Strength in Diversity: Nuclear Export of Viral RNAs
Abstract
1. Introduction
Nucleocytoplasmic Transport of Macromolecules
2. Nuclear Export of Viral mRNAs
2.1. The Success of Retroviruses
2.1.1. HIV-1 Research Paved the Way for Nuclear Export Pathway Discovery
2.1.2. From HIV-1 to the Simple Mason–Pfizer Monkey Virus
2.1.3. Diversity within the Retroviridae Family
2.2. mRNA Nuclear Export of Pararetroviruses
2.3. Nuclear Replicating DNA Viruses
2.3.1. Adenovirus mRNA Nuclear Export
2.3.2. Herpesvirus mRNA Export—A Shared Strategy with Different Tools
2.3.3. Small DNA Virus mRNA Export
2.4. The One Exception—The Nuclear Export of Influenza Virus RNAs
2.4.1. Export of Influenza A Virus mRNAs
2.4.2. Export of Influenza A Virus vRNPs
3. Concluding Comments
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Callan, H.G.; Tomlin, S.G. Experimental studies on amphibian oocyte nuclei I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc. R. Soc. Lond. Ser. B Biol. Sci. 1950, 137, 367–378. [Google Scholar] [CrossRef]
- Gall, J. Observations on the nuclear membrane with the electron microscope. Exp. Cell Res. 1954, 7, 197–200. [Google Scholar] [CrossRef]
- Lin, D.H.; Hoelz, A. The Structure of the Nuclear Pore Complex (An Update). Annu. Rev. Biochem. 2019, 88, 725–783. [Google Scholar] [CrossRef] [PubMed]
- Edwards, R.A.; Rohwer, F. Viral metagenomics. Nat. Rev. Genet. 2005, 3, 504–510. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M. Origins and evolution of viruses of eukaryotes: The ultimate modularity. Virology 2015, 479, 2–25. [Google Scholar] [CrossRef] [PubMed]
- Kobiler, O.; Drayman, N.; Butin-Israeli, V.; Oppenheim, A. Virus strategies for passing the nuclear envelope barrier. Nucleus 2012, 3, 526–539. [Google Scholar] [CrossRef]
- Fay, N.; Panté, N. Nuclear entry of DNA viruses. Front. Microbiol. 2015, 6, 467. [Google Scholar] [CrossRef]
- Tessier, T.M.; Dodge, M.J.; Prusinkiewicz, M.A.; Mymryk, J.S. Viral Appropriation: Laying Claim to Host Nuclear Transport Machinery. Cells 2019, 8, 559. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Origin of the cell nucleus, mitosis and sex: Roles of intracellular coevolution. Biol. Direct 2010, 5, 7. [Google Scholar] [CrossRef]
- Ribbeck, K.; Görlich, D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001, 20, 1320–1330. [Google Scholar] [CrossRef]
- Fabergé, A.C. Direct demonstration of eight-fold symmetry in nuclear pores. Zeitschrift für Zellforschung und Mikroskopische Anatomie 1973, 136, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Akey, C.W.; Radermacher, M. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 1993, 122, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Beck, M.; Förster, F.; Ecke, M.; Plitzko, J.M.; Melchior, F.; Gerisch, G.; Baumeister, W.; Medalia, O. Nuclear Pore Complex Structure and Dynamics Revealed by Cryoelectron Tomography. Science 2004, 306, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Elad, N.; Maimon, T.; Frenkiel-Krispin, D.; Lim, R.Y.; Medalia, O. Structural analysis of the nuclear pore complex by integrated approaches. Curr. Opin. Struct. Biol. 2009, 19, 226–232. [Google Scholar] [CrossRef]
- Goldberg, M.W.; Allen, T.D. High resolution scanning electron microscopy of the nuclear envelope: Demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol. 1992, 119, 1429–1440. [Google Scholar] [CrossRef] [PubMed]
- Jarnik, M.; Aebi, U. Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol. 1991, 107, 291–308. [Google Scholar] [CrossRef]
- Denning, D.P.; Patel, S.S.; Uversky, V.; Fink, A.L.; Rexach, M. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc. Natl. Acad. Sci. USA 2003, 100, 2450–2455. [Google Scholar] [CrossRef]
- Lim, R.Y.; Fahrenkrog, B.; Köser, J.; Schwarz-Herion, K.; Deng, J.; Aebi, U. Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science 2007, 318, 640–643. [Google Scholar] [CrossRef]
- Lim, R.Y.; Köser, J.; Huang, N.-P.; Schwarz-Herion, K.; Aebi, U. Nanomechanical interactions of phenylalanine–glycine nucleoporins studied by single molecule force–volume spectroscopy. J. Struct. Biol. 2007, 159, 277–289. [Google Scholar] [CrossRef]
- Frey, S.; Görlich, D. A Saturated FG-Repeat Hydrogel Can Reproduce the Permeability Properties of Nuclear Pore Complexes. Cell 2007, 130, 512–523. [Google Scholar] [CrossRef]
- Frey, S.; Görlich, D. FG/FxFG as well as GLFG repeats form a selective permeability barrier with self-healing properties. EMBO J. 2009, 28, 2554–2567. [Google Scholar] [CrossRef] [PubMed]
- Paine, P.L.; Moore, L.C.; Horowitz, S.B. Nuclear envelope permeability. Nature 1975, 254, 109–114. [Google Scholar] [CrossRef]
- Bonner, W.M. Protein migration into nuclei. I. Frog oocyte nuclei in vivo accumulate microinjected histones, allow entry to small proteins, and exclude large proteins. J. Cell Biol. 1975, 64, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Keminer, O.; Peters, R. Permeability of Single Nuclear Pores. Biophys. J. 1999, 77, 217–228. [Google Scholar] [CrossRef]
- Milles, S.; Mercadante, D.; Aramburu, I.V.; Jensen, M.R.; Banterle, N.; Koehler, C.; Tyagi, S.; Clarke, J.; Shammas, S.L.; Blackledge, M.; et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell 2015, 163, 734–745. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.K.; Ford, I.J.; Šarić, A.; Hoogenboom, B.W. Intrinsically disordered nuclear pore proteins show ideal-polymer morphologies and dynamics. Phys. Rev. E 2020, 101, 022420. [Google Scholar] [CrossRef]
- Pemberton, L.F.; Paschal, B.M. Mechanisms of Receptor-Mediated Nuclear Import and Nuclear Export. Traffic 2005, 6, 187–198. [Google Scholar] [CrossRef]
- Stewart, M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007, 8, 195–208. [Google Scholar] [CrossRef]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (Crm1p) Is an Essential Nuclear Export Factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef]
- Cokol, M.; Nair, R.; Rost, B. Finding nuclear localization signals. EMBO Rep. 2000, 1, 411–415. [Google Scholar] [CrossRef]
- Kırlı, K.; Karaca, S.; Dehne, H.J.; Samwer, M.; Pan, K.-T.; Lenz, C.; Urlaub, H.; Görlich, D. A deep proteomics perspective on CRM1-mediated nuclear export and nucleocytoplasmic partitioning. Elife 2015, 4, e11466. [Google Scholar] [CrossRef] [PubMed]
- Tinland, B.; Koukolíková-Nicola, Z.; Hall, M.N.; Hohn, B. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl. Acad. Sci. USA 1992, 89, 7442–7446. [Google Scholar] [CrossRef] [PubMed]
- Moede, T.; Leibiger, B.; Pour, H.G.; Berggren, P.-O.; Leibiger, I.B. Identification of a nuclear localization signal, RRMKWKK, in the homeodomain transcription factor PDX-1. FEBS Lett. 1999, 461, 229–234. [Google Scholar] [CrossRef]
- Imamoto, N.; Shimamoto, T.; Kose, S.; Takao, T.; Tachibana, T.; Matsubae, M.; Sekimoto, T.; Shimonishi, Y.; Yoneda, Y.; Naoko, I.; et al. The nuclear pore-targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 1995, 368, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Radu, A.; Blobel, G.; Moore, M.S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl. Acad. Sci. USA 1995, 92, 1769–1773. [Google Scholar] [CrossRef] [PubMed]
- Görlich, D.; Henklein, P.; Laskey, R.A.; Hartmann, E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996, 15, 1810–1817. [Google Scholar] [CrossRef]
- Görlich, D.; Kostka, S.; Kraft, R.; Dingwall, C.; Laskey, R.A.; Hartmann, E.; Prehn, S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr. Biol. 1995, 5, 383–392. [Google Scholar] [CrossRef]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997, 390, 308–311. [Google Scholar] [CrossRef]
- Arnaoutov, A.; Azuma, Y.; Ribbeck, K.; Joseph, J.; Boyarchuk, Y.; Karpova, T.S.; McNally, J.; Dasso, M. Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 2005, 7, 626–632. [Google Scholar] [CrossRef]
- Bischoff, F.; Görlich, D. RanBP1 is crucial for the release of RanGTP from importin β-related nuclear transport factors. FEBS Lett. 1997, 419, 249–254. [Google Scholar] [CrossRef]
