | A |
| Amino Acid (AA) Variations | |||||||||||||
| SARS-CoV-2 Involved Genes | ORF1ab | S (Spike) | ORF3a | ORF8 | N | 3′UTR | |||||||
| Reference (NC_045512.2) | A | G | G | L | S | ||||||||
| ITM C2181 (EPI_ISL_487433) | M4 | Silent | Silent | Silent | V | Silent | R | S | N | Untranslated | |||
| ITM C2185 (EPI_ISL_487436) | M7 | Silent | Silent | Silent | V | Silent | R | S | N | ||||
| ITM C2183 (EPI_ISL_487435) | M6 | Silent | Silent | Silent | V | V | Silent | R | S | N | |||
| ITM C2182 (EPI_ISL_487434) | M5 | Silent | Silent | Silent | Silent | V | Silent | Silent | R | S | N | ||
| Weight of AA Variations (BLOSUM62 Matrix) | |||||||||||||
| AA Variation | A/V | G/V | G/R | L/S | S/N | ||||||||
| Weight | 0 | −3 | −2 | −2 | 1 | ||||||||
| Clade signature mutations | S | S | |||||||||||
| Variation | Genome Name | GISAID ID | Collection Date |
|---|---|---|---|
| A361G, shared by all four genomes | hCoV-19/USA/VA_6171/2020 | EPI_ISL_424907 | 06/03/2020 |
| hCoV-19/USA/VA-DCLS-0021/2020 | EPI_ISL_419713 | 11/03/2020 | |
| hCoV-19/Mali/M002644/2020 | EPI_ISL_487450 | 08/04/2020 | |
| hCoV-19/Benin/2012/2020 | EPI_ISL_476831 | 15/04/2020 | |
| hCoV-19/Egypt/MASRI-009/2020 | EPI_ISL_483035 | 30/04/2020 | |
| hCoV-19/Egypt/MASRI-3/2020 | EPI_ISL_475746 | May 2020 | |
| hCoV-19/Nigeria/KW298-CV48/2020 | EPI_ISL_487107 | 08/05/2020 | |
| C3589T, shared by all four genomes | SARS-CoV-2/human/USA/VA-DCLS-0543/2020 | EPI_ISL_485844 | 12/03/2020 |
| SARS-CoV-2/human/USA/VA-DCLS-0002/2020 | EPI_ISL_418956 | 17/03/2020 | |
| SARS-CoV-2/human/USA/VA-DCLS-0003/2020 | EPI_ISL_418957 | 17/03/2020 | |
| SARS-CoV-2/human/USA/VA-DCLS-0004/2020 | EPI_ISL_418958 | 14/03/2020 | |
| SARS-CoV-2/human/USA/UNC_200323/2020 | Not applicable | April 2020 | |
| SARS-CoV-2/human/USA/UNC_200300/2020 | Not applicable | April 2020 | |
| G20477T, specific for genome C2183 | hCoV-19/France/10063BI/2020 | EPI_ISL_447679 | March 2020 |
| C19164T, specific for genome C2182 | hCoV-19/USA/WI-GMF-M00002/2020 | EPI_ISL_455574 | 17/05/2020 |
| hCoV-19/USA/WI-UW-442/2020 | EPI_ISL_484817 | 14/06/2020 |
| Time Period | Externs | Type of Contact |
|---|---|---|
| Arrival between 17 and 27 December 2019 | IQARUS Damage Control Surgery Team, consisting of medics from Bulgaria (n = 2), France (n = 1), Germany (n = 1) and South Africa (n = 1) | The team stayed at the Maradi training center and shared most of the camp’s facilities with the Belgian military |
| 4 to 5 February 2020 | Visit of two Belgian senior officers | The officers stayed at the Maradi training center and shared most of the camp’s facilities with the Belgian military |
| Arrival between 17 February and 10 March 2020 | IQARUS Damage Control Surgery Team, consisting of medics from South Africa (n = 2), UK (n = 2), and France (n = 1) | The team stayed at the Maradi training center and shared most of the camp’s facilities with the Belgian military |
| 27 February 2020 | Visit of USA military stationed in Niamey, Niger | Contact with Belgian medics |
| 10 to 11 March | Visit of two Belgian senior officers | The officers stayed at the Maradi training center and shared most of the camp’s facilities with the Belgian military |
| During the entire stay | Nigerien military trainees | Frequent contacts with several Belgian military service men, especially with the military trainers |
| During the entire stay | Nigerien civilians | Several Belgian military service men had sporadic contacts with Nigerien civilians during visits to the city of Maradi |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pirnay, J.-P.; Selhorst, P.; Cochez, C.; Petrillo, M.; Claes, V.; Van der Beken, Y.; Verbeken, G.; Degueldre, J.; T’Sas, F.; Van den Eede, G.; et al. Study of a SARS-CoV-2 Outbreak in a Belgian Military Education and Training Center in Maradi, Niger. Viruses 2020, 12, 949. https://doi.org/10.3390/v12090949
Pirnay J-P, Selhorst P, Cochez C, Petrillo M, Claes V, Van der Beken Y, Verbeken G, Degueldre J, T’Sas F, Van den Eede G, et al. Study of a SARS-CoV-2 Outbreak in a Belgian Military Education and Training Center in Maradi, Niger. Viruses. 2020; 12(9):949. https://doi.org/10.3390/v12090949
Chicago/Turabian StylePirnay, Jean-Paul, Philippe Selhorst, Christel Cochez, Mauro Petrillo, Vincent Claes, Yolien Van der Beken, Gilbert Verbeken, Julie Degueldre, France T’Sas, Guy Van den Eede, and et al. 2020. "Study of a SARS-CoV-2 Outbreak in a Belgian Military Education and Training Center in Maradi, Niger" Viruses 12, no. 9: 949. https://doi.org/10.3390/v12090949
APA StylePirnay, J.-P., Selhorst, P., Cochez, C., Petrillo, M., Claes, V., Van der Beken, Y., Verbeken, G., Degueldre, J., T’Sas, F., Van den Eede, G., Weuts, W., Smets, C., Mertens, J., Geeraerts, P., Ariën, K. K., Neirinckx, P., & Soentjens, P. (2020). Study of a SARS-CoV-2 Outbreak in a Belgian Military Education and Training Center in Maradi, Niger. Viruses, 12(9), 949. https://doi.org/10.3390/v12090949