- Kim, Y.-H.; Han, M.-E.; Oh, S.-O. The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 2017, 50, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Björk, P.; Wieslander, L. Integration of mRNP formation and export. Cell Mol. Life Sci. 2017, 74, 2875–2897. [Google Scholar] [CrossRef] [PubMed]
- Clouse, K.N.; Luo, M.-J.; Zhou, Z.; Reed, R. A Ran-independent pathway for export of spliced mRNA. Nature 2001, 3, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Viphakone, N.; Hautbergue, G.M.; Walsh, M.; Chang, C.-T.; Holland, A.; Folco, E.G.; Reed, R.; Wilson, S.A. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012, 3, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sträßer, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodríguez-Navarro, S.; Rondón, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Gatfield, D.; Le Hir, H.; Schmitt, C.; Braun, I.C.; Köcher, T.; Wilm, M.; Izaurralde, E.; Köcher, T. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 2001, 11, 1716–1721. [Google Scholar] [CrossRef]
- Hautbergue, G.M.; Hung, M.-L.; Walsh, M.J.; Snijders, A.P.; Chang, C.-T.; Jones, R.; Ponting, C.P.; Dickman, M.J.; Wilson, S.A. UIF, a New mRNA Export Adaptor that Works Together with REF/ALY, Requires FACT for Recruitment to mRNA. Curr. Biol. 2009, 19, 1918–1924. [Google Scholar] [CrossRef]
- Viphakone, N.; Sudbery, I.; Griffith, L.; Heath, C.G.; Sims, D.; Wilson, H.L. Co-transcriptional Loading of RNA Export Factors Shapes the Human Transcriptome. Mol. Cell 2019, 75, 310–323.e8. [Google Scholar] [CrossRef]
- Ghosh, A.; Lima, C.D. Enzymology of RNA cap synthesis. Wiley Interdiscip. Rev. RNA 2010, 1, 152–172. [Google Scholar] [CrossRef]
- Rozen, F.; Sonenberg, N. Identification of nuclear cap specific proteins in HeLa cells. Nucleic Acids Res. 1987, 15, 6489–6500. [Google Scholar] [CrossRef]
- Cheng, H.; Dufu, K.; Lee, C.-S.; Hsu, J.L.; Dias, A.; Reed, R. Human mRNA Export Machinery Recruited to the 5′ End of mRNA. Cell 2006, 127, 1389–1400. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Barman, P.; Kaja, A.; Ferdoush, J.; Lahudkar, S.; Roy, A.; Bhaumik, S.R. Distinct Functions of the Cap-Binding Complex in Stimulation of Nuclear mRNA Export. Mol. Cell. Biol. 2019, 39, e00540-18. [Google Scholar] [CrossRef] [PubMed]
- McCloskey, A.; Taniguchi, I.; Shinmyozu, K.; Ohno, M. hnRNP C Tetramer Measures RNA Length to Classify RNA Polymerase II Transcripts for Export. Science 2012, 335, 1643–1646. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Rio, D.C. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015, 84, 291–323. [Google Scholar] [CrossRef]
- Huang, Y.; Steitz, J.A. Splicing Factors SRp20 and 9G8 Promote the Nucleocytoplasmic Export of mRNA. Mol. Cell 2001, 7, 899–905. [Google Scholar] [CrossRef]
- Müller-McNicoll, M.; Botti, V.; Domingues, A.M.; Brandl, H.; Schwich, O.D.; Steiner, M.C.; Curk, T.; Poser, I.; Zarnack, K.; Neugebauer, K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016, 30, 553–566. [Google Scholar] [CrossRef]
- Shen, H. UAP56—A key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep. 2009, 42, 185–188. [Google Scholar] [CrossRef]
- Edmonds, M.; Abrams, R. Polynucleotide biosynthesis: Formation of a sequence of adenylate units from adenosine triphosphate by an enzyme from thymus nuclei. J. Biol. Chem. 1960, 235, 1142–1149. [Google Scholar]
- Dantonel, J.-C.; Murthy, K.G.; Manley, J.L.; Tora, L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 1997, 389, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Elkon, R.; Ugalde, A.P.; Agami, R. Alternative cleavage and polyadenylation: Extent, regulation and function. Nat. Rev. Genet. 2013, 14, 496–506. [Google Scholar] [CrossRef]
- Johnson, S.A.; Cubberley, G.; Bentley, D.L. Cotranscriptional Recruitment of the mRNA Export Factor Yra1 by Direct Interaction with the 3′ End Processing Factor Pcf11. Mol. Cell 2009, 33, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Lykke-Andersen, S.; Nasser, T.; Saguez, C.; Bertrand, E.; Jensen, T.H.; Moore, C.L. Assembly of an Export-Competent mRNP Is Needed for Efficient Release of the 3′-End Processing Complex after Polyadenylation—IGMM. Mol. Cell. Biol. 2009, 29, 5327–5338. [Google Scholar] [CrossRef] [PubMed]
- Wahle, E. Poly(A) Tail Length Control Is Caused by Termination of Processive Synthesis. J. Biol. Chem. 1995, 270, 2800–2808. [Google Scholar] [CrossRef]
- Iglesias, N.; Tutucci, E.; Gwizdek, C.; Vinciguerra, P.; Von Dach, E.; Corbett, A.H.; Dargemont, C.; Stutz, F. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010, 24, 1927–1938. [Google Scholar] [CrossRef] [PubMed]
- Hautbergue, G.M.; Hung, M.-L.; Golovanov, A.P.; Lian, L.-Y.; Wilson, S.A. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. USA 2008, 105, 5154–5159. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; Suliman, S.; Ben-Yishay, R.; Yunger, S.; Brody, Y.; Shav-Tal, Y. Dynamics of single mRNP nucleocytoplasmic transport and export through the nuclear pore in living cells. Nat. Cell Biol. 2010, 12, 543–552. [Google Scholar] [CrossRef]
- Dilworth, D.J.; Tackett, A.J.; Rogers, R.S.; Yi, E.C.; Christmas, R.H.; Smith, J.J.; Siegel, A.F.; Chait, B.T.; Wozniak, R.W.; Aitchison, J.D. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J. Cell Biol. 2005, 171, 955–965. [Google Scholar] [CrossRef]
- Ishii, K.; Arib, G.; Lin, C.; Van Houwe, G.; Laemmli, U.K. Chromatin Boundaries in Budding Yeast: The nuclear pore connection. Cell 2002, 109, 551–562. [Google Scholar] [CrossRef]
- Burns, L.T.; Wente, S.R. From Hypothesis to Mechanism: Uncovering Nuclear Pore Complex Links to Gene Expression. Mol. Cell. Biol. 2014, 34, 2114–2120. [Google Scholar] [CrossRef]
- Jani, D.; Valkov, E.; Stewart, M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Res. 2014, 42, 6686–6697. [Google Scholar] [CrossRef]
- Fischer, T.; Strässer, K.; Rácz, A.; Rodríguez-Navarro, S.; Oppizzi, M.; Ihrig, P.; Lechner, J.; Hurt, E. The mRNA export machinery requires the novel Sac3p–Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 2002, 21, 5843–5852. [Google Scholar] [CrossRef] [PubMed]
- Kurshakova, M.M.; Krasnov, A.N.; Kopytova, D.V.; Shidlovskii, Y.V.; Nikolenko, J.V.; Nabirochkina, E.N.; Spehner, D.; Schultz, P.; Tora, L.; Georgieva, S.G. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J. 2007, 26, 4956–4965. [Google Scholar] [CrossRef]
- Jani, D.; Lutz, S.; Marshall, N.J.; Fischer, T.; Köhler, A.; Ellisdon, A.M.; Hurt, E.; Stewart, M. Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform that Promotes Nuclear Pore Association and mRNA Export. Mol. Cell 2009, 33, 727–737. [Google Scholar] [CrossRef]
- Wickramasinghe, V.O.; McMurtrie, P.I.; Mills, A.D.; Takei, Y.; Penrhyn-Lowe, S.; Amagase, Y.; Main, S.; Marr, J.; Stewart, M.; Laskey, R.A. mRNA Export from Mammalian Cell Nuclei Is Dependent on GANP. Curr. Biol. 2010, 20, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Alcázar-Román, A.R.; Tran, E.J.; Guo, S.; Wente, S.R. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat. Cell Biol. 2006, 8, 711–716. [Google Scholar] [CrossRef]
- Lin, D.H.; Correia, A.R.; Cai, S.W.; Huber, F.M.; Jette, C.A.; Hoelz, A. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nat. Commun. 2018, 9, 2319. [Google Scholar] [CrossRef] [PubMed]
- Folkmann, A.W.; Noble, K.N.; Cole, C.N.; Wente, S.R. Dbp5, Gle1-IP6 and Nup. Nucleus 2011, 2, 540–548. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yang, Y.; Sun, B.-F.; Chen, Y.-S.; Xu, J.-W.; Lai, W.-Y.; Li, A.; Wang, X.; Bhattarai, D.P.; Xiao, W.; et al. 5-methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017, 27, 606–625. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.-H.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Lesbirel, S.; Wilson, S.A. The m6A-methylase complex and mRNA export. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 319–328. [Google Scholar] [CrossRef]
- Zuckerman, B.; Ron, M.; Mikl, M.; Segal, E.; Ulitsky, I. Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol. Cell 2020, 79, 251–267.e6. [Google Scholar] [CrossRef] [PubMed]
- Palazzo, A.F.; Springer, M.; Shibata, Y.; Lee, C.-S.; Dias, A.P.; Rapoport, T.A. The signal sequence coding region promotes nuclear export of mRNA. PLoS Biol. 2007, 5, e322. [Google Scholar] [CrossRef] [PubMed]
- Akef, A.; Zhang, H.; Masuda, S.; Palazzo, A.F. Trafficking of mRNAs containing ALREX-promoting elements through nuclear speckles. Nucleus 2013, 4, 326–340. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.; Dias, A.P.; Reed, R. Export and stability of naturally intronless mRNAs require specific coding region sequences and the TREX mRNA export complex. Proc. Natl. Acad. Sci. USA 2011, 108, 17985–17990. [Google Scholar] [CrossRef]
- Hutten, S.; Kehlenbach, R.H. CRM1-mediated nuclear export: To the pore and beyond. Trends Cell Biol. 2007, 17, 193–201. [Google Scholar] [CrossRef]
- Delaleau, M.; Borden, K.L. Multiple Export Mechanisms for mRNAs. Cells 2015, 4, 452–473. [Google Scholar] [CrossRef]
- Brennan, C.M.; Gallouzi, I.-E.; Steitz, J.A. Protein Ligands to Hur Modulate Its Interaction with Target Mrnas in Vivo. J. Cell Biol. 2000, 151, 1–14. [Google Scholar] [CrossRef]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef]
- Gallouzi, I.-E.; Steitz, J.A. Delineation of mRNA Export Pathways by the Use of Cell-Permeable Peptides. Science 2001, 294, 1895–1901. [Google Scholar] [CrossRef]
- Fries, B.; Heukeshoven, J.; Hauber, I.; Grüttner, C.; Stocking, C.; Kehlenbach, R.H.; Hauber, J.; Chemnitz, J. Analysis of Nucleocytoplasmic Trafficking of the HuR Ligand APRIL and Its Influence on CD83 Expression. J. Biol. Chem. 2007, 282, 4504–4515. [Google Scholar] [CrossRef]
- Kimura, T.; Fujisawa, J.-I.; Hashimoto, I.; Nagase, T. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-alpha1 mRNA. J. Cell Sci. 2004, 117, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Culjkovic, B.; Topisirovic, I.; Skrabanek, L.; Ruiz-Gutierrez, M.; Borden, K.L. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 2006, 175, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Rong, L.; Livingstone, M.; Sukarieh, R.; Petroulakis, E.; Gingras, A.-C.; Crosby, K.; Smith, B.; Polakiewicz, R.D.; Pelletier, J.; Ferraiuolo, M.A.; et al. Control of eIF4E cellular localization by eIF4E-binding proteins, 4E-BPs. RNA 2008, 14, 1318–1327. [Google Scholar] [CrossRef] [PubMed]
- Topisirovic, I.; Siddiqui, N.; Lapointe, V.L.; Trost, M.; Thibault, P.; Bangeranye, C.; Piñol-Roma, S.; Borden, K.L. Molecular dissection of the eukaryotic initiation factor 4E (eIF4E) export-competent RNP. EMBO J. 2009, 28, 1087–1098. [Google Scholar] [CrossRef]
- Volpon, L.; Culjkovic-Kraljacic, B.; Sohn, H.S.; Blanchet-Cohen, A.; Osborne, M.J.; Borden, K.L. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA 2017, 23, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Bresson, S.; Tollervey, D. Surveillance-ready transcription: Nuclear RNA decay as a default fate. Open Biol. 2018, 8, 170270. [Google Scholar] [CrossRef]
- Toro-Ascuy, D.; Rojas-Araya, B.; Valiente-Echeverría, F.; Rifo, R.S. Interactions between the HIV-1 Unspliced mRNA and Host mRNA Decay Machineries. Viruses 2016, 8, 320. [Google Scholar] [CrossRef]
- Balagopal, V.; Beemon, K.L. Rous Sarcoma Virus RNA Stability Element Inhibits Deadenylation of mRNAs with Long 3′UTRs. Viruses 2017, 9, 204. [Google Scholar] [CrossRef]
- Balistreri, G.; Bognanni, C.; Mühlemann, O. Virus Escape and Manipulation of Cellular Nonsense-Mediated mRNA Decay. Viruses 2017, 9, 24. [Google Scholar] [CrossRef]
- Temin, H.M. Nature of the provirus of Rous sarcoma. Nat. Cancer Inst. Monogr. 1964, 17, 557–570. [Google Scholar]
- Baltimore, D. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of RNA Tumour Viruses. Nature 1970, 226, 1209–1211. [Google Scholar] [CrossRef] [PubMed]
- Temin, H.M.; Mizutani, S. Viral RNA-dependent DNA Polymerase: RNA-dependent DNA Polymerase in Virions of Rous Sarcoma Virus. Nature 1970, 226, 1211–1213. [Google Scholar] [CrossRef] [PubMed]
- Brahic, M.; Stowring, L.; Ventura, P.; Haase, A.T. Gene expression in visna virus infection in sheep. Nature 1981, 292, 240–242. [Google Scholar] [CrossRef] [PubMed]
- Folks, T.; Powell, D.; Lightfoote, M.; Benn, S.; Martin, M.; Fauci, A. Induction of HTLV-III/LAV from a nonvirus-producing T-cell line: Implications for latency. Science 1986, 231, 600–602. [Google Scholar] [CrossRef]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. Genetic Organization; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; ISBN 978-0-87969-497-5. [Google Scholar]
- Swanstrom, R.; Wills, J.W. Synthesis, Assembly, and Processing of Viral Proteins. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; ISBN 978-0-87969-571-2. [Google Scholar]
- Rabson, A.B.; Graves, B.J. Synthesis and Processing of Viral RNA. In Retroviruses; Coffin, J.M., Hughes, S.H., Varmus, H.E., Eds.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; ISBN 978-0-87969-571-2. [Google Scholar]
- Coffin, J.M.; Hughes, S.H.; Varmus, H.E. Retroviral “Lifestyles”: Simple Versus Complex; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1997; ISBN 978-0-87969-497-5. [Google Scholar]
- Barré-Sinoussi, F.; Chermann, J.; Rey, F.; Nugeyre, M.; Chamaret, S.; Gruest, J.; Dauguet, C.; Axler-Blin, C.; Vezinet-Brun, F.; Rouzioux, C.; et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 1983, 220, 868–871. [Google Scholar] [CrossRef]
- Vilmer, E.; Barré-Sinoussi, F.; Rouzioux, C.; Gazengel, C.; Brun, F.V.; Dauguet, C.; Fischer, A.; Manigne, P.; Chermann, J.; Griscelli, C.; et al. Isolation of new lymphotropic retrovirus from two siblings with haemophilia B, one with AIDS. Lancet 1984, 323, 753–757. [Google Scholar] [CrossRef]
- Vahlne, A. A historical reflection on the discovery of human retroviruses. Retrovirology 2009, 6, 40. [Google Scholar] [CrossRef]
- Lai, C.J.; Khoury, G. Deletion mutants of simian virus 40 defective in biosynthesis of late viral mRNA. Proc. Natl. Acad. Sci. USA 1979, 76, 71–75. [Google Scholar] [CrossRef]
- Valencia, P.; Dias, A.P.; Reed, R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl. Acad. Sci. USA 2008, 105, 3386–3391. [Google Scholar] [CrossRef]
- Ocwieja, K.E.; Sherrill-Mix, S.; Mukherjee, R.; Custers-Allen, R.; David, P.; Brown, M.; Wang, S.; Link, D.R.; Olson, J.; Travers, K.; et al. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res. 2012, 40, 10345–10355. [Google Scholar] [CrossRef]
- Mahiet, C.; Swanson, C.M. Control of HIV-1 gene expression by SR proteins. Biochem. Soc. Trans. 2016, 44, 1417–1425. [Google Scholar] [CrossRef] [PubMed]
- Hadzopoulou-Cladaras, M.; Felber, B.K.; Cladaras, C.; Athanassopoulos, A.; Tse, A.; Pavlakis, G.N. The rev (trs/art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the env region. J. Virol. 1989, 63, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Felber, B.K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G.N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.-Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 1989, 342, 816–819. [Google Scholar] [CrossRef]
- Kjems, J.; Brown, M.; Chang, D.D.; Sharp, P.A. Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl. Acad. Sci. USA 1991, 88, 683–687. [Google Scholar] [CrossRef]
- Wen, W.; Meinkotht, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- Fritz, C.C.; Zapp, M.L.; Green, M.R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 1995, 376, 530–533. [Google Scholar] [CrossRef]
- Dicks, M.D.J.; Betancor, G.; Jimenez-Guardeño, J.M.; Pessel-Vivares, L.; Apolonia, L.; Goujon, C.; Malim, M.H. Multiple components of the nuclear pore complex interact with the amino-terminus of MX2 to facilitate HIV-1 restriction. PLoS Pathog. 2018, 14, e1007408. [Google Scholar] [CrossRef]
- Fritz, C.C.; Green, M.R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr. Biol. 1996, 6, 848–854. [Google Scholar] [CrossRef][Green Version]
- Wolff, B.; Sanglier, J.-J.; Wang, Y. Leptomycin B is an inhibitor of nuclear export: Inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 1997, 4, 139–147. [Google Scholar] [CrossRef]
- Nishi, K.; Yoshida, M.; Fujiwara, D.; Nishikawa, M.; Horinouchi, S.; Beppu, T. Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J. Biol. Chem. 1994, 269, 6320–6324. [Google Scholar] [PubMed]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Ho, J.H.-N.; Kallstrom, G.; Johnson, A.W. Nmd3p Is a Crm1p-Dependent Adapter Protein for Nuclear Export of the Large Ribosomal Subunit. J. Cell Biol. 2000, 151, 1057–1066. [Google Scholar] [CrossRef]
- Sánchez-Velar, N.; Udofia, E.B.; Yu, Z.; Zapp, M.L. hRIP, a cellular cofactor for Rev function, promotes release of HIV RNAs from the perinuclear region. Genes Dev. 2004, 18, 23–34. [Google Scholar] [CrossRef]
- Edgcomb, S.P.; Carmel, A.B.; Naji, S.; Ambrus-Aikelin, G.; Reyes, J.R.; Saphire, A.C.; Gerace, L.; Williamson, J.R. DDX1 Is an RNA-Dependent ATPase Involved in HIV-1 Rev Function and Virus Replication. J. Mol. Biol. 2012, 415, 61–74. [Google Scholar] [CrossRef]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.-H.; Kleiman, L.; Jeang, K.-T. Requirement of DDX3 DEAD Box RNA Helicase for HIV-1 Rev-RRE Export Function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Garbelli, A.; Beermann, S.; Di Cicco, G.; Dietrich, U.; Maga, G. A Motif Unique to the Human Dead-Box Protein DDX3 Is Important for Nucleic Acid Binding, ATP Hydrolysis, RNA/DNA Unwinding and HIV-1 Replication. PLoS ONE 2011, 6, e19810. [Google Scholar] [CrossRef]
- Askjaer, P.; Jensen, T.H.; Nilsson, J.; Englmeier, L.; Kjems, J. The Specificity of the CRM1-Rev Nuclear Export Signal Interaction Is Mediated by RanGTP. J. Biol. Chem. 1998, 273, 33414–33422. [Google Scholar] [CrossRef]
- Yue, Y.; Coskun, A.K.; Jawanda, N.; Auer, J.; Sutton, R.E. Differential interaction between human and murine Crm1 and lentiviral Rev proteins. Virology 2018, 513, 1–10. [Google Scholar] [CrossRef]
- Budhiraja, S.; Liu, H.; Couturier, J.; Malovannaya, A.; Qin, J.; Lewis, D.E.; Rice, A.P. Mining the Human Complexome Database Identifies RBM14 as an XPO1-Associated Protein Involved in HIV-1 Rev Function. J. Virol. 2015, 89, 3557–3567. [Google Scholar] [CrossRef]
- Liu, H.; Hu, P.-W.; Budhiraja, S.; Misra, A.; Couturier, J.; Lloyd, R.E.; Lewis, D.E.; Kimata, J.T.; Rice, A.P. PACS1 is an HIV-1 cofactor that functions in Rev-mediated nuclear export of viral RNA. Virology 2019, 540, 88–96. [Google Scholar] [CrossRef]
- Bevec, D.; Jaksche, H.; Oft, M.; Wöhl, T.; Himmelspach, M.; Pacher, A.; Schebesta, M.; Koettnitz, K.; Dobrovnik, M.; Csonga, R.; et al. Inhibition of HIV-1 Replication in Lymphocytes by Mutants of the Rev Cofactor eIF-5A. Science 1996, 271, 1858–1860. [Google Scholar] [CrossRef] [PubMed]
- Modem, S.; Badri, K.R.; Holland, T.C.; Reddy, T.R. Sam68 is absolutely required for Rev function and HIV-1 production. Nucleic Acids Res. 2005, 33, 873–879. [Google Scholar] [CrossRef] [PubMed]
- Suhasini, M.; Reddy, T.R. Cellular proteins and HIV-1 Rev function. Curr. HIV Res. 2009, 7, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Hulver, M.J.; Trautman, J.P.; Goodwin, A.P.; Roszczenko, S.K.; Fogarty, K.H.; Miller, H.B. Human Tat-specific factor 1 binds the HIV-1 genome and selectively transports HIV-1 RNAs. Mol. Biol. Rep. 2020, 47, 1759–1772. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Prasad, S.; Dubay, J.W.; Hunter, E.; Jeang, K.T.; Rekosh, D.; Hammarskjold, M.L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Natl. Acad. Sci. USA 1994, 91, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Zolotukhin, A.S.; Valentin, A.; Pavlakis, G.N.; Felber, B.K. Continuous propagation of RRE(-) and Rev(-)RRE(-) human immunodeficiency virus type 1 molecular clones containing a cis-acting element of simian retrovirus type 1 in human peripheral blood lymphocytes. J. Virol. 1994, 68, 7944–7952. [Google Scholar] [CrossRef] [PubMed]
- Van Regenmortel, M.; Fauquet, C.; Bishop, D.; Carstens, E.; Estes, M.; Lemon, S.; Maniloff, J.; Mayo, M.; McGeoch, D.; Pringle, C.; et al. Virus Taxonomy: Classification and Nomenclature of Viruses. Seventh Report of the International Committee on Taxonomy of Viruses; London Academic Press: London, UK, 2000; ISBN 0-12-370200-3. [Google Scholar]
- Tabernero, C.; Zolotukhin, A.S.; Valentin, A.; Pavlakis, G.N.; Felber, B.K. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J. Virol. 1996, 70, 5998–6011. [Google Scholar] [CrossRef]
- Ernst, R.K.; Bray, M.; Rekosh, D.; Hammarskjold, M.L. Secondary structure and mutational analysis of the Mason-Pfizer monkey virus RNA constitutive transport element. RNA 1997, 3, 210–222. [Google Scholar]
- Ernst, R.K.; Bray, M.; Rekosh, D.; Hammarskjöld, M.L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 1997, 17, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, C.; Felber, B.; Izaurralde, E. The simian retrovirus-1 constitutive transport element, unlike the HIV-1 RRE, uses factors required for cellular mRNA export. Curr. Biol. 1997, 7, 619–628. [Google Scholar] [CrossRef]
- Pasquinelli, A.E.; Ernst, R.K.; Lund, E.; Grimm, C.; Zapp, M.L.; Rekosh, D.; Hammarskjöld, M.; Dahlberg, J.E. The constitutive transport element (CTE) of Mason-Pfizer monkey virus (MPMV) accesses a cellular mRNA export pathway. EMBO J. 1997, 16, 7500–7510. [Google Scholar] [CrossRef] [PubMed]
- Grüter, P.; Tabernero, C.; Von Kobbe, C.; Schmitt, C.; Saavedra, C.; Bachi, A.; Wilm, M.; Felber, B.K.; Izaurralde, E. TAP, the Human Homolog of Mex67p, Mediates CTE-Dependent RNA Export from the Nucleus. Mol. Cell 1998, 1, 649–659. [Google Scholar] [CrossRef]
- Segref, A.; Sharma, K.; Doye, V.; Hellwig, A.; Huber, J.; Lührmann, R.; Hurt, E. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997, 16, 3256–3271. [Google Scholar] [CrossRef]
- Santos-Rosa, H.; Moreno, H.; Simos, G.; Segref, A.; Fahrenkrog, B.; Panté, N.; Hurt, E. Nuclear mRNA Export Requires Complex Formation between Mex67p and Mtr2p at the Nuclear Pores. Mol. Cell. Biol. 1998, 18, 6826–6838. [Google Scholar] [CrossRef]
- Bachi, A.; Braun, I.C.; Rodrigues, J.P.; Panté, N.; Ribbeck, K.; Von Kobbe, C.; Kutay, U.; Wilm, M.; Görlich, D.; Carmo-Fonseca, M.; et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA 2000, 6, 136–158. [Google Scholar] [CrossRef]
- Mertz, J.A.; Simper, M.S.; Lozano, M.M.; Payne, S.M.; Dudley, J.P. Mouse Mammary Tumor Virus Encodes a Self-Regulatory RNA Export Protein and Is a Complex Retrovirus. J. Virol. 2005, 79, 14737–14747. [Google Scholar] [CrossRef]
- Müllner, M.; Salmons, B.; Günzburg, W.H.; Indik, S. Identification of the Rem-responsive element of mouse mammary tumor virus. Nucleic Acids Res. 2008, 36, 6284–6294. [Google Scholar] [CrossRef]
- Hofacre, A.; Nitta, T.; Fan, H. Jaagsiekte Sheep Retrovirus Encodes a Regulatory Factor, Rej, Required for Synthesis of Gag Protein. J. Virol. 2009, 83, 12483–12498. [Google Scholar] [CrossRef]
- Yang, J.; Bogerd, H.P.; Peng, S.; Wiegand, H.; Truant, R.; Cullen, B.R. An ancient family of human endogenous retroviruses encodes a functional homolog of the HIV-1 Rev protein. Proc. Natl. Acad. Sci. USA 1999, 96, 13404–13408. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, M.; Inoue, J.; Yoshida, M.; Seiki, M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988, 7, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Hanly, S.M.; Rimsky, L.T.; Malim, M.H.; Kim, J.H.; Hauber, J.; Dodon, M.D.; Le, S.Y.; Maizel, J.V.; Cullen, B.R.; Greene, W.C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989, 3, 1534–1544. [Google Scholar] [CrossRef]
- Seiki, M.; Inoue, J.; Hidaka, M.; Yoshida, M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 1988, 85, 7124–7128. [Google Scholar] [CrossRef] [PubMed]
- Seiki, M.; Hattori, S.; Hirayama, Y.; Yoshida, M. Human adult T-cell leukemia virus: Complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 1983, 80, 3618–3622. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Y.F.; Gilmartin, G.M.; Hanly, S.M.; Nevins, J.R.; Greene, W.C. The HTLV-I rex response element mediates a novel form of mRNA polyadenylation. Cell 1991, 64, 727–737. [Google Scholar] [CrossRef]
- Sinha-Datta, U.; Datta, A.; Ghorbel, S.; Dodon, M.D.; Nicot, C. Human T-cell Lymphotrophic Virus Type I Rex and p30 Interactions Govern the Switch between Virus Latency and Replication. J. Biol. Chem. 2007, 282, 14608–14615. [Google Scholar] [CrossRef]
- Moles, R.; Sarkis, S.; Galli, V.; Omsland, M.; Purcell, D.F.J.; Yurick, D.; Khoury, G.; Pise-Masison, C.A.; Franchini, G. p30 protein: A critical regulator of HTLV-1 viral latency and host immunity. Retrovirology 2019, 16, 1–15. [Google Scholar] [CrossRef]
- Martin, G.S. Rous Sarcoma Virus: A Function required for the Maintenance of the Transformed State. Nature 1970, 227, 1021–1023. [Google Scholar] [CrossRef]
- Bernstein, A.; MacCormick, R.; Martin, G. Transformation-defective mutants of avian sarcoma viruses: The genetic relationship between conditional and nonconditional mutants. Virology 1976, 70, 206–209. [Google Scholar] [CrossRef]
- Paca, R.E.; Ogert, R.A.; Hibbert, C.S.; Izaurralde, E.; Beemon, K.L. Rous Sarcoma Virus DR Posttranscriptional Elements Use a Novel RNA Export Pathway. J. Virol. 2000, 74, 9507–9514. [Google Scholar] [CrossRef] [PubMed]
- Leblanc, J.J.; Uddowla, S.; Abraham, B.; Clatterbuck, S.; Beemon, K.L. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology 2007, 363, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, R.J.K.; Rice, B.; Chen, E.C.; Tuffy, K.M.; Chiari, E.F.; Fahrbach, K.M.; Hope, T.J.; Parent, L.J. Visualizing Association of the Retroviral Gag Protein with Unspliced Viral RNA in the Nucleus. Mbio 2020, 11, e00524-20. [Google Scholar] [CrossRef]
- Yu, K.L.; Lee, S.H.; Lee, E.S.; You, J.C. HIV-1 nucleocapsid protein localizes efficiently to the nucleus and nucleolus. Virology 2016, 492, 204–212. [Google Scholar] [CrossRef]
- Weldon, R.A.; Sarkar, P.; Brown, S.M.; Weldon, S.K. Mason–Pfizer monkey virus Gag proteins interact with the human sumo conjugating enzyme, hUbc. Virology 2003, 314, 62–73. [Google Scholar] [CrossRef]
- Maldonado, R.K.; Parent, L.J. Orchestrating the Selection and Packaging of Genomic RNA by Retroviruses: An Ensemble of Viral and Host Factors. Viruses 2016, 8, 257. [Google Scholar] [CrossRef]
- Pessel-Vivares, L.; Houzet, L.; Lainé, S.; Mougel, M. Insights into the nuclear export of murine leukemia virus intron-containing RNA. RNA Biol. 2015, 12, 942–949. [Google Scholar] [CrossRef]
- Pessel-Vivares, L.; Ferrer, M.; Laine, S.; Mougel, M. MLV requires Tap/NXF1-dependent pathway to export its unspliced RNA to the cytoplasm and to express both spliced and unspliced RNAs. Retrovirology 2014, 11, 21. [Google Scholar] [CrossRef]
- Sakuma, T.; Tonne, J.M.; Ikeda, Y. Murine Leukemia Virus Uses TREX Components for Efficient Nuclear Export of Unspliced Viral Transcripts. Viruses 2014, 6, 1135–1148. [Google Scholar] [CrossRef]
- Bartels, H.; Luban, J. Gammaretroviral pol sequences act in cis to direct polysome loading and NXF1/NXT-dependent protein production by gag-encoded RNA. Retrovirology 2014, 11, 73. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mougel, M.; Akkawi, C.; Chamontin, C.; Feuillard, J.; Pessel-Vivares, L.; Socol, M.; Laine, S. NXF1 and CRM1 nuclear export pathways orchestrate nuclear export, translation and packaging of murine leukaemia retrovirus unspliced RNA. RNA Biol. 2020, 17, 528–538. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.F.; Baldwin, D.N.; Gwynn, S.R.; Yendapalli, S.; Linial, M.L. Human Foamy Virus Replication: A Pathway Distinct from That of Retroviruses and Hepadnaviruses. Science 1996, 271, 1579–1582. [Google Scholar] [CrossRef] [PubMed]
- Löchelt, M.; Muranyi, W.; Flügel, R.M. Human foamy virus genome possesses an internal, Bel-1-dependent and functional promoter. Proc. Natl. Acad. Sci. USA 1993, 90, 7317–7321. [Google Scholar] [CrossRef]
- Moebes, A.; Enssle, J.; Bieniasz, P.D.; Heinkelein, M.; Lindemann, D.; Bock, M.; McClure, M.O.; Rethwilm, A. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 1997, 71, 7305–7311. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.F.; Sullivan, M.D.; Linial, M.L. Evidence that the Human Foamy Virus Genome Is DNA. J. Virol. 1999, 73, 1565–1572. [Google Scholar] [CrossRef]
- Goepfert, P.A.; Shaw, K.L.; Ritter, G.D.; Mulligan, M.J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J. Virol. 1997, 71, 778–784. [Google Scholar] [CrossRef]
- Bodem, J.; Schied, T.; Gabriel, R.; Rammling, M.; Rethwilm, A. Foamy Virus Nuclear RNA Export Is Distinct from That of Other Retroviruses. J. Virol. 2010, 85, 2333–2341. [Google Scholar] [CrossRef]
- Rethwilm, A.; Bodem, J. Evolution of Foamy Viruses: The Most Ancient of All Retroviruses. Viruses 2013, 5, 2349–2374. [Google Scholar] [CrossRef]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar] [CrossRef]
- Renault, N.; Tobaly-Tapiero, J.; Paris, J.; Giron, M.-L.; Coiffic, A.; Roingeard, P.; Saïb, A. A nuclear export signal within the structural Gag protein is required for prototype foamy virus replication. Retrovirology 2011, 8, 6. [Google Scholar] [CrossRef]
- Summers, J.; Mason, W.S. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 1982, 29, 403–415. [Google Scholar] [CrossRef]
- Chen, Y.; Michitaka, K.; Matsubara, H.; Yamamoto, K.; Horiike, N.; Onji, M. Complete genome sequence of hepatitis B virus (HBV) from a patient with fulminant hepatitis without precore and core promoter mutations: Comparison with HBV from a patient with acute hepatitis infected from the same infectious source. J. Hepatol. 2003, 38, 84–90. [Google Scholar] [CrossRef]
- Kitamura, K.; Que, L.; Shimadu, M.; Koura, M.; Ishihara, Y.; Wakae, K.; Nakamura, T.; Watashi, K.; Wakita, T.; Muramatsu, M. Flap endonuclease 1 is involved in cccDNA formation in the hepatitis B virus. PLoS Pathog. 2018, 14, e1007124. [Google Scholar] [CrossRef] [PubMed]
- Werle–Lapostolle, B.; Bowden, S.; Locarnini, S.; Wursthorn, K.; Petersen, J.; Lau, G.; Trepo, C.; Marcellin, P.; Goodman, Z.; Delaney, W.E.; et al. Persistence of cccDNA during the natural history of chronic hepatitis B and decline during adefovir dipivoxil therapy. Gastroenterology 2004, 126, 1750–1758. [Google Scholar] [CrossRef]
- Quasdorff, M.; Protzer, U. Control of hepatitis B virus at the level of transcription. J. Viral Hepat. 2010, 17, 527–536. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Aoyama, K.; Ohno, N.; Iwata, K.; Akahane, Y.; Baba, K.; Yoshizawa, H.; Mishiro, S. The precore/core promoter mutant (T1762A1764) of hepatitis B virus: Clinical significance and an easy method for detection. J. Gen. Virol. 1995, 76, 3159–3164. [Google Scholar] [CrossRef]
- Cattaneo, R.; Will, H.; Schaller, H. Hepatitis B virus transcription in the infected liver. EMBO J. 1984, 3, 2191–2196. [Google Scholar] [CrossRef]
- Lucifora, J.; Arzberger, S.; Durantel, D.; Belloni, L.; Strubin, M.; Levrero, M.; Zoulim, F.; Hantz, O.; Protzer, U. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J. Hepatol. 2011, 55, 996–1003. [Google Scholar] [CrossRef]
- Soussan, P.; Tuveri, R.; Nalpas, B.; Garreau, F.; Zavala, F.; Masson, A.; Pol, S.; Brechot, C.; Kremsdorf, D. The expression of hepatitis B spliced protein (HBSP) encoded by a spliced hepatitis B virus RNA is associated with viral replication and liver fibrosis. J. Hepatol. 2003, 38, 343–348. [Google Scholar] [CrossRef]
- Chen, J.-Y.; Chen, W.-N.; Liu, L.-L.; Lin, W.-S.; Jiao, B.-Y.; Wu, Y.-L.; Lin, J.-Y.; Lin, X. Hepatitis B spliced protein (HBSP) generated by a spliced hepatitis B virus RNA participates in abnormality of fibrin formation and functions by binding to fibrinogen γ chain. J. Med. Virol. 2010, 82, 2019–2026. [Google Scholar] [CrossRef]
- Wang, J.C.-Y.; Nickens, D.G.; Lentz, T.B.; Loeb, D.D.; Zlotnick, A. Encapsidated hepatitis B virus reverse transcriptase is poised on an ordered RNA lattice. Proc. Natl. Acad. Sci. USA 2014, 111, 11329–11334. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Liang, T.J. A novel hepatitis B virus (HBV) genetic element with Rev response element-like properties that is essential for expression of HBV gene products. Mol. Cell. Biol. 1993, 13, 7476–7486. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.M.; Yen, T.S. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol. Cell. Biol. 1995, 15, 3864–3869. [Google Scholar] [CrossRef] [PubMed]
- Zang, W.-Q.; Yen, T.B. Distinct Export Pathway Utilized by the Hepatitis B Virus Posttranscriptional Regulatory Element. Virology 1999, 259, 299–304. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Smith, G.J.; Donello, J.E.; Hope, T.J.; Lück, R.; Steger, G. The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops which are required for function. Nucleic Acids Res. 1998, 26, 4818–4827. [Google Scholar] [CrossRef] [PubMed]
- Zang, W.-Q.; Li, B.; Huang, P.-Y.; Lai, M.M.C.; Yen, T.B. Role of Polypyrimidine Tract Binding Protein in the Function of the Hepatitis B Virus Posttranscriptional Regulatory Element. J. Virol. 2001, 75, 10779–10786. [Google Scholar] [CrossRef]
- Li, Y.; Huang, T.; Zhang, X.; Wan, T.; Hu, J.; Huang, A.; Tang, H. Role of glyceraldehyde-3-phosphate dehydrogenase binding to hepatitis B virus posttranscriptional regulatory element in regulating expression of HBV surface antigen. Arch. Virol. 2009, 154, 519–524. [Google Scholar] [CrossRef]
- Horke, S.; Reumann, K.; Rang, A.; Heise, T. Molecular Characterization of the Human La Protein·Hepatitis B Virus RNA.B Interactionin Vitro. J. Biol. Chem. 2002, 277, 34949–34958. [Google Scholar] [CrossRef]
- Kamath, R.V.; Leary, D.J.; Huang, S. Nucleocytoplasmic Shuttling of Polypyrimidine Tract-binding Protein Is Uncoupled from RNA Export. Mol. Biol. Cell 2001, 12, 3808–3820. [Google Scholar] [CrossRef]
- Chi, B.; Wang, K.; Du, Y.; Gui, B.; Chang, X.; Wang, L.; Fan, J.; Chen, S.; Wu, X.; Li, G.; et al. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Res. 2014, 42, 7305–7318. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-C.; Huang, E.-Y.; Su, P.-Y.; Wu, S.-Y.; Yang, C.-C.; Lin, Y.-S.; Chang, W.-C.; Shih, C. Nuclear Export and Import of Human Hepatitis B Virus Capsid Protein and Particles. PLoS Pathog. 2010, 6, e1001162. [Google Scholar] [CrossRef] [PubMed]
- Porterfield, J.Z.; Dhason, M.S.; Loeb, D.D.; Nassal, M.; Stray, S.J.; Zlotnick, A. Full-Length Hepatitis B Virus Core Protein Packages Viral and Heterologous RNA with Similarly High Levels of Cooperativity. J. Virol. 2010, 84, 7174–7184. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.-C.; Huang, E.-Y.; Li, H.-C.; Su, P.-Y.; Shih, C. Nuclear Export of Human Hepatitis B Virus Core Protein and Pregenomic RNA Depends on the Cellular NXF1-p15 Machinery. PLoS ONE 2014, 9, e106683. [Google Scholar] [CrossRef]
- Makokha, G.N.; Abe-Chayama, H.; Chowdhury, S.; Hayes, C.N.; Tsuge, M.; Yoshima, T.; Ishida, Y.; Zhang, Y.; Uchida, T.; Tateno, C.; et al. Regulation of the Hepatitis B virus replication and gene expression by the multi-functional protein TARDBP. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef]
- Hu, B.; Yu, L.; Zhu, N.-S.; Xie, J. Cellular UAP56 interacts with the HBx protein of the hepatitis B virus and is involved in viral RNA nuclear export in hepatocytes. Exp. Cell Res. 2020, 390, 111929. [Google Scholar] [CrossRef]
- Zoulim, F. Hepatitis B virus resistance to antiviral drugs: Where are we going? Liver Int. 2011, 31, 111–116. [Google Scholar] [CrossRef]
- Gale, M.; Tan, S.-L.; Katze, M.G. Translational Control of Viral Gene Expression in Eukaryotes. Microbiol. Mol. Biol. Rev. 2000, 64, 239–280. [Google Scholar] [CrossRef]
- Rowe, W.P.; Huebner, R.J.; Gilmore, L.K.; Parrott, R.H.; Ward, T.G. Isolation of a Cytopathogenic Agent from Human Adenoids Undergoing Spontaneous Degeneration in Tissue Culture. Proc. Soc. Exp. Biol. Med. 1953, 84, 570–573. [Google Scholar] [CrossRef]
- Lion, T. Adenovirus Infections in Immunocompetent and Immunocompromised Patients. Clin. Microbiol. Rev. 2014, 27, 441–462. [Google Scholar] [CrossRef]
- Sprengel, J.; Schmitz, B.; Heuss-Neitzel, D.; Zock, C.; Doerfler, W. Nucleotide sequence of human adenovirus type 12 DNA: Comparative functional analysis. J. Virol. 1994, 68, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J. Adenoviridae. In Fields Virology; Knipe, D.M., Howley, P.M., Eds.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 978-1-4511-0563-6. [Google Scholar]
- Crisostomo, L.; Soriano, A.M.; Mendez, M.; Graves, D.; Pelka, P. Temporal dynamics of adenovirus 5 gene expression in normal human cells. PLoS ONE 2019, 14, e0211192. [Google Scholar] [CrossRef] [PubMed]
- Fessler, S.P.; Young, C.S.H. Control of Adenovirus Early Gene Expression during the Late Phase of Infection. J. Virol. 1998, 72, 4049–4056. [Google Scholar] [CrossRef] [PubMed]
- Biasiotto, R.; Akusjärvi, G. Regulation of Human Adenovirus Alternative RNA Splicing by the Adenoviral L4-33K and L4-22K Proteins. Int. J. Mol. Sci. 2015, 16, 2893–2912. [Google Scholar] [CrossRef] [PubMed]
- Pilder, S.; Moore, M.; Logan, J.; Shenk, T. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 1986, 6, 470–476. [Google Scholar] [CrossRef]
- Leppard, K.N.; Shenk, T. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989, 8, 2329–2336. [Google Scholar] [CrossRef]
- Leppard, K.N.; Everett, R.D. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 1999, 80, 997–1008. [Google Scholar] [CrossRef]
- Ornelles, D.A.; Shenk, T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: Association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 1991, 65, 424–429. [Google Scholar] [CrossRef]
- Beltz, G.A.; Flint, S.J. Inhibition of HeLa cell protein synthesis during adenovirus infection: Restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 1979, 131, 353–373. [Google Scholar] [CrossRef]
- Blanchette, P.; Kindsmüller, K.; Groitl, P.; Dallaire, F.; Speiseder, T.; Branton, P.E.; Dobner, T. Control of mRNA Export by Adenovirus E4orf6 and E1B55K Proteins during Productive Infection Requires E4orf6 Ubiquitin Ligase Activity. J. Virol. 2008, 82, 2642–2651. [Google Scholar] [CrossRef]
- Woo, J.L.; Berk, A.J. Adenovirus Ubiquitin-Protein Ligase Stimulates Viral Late mRNA NuclearExport. J. Virol. 2007, 81, 575–587. [Google Scholar] [CrossRef]
- Hidalgo, P.; Ip, W.H.; Dobner, T.; Gonzalez, R.A. The biology of the adenovirus E1B 55K protein. FEBS Lett. 2019, 593, 3504–3517. [Google Scholar] [CrossRef] [PubMed]
- Gabler, S.; Schütt, H.; Groitl, P.; Wolf, H.; Shenk, T.; Dobner, T. E1B 55-Kilodalton-Associated Protein: A Cellular Protein with RNA-Binding Activity Implicated in Nucleocytoplasmic Transport of Adenovirus and Cellular mRNAs. J. Virol. 1998, 72, 7960–7971. [Google Scholar] [CrossRef] [PubMed]
- Yatherajam, G.; Huang, W.; Flint, S.J. Export of Adenoviral Late mRNA from the Nucleus Requires the Nxf1/Tap Export Receptor. J. Virol. 2011, 85, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Muik, A.; Stubbert, L.J.; Jahedi, R.Z.; Geiβ, Y.; Kimpel, J.; Dold, C.; Tober, R.; Volk, A.; Klein, S.; Dietrich, U.; et al. Re-engineering Vesicular Stomatitis Virus to Abrogate Neurotoxicity, Circumvent Humoral Immunity, and Enhance Oncolytic Potency. Cancer Res. 2014, 74, 3567–3578. [Google Scholar] [CrossRef]
- Heo, J.; Reid, T.; Ruo, L.; Breitbach, C.J.; Rose, S.; Bloomston, M.; Cho, M.; Lim, H.Y.; Chung, H.C.; Kim, C.W.; et al. Randomized dose-finding clinical trial of oncolytic immunotherapeutic vaccinia JX-594 in liver cancer. Nat. Med. 2013, 19, 329–336. [Google Scholar] [CrossRef]
- Sauthoff, H.; Pipiya, T.; Heitner, S.; Chen, S.; Bleck, B.; Reibman, J.; Chang, W.; Norman, R.G.; Rom, W.N.; Hay, J.G. Impact of E1a Modifications on Tumor-Selective Adenoviral Replication and Toxicity. Mol. Ther. 2004, 10, 749–757. [Google Scholar] [CrossRef]
- Zheng, M.; Huang, J.; Tong, A.; Yang, H. Oncolytic Viruses for Cancer Therapy: Barriers and Recent Advances. Mol. Ther. Oncolytics 2019, 15, 234–247. [Google Scholar] [CrossRef]
- O’Shea, C.C.; Johnson, L.; Bagus, B.; Choi, S.; Nicholas, C.; Shen, A.; Boyle, L.; Pandey, K.; Soria, C.; Kunich, J.; et al. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 2004, 6, 611–623. [Google Scholar] [CrossRef]
- Khuri, F.R.; Nemunaitis, J.; Ganly, I.; Arseneau, J.; Tannock, I.F.; Romel, L.; Gore, M.; Ironside, J.; MacDougall, R.; Heise, C.; et al. A controlled trial of intratumoral ONYX-015, a selectively-replicating adenovirus, in combination with cisplatin and 5-fluorouracil in patients with recurrent head and neck cancer. Nat. Med. 2000, 6, 879–885. [Google Scholar] [CrossRef]
- Liu, F.; Zhou, Z.H. Comparative virion structures of human herpesviruses. In Human Herpesviruses; Cambridge University Press: Cambridge, UK, 2010; pp. 27–43. [Google Scholar]
- Singh, N.; Tscharke, D.C. Herpes Simplex Virus Latency Is Noisier the Closer We Look. J. Virol. 2020, 94, e01701-19. [Google Scholar] [CrossRef] [PubMed]
- McGeoch, D.J.; Dolan, A.; Ralph, A.C. Toward a Comprehensive Phylogeny for Mammalian and Avian Herpesviruses. J. Virol. 2000, 74, 10401–10406. [Google Scholar] [CrossRef] [PubMed]
- Wald, A.; Corey, L. HSV: Persistence in the population: Epidemiology, transmission. In Human Herpesviruses; Cambridge University Press: Cambridge, UK, 2010; pp. 656–672. [Google Scholar]
- Tang, S.; Patel, A.; Krause, P.R. Hidden regulation of herpes simplex virus 1 pre-mRNA splicing and polyadenylation by virally encoded immediate early gene ICP27. PLoS Pathog. 2019, 15, e1007884. [Google Scholar] [CrossRef] [PubMed]
- Mears, W.E.; Lam, V.; Rice, S.A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 1995, 69, 935–947. [Google Scholar] [CrossRef] [PubMed]
- Mears, W.E.; Rice, S.A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 1996, 70, 7445–7453. [Google Scholar] [CrossRef] [PubMed]
- Mears, W.E.; Rice, S.A. The Herpes Simplex Virus Immediate-Early Protein ICP27 Shuttles between Nucleus and Cytoplasm. Virology 1998, 242, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Phelan, A.; Clements, J.B. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 1997, 78, 3327–3331. [Google Scholar] [CrossRef]
- Sandri-Goldin, R.M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998, 12, 868–879. [Google Scholar] [CrossRef]
- Koffa, M.D.; Clements, J.B.; Izaurralde, E.; Wadd, S.; Wilson, S.A.; Mattaj, I.W.; Kuersten, S. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 2001, 20, 5769–5778. [Google Scholar] [CrossRef]
- Chen, I.-H.B.; Sciabica, K.S.; Sandri-Goldin, R.M. ICP27 Interacts with the RNA Export Factor Aly/REF To Direct Herpes Simplex Virus Type 1 Intronless mRNAs to the TAP Export Pathway. J. Virol. 2002, 76, 12877–12889. [Google Scholar] [CrossRef]
- Hernandez, F.P.; Sandri-Goldin, R.M. Head-to-Tail Intramolecular Interaction of Herpes Simplex Virus Type 1 Regulatory Protein ICP27 Is Important for Its Interaction with Cellular mRNA Export Receptor TAP/NXF1. Mbio 2010, 1, e00268-10. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.A.; Sandri-Goldin, R.M. Efficient Nuclear Export of Herpes Simplex Virus 1 Transcripts Requires both RNA Binding by ICP27 and ICP27 Interaction with TAP/NXF1. J. Virol. 2009, 83, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Tunnicliffe, R.B.; Hautbergue, G.M.; Kalra, P.; Jackson, B.R.; Whitehouse, A.; Wilson, S.A.; Golovanov, A.P. Structural Basis for the Recognition of Cellular mRNA Export Factor REF by Herpes Viral Proteins HSV-1 ICP27 and HVS ORF. PLoS Pathog. 2011, 7, e1001244. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Devi-Rao, G.; Golovanov, A.P.; Sandri-Goldin, R.M. The Interaction of the Cellular Export Adaptor Protein Aly/REF with ICP27 Contributes to the Efficiency of Herpes Simplex Virus 1 mRNA Export. J. Virol. 2013, 87, 7210–7217. [Google Scholar] [CrossRef] [PubMed]
- Tunnicliffe, R.B.; Hautbergue, G.M.; Wilson, S.A.; Kalra, P.; Golovanov, A.P. Competitive and Cooperative Interactions Mediate RNA Transfer from Herpesvirus Saimiri ORF57 to the Mammalian Export Adaptor ALYREF. PLoS Pathog. 2014, 10, e1003907. [Google Scholar] [CrossRef] [PubMed]
- Malik, P.; Tabarraei, A.; Kehlenbach, R.H.; Korfali, N.; Iwasawa, R.; Graham, S.V.; Schirmer, E.C. Herpes Simplex Virus ICP27 Protein Directly Interacts with the Nuclear Pore Complex through Nup62, Inhibiting Host Nucleocytoplasmic Transport Pathways. J. Biol. Chem. 2012, 287, 12277–12292. [Google Scholar] [CrossRef]
- Tunnicliffe, R.B.; Tian, X.; Storer, J.; Sandri-Goldin, R.M.; Golovanov, A.P. Overlapping motifs on the herpes viral proteins ICP27 and ORF57 mediate interactions with the mRNA export adaptors ALYREF and UIF. Sci. Rep. 2018, 8, 15005. [Google Scholar] [CrossRef]
- Davison, A.J.; Scott, J.E. The Complete DNA Sequence of Varicella-Zoster Virus. J. Gen. Virol. 1986, 67, 1759–1816. [Google Scholar] [CrossRef]
- Winkler, M.; Rice, S.A.; Stamminger, T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 1994, 68, 3943–3954. [Google Scholar] [CrossRef]
- Ote, I.; Piette, J.; Sadzot-Delvaux, C. The Varicella-Zoster virus IE4 protein: A conserved member of the herpesviral mRNA export factors family and a potential alternative target in antiherpetic therapies. Biochem. Pharmacol. 2010, 80, 1973–1980. [Google Scholar] [CrossRef]
- Gruffat, H.; Batisse, J.; Pich, D.; Neuhierl, B.; Manet, E.; Hammerschmidt, W.; Sergeant, A. Epstein-Barr Virus mRNA Export Factor EB2 Is Essential for Production of Infectious Virus. J. Virol. 2002, 76, 9635–9644. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, M.; Kuhn, J.H. Create a Megataxonomic Framework, Filling All Principal Taxonomic Ranks, for ssDNA Viruses; ICTV TaxoProp: Berlin, Germany, 2019. [Google Scholar]
- De Martel, C.; Georges, D.; Bray, F.; Ferlay, J.; Clifford, G.M. Global burden of cancer attributable to infections in 2018: A worldwide incidence analysis. Lancet Glob. Health 2020, 8, e180–e190. [Google Scholar] [CrossRef]
- Baker, C.; Calef, C. Maps of Papillomavirus mRNA Transcripts. In Human Papillomaviruses 1996: A Compilation and Analysis of Nucleic Acid and Amino Acid Sequences; Theoretical Biology and Biophysics: Los Alamos, NM, USA, 1996; pp. 1–20. [Google Scholar]
- Van Der Meijden, E.; Kazem, S.; Dargel, C.A.; Van Vuren, N.; Hensbergen, P.J.; Feltkamp, M.C. Characterization of T Antigens, Including Middle T and Alternative T, Expressed by the Human Polyomavirus Associated with Trichodysplasia Spinulosa. J. Virol. 2015, 89, 9427–9439. [Google Scholar] [CrossRef]
- Abend, J.R.; Joseph, A.E.; Das, D.; Campbell-Cecen, D.B.; Imperiale, M.J. A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. J. Gen. Virol. 2009, 90, 1238–1245. [Google Scholar] [CrossRef] [PubMed]
- Trowbridge, P.W.; Frisque, R.J. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J. Neurovirol. 1995, 1, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Graham, S.V.; Faizo, A.A.A. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2017, 231, 83–95. [Google Scholar] [CrossRef]
- Huang, Y.; Carmichael, G.G. RNA processing in the polyoma virus life cycle. Front. Biosci. 2009, 14, 4968–4977. [Google Scholar] [CrossRef]
- Gardner, S.D.; Field, A.M.; Coleman, D.V.; Hulme, B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet 1971, 297, 1253–1257. [Google Scholar] [CrossRef]
- Padgett, B.L.; Walker, D.L.; ZuRhein, G.M.; Eckroade, R.J.; Dessel, B.H. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet 1971, 1, 1257–1260. [Google Scholar] [CrossRef]
- Miller, M.A.; Weibel, C.; Ferguson, D.; Landry, M.L.; Kahn, J.S. WU Polyomavirus in Patients Infected with HIV or Hepatitis C Virus, Connecticut, USA, 2007. Emerg. Infect. Dis. 2009, 15, 1095–1097. [Google Scholar] [CrossRef]
- Cedeno-Laurent, F.; Penalva de Oliveira, A.C.; Vidal, J.E.; Trujillo, J.R. Human Polyomavirus-Associated Cerebral Disorders in the Post-HAART Era. Available online: https://www.hindawi.com/journals/pri/2011/562427/ (accessed on 19 August 2020).
- Di Taranto, C.; Pietropaolo, V.; Orsi, G.; Jin, L.; Sinibaldi, L.; Degener, A.; Lin, L. Detection of BK polyomavirus genotypes in healthy and HIV-positive children. Eur. J. Epidemiol. 1997, 13, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Maglennon, G.A.; McIntosh, P.B.; Doorbar, J. Immunosuppression Facilitates the Reactivation of Latent Papillomavirus Infections. J. Virol. 2014, 88, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Cutrone, R.; Lednicky, J.; Dunn, G.; Rizzo, P.; Bocchetta, M.; Chumakov, K.; Minor, P.; Carbone, M. Some Oral Poliovirus Vaccines Were Contaminated with Infectious SV40 after 1961. Cancer Res. 2005, 65, 10273–10279. [Google Scholar] [CrossRef] [PubMed]
- Testa, J.R.; Carbone, M.; Hirvonen, A.; Khalili, K.; Krynska, B.; Linnainmaa, K.; Pooley, F.D.; Rizzo, P.; Rusch, V.; Xiao, G.H. A multi-institutional study confirms the presence and expression of simian virus 40 in human malignant mesotheliomas. Cancer Res. 1998, 58, 4505–4509. [Google Scholar]
- Lopez-Rios, F.; Illei, P.B.; Rusch, V.W.; Ladanyi, M. Evidence against a role for SV40 infection in human mesotheliomas and high risk of false-positive PCR results owing to presence of SV40 sequences in common laboratory plasmids. Lancet 2004, 364, 1157–1166. [Google Scholar] [CrossRef]
- Strickler, H.D. A multicenter evaluation of assays for detection of SV40 DNA and results in masked mesothelioma specimens. Cancer Epidemiol. Biomarker. Prev. 2001, 10, 523–532. [Google Scholar]
- Engels, E.A.; Sarkar, C.; Daniel, R.W.; Gravitt, P.E.; Verma, K.; Quezado, M.; Shah, K.V. Absence of simian virus 40 in human brain tumors from northern India. Int. J. Cancer 2002, 101, 348–352. [Google Scholar] [CrossRef]
- De Rienzo, A.; Tor, M.; Sterman, D.H.; Aksoy, F.; Albelda, S.M.; Testa, J.R. Detection of SV40 DNA sequences in malignant mesothelioma specimens from the United States, but not from Turkey. J. Cell. Biochem. 2002, 84, 455–459. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Mazzoni, E.; Bononi, I.; Tognon, M.; Martini, F. Association between Simian Virus 40 and Human Tumors. Front. Oncol. 2019, 9, 670. [Google Scholar] [CrossRef]
- Ryu, W.-S.; Mertz, J.E. Simian virus 40 late transcripts lacking excisable intervening sequences are defective in both stability in the nucleus and transport to the cytoplasm. J. Virol. 1989, 63, 4386–4394. [Google Scholar] [CrossRef]
- Barrett, N.L.; Carmichael, G.G.; Luo, Y. Splice site requirement for the efficient accumulation of polyoma virus late mRNAs. Nucleic Acids Res. 1991, 19, 3011–3017. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Carmichael, G.G. A suboptimal 5′ splice site is a cis-acting determinant of nuclear export of polyomavirus late mRNAs. Mol. Cell. Biol. 1996, 16, 6046–6054. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Genital HPV Infection—CDC Fact Sheet; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2017.
- Ozbun, M.A.; Meyers, C. Temporal Usage of Multiple Promoters during the Life Cycle of Human Papillomavirus Type 31b. J. Virol. 1998, 72, 2715–2722. [Google Scholar] [CrossRef] [PubMed]
- Geisen, C.; Kahn, T. Promoter Activity of Sequences Located Upstream of the Human Papillomavirus Types 16 and 18 Late Regions. J. Gen. Virol. 1996, 77, 2193–2200. [Google Scholar] [CrossRef]
- Braunstein, T.H.; Madsen, B.S.; Gavnholt, B.; Rosenstierne, M.W.; Johnsen, C.K.; Norrild, B. Identification of a new promoter in the early region of the human papillomavirus type 16 genome. J. Gen. Virol. 1999, 80, 3241–3250. [Google Scholar] [CrossRef]
- Pyeon, D.; Pearce, S.M.; Lank, S.M.; Ahlquist, P.; Lambert, P.F. Establishment of Human Papillomavirus Infection Requires Cell Cycle Progression. PLoS Pathog. 2009, 5, e1000318. [Google Scholar] [CrossRef]
- Shafti-Keramat, S.; Handisurya, A.; Kriehuber, E.; Meneguzzi, G.; Slupetzky, K.; Kirnbauer, R. Different Heparan Sulfate Proteoglycans Serve asCellular Receptors for HumanPapillomaviruses. J. Virol. 2003, 77, 13125–13135. [Google Scholar] [CrossRef]
- Ozbun, M.A. Human Papillomavirus Type 31b Infection of Human Keratinocytes and the Onset of Early Transcription. J. Virol. 2002, 76, 11291–11300. [Google Scholar] [CrossRef]
- Nakahara, T.; Nishimura, A.; Tanaka, M.; Ueno, T.; Ishimoto, A.; Sakai, H. Modulation of the Cell Division Cycle by Human Papillomavirus Type 18 E4. J. Virol. 2002, 76, 10914–10920. [Google Scholar] [CrossRef]
- Hansen, C.N.; Nielsen, L.; Norrild, B. Activities of E7 promoters in the human papillomavirus type 16 genome during cell differentiation. Virus Res. 2010, 150, 34–42. [Google Scholar] [CrossRef]
- Ozbun, M.A.; Meyers, C. Two Novel Promoters in the Upstream Regulatory Region of Human Papillomavirus Type 31b Are Negatively Regulated by Epithelial Differentiation. J. Virol. 1999, 73, 3505–3510. [Google Scholar] [CrossRef] [PubMed]
- Hummel, M.; Hudson, J.B.; Laimins, L.A. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 1992, 66, 6070–6080. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Ryan, G.B.; Knight, G.L.; Laimins, L.A.; Roberts, S. The full-length E1∧E4 protein of human papillomavirus type 18 modulates differentiation-dependent viral DNA amplification and late gene expression. Virology 2007, 362, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Genther, S.M.; Sterling, S.; Duensing, S.; Munger, K.; Sattler, C.; Lambert, P.F. Quantitative Role of the Human Papillomavirus Type 16 E5 Gene during the Productive Stage of the Viral Life Cycle. J. Virol. 2003, 77, 2832–2842. [Google Scholar] [CrossRef]
- Bryan, J.T.; Brown, D.R. Association of the Human Papillomavirus Type 11 E1∧E4 Protein with Cornified Cell Envelopes Derived from Infected Genital Epithelium. Virology 2000, 277, 262–269. [Google Scholar] [CrossRef]
- Wang, X.; Meyers, C.; Wang, H.-K.; Chow, L.T.; Zheng, Z.-M. Construction of a Full Transcription Map of Human Papillomavirus Type 18 during Productive Viral Infection. J. Virol. 2011, 85, 8080–8092. [Google Scholar] [CrossRef]
- Graham, S.V. Keratinocyte Differentiation-Dependent Human Papillomavirus Gene Regulation. Viruses 2017, 9, 245. [Google Scholar] [CrossRef]
- Nilsson, K.; Wu, C.; Schwartz, S. Role of the DNA Damage Response in Human Papillomavirus RNA Splicing and Polyadenylation. Int. J. Mol. Sci. 2018, 19, 1735. [Google Scholar] [CrossRef]
- Crum, C.P.; Nuovo, G.; Friedman, D.; Silverstein, S.J. Accumulation of RNA homologous to human papillomavirus type 16 open reading frames in genital precancers. J. Virol. 1988, 62, 84–90. [Google Scholar] [CrossRef]
- Stoler, M.H.; Wolinsky, S.; Whitbeck, A.; Broker, T.R.; Chow, L.T. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 1989, 172, 331–340. [Google Scholar] [CrossRef]
- Beyer-Finkler, E.; Stoler, M.; Girardi, F.; Pfister, H. Cell differentiation-related gene expression of human papillomavirus. Med. Microbiol. Immunol. 1990, 179, 185–192. [Google Scholar] [CrossRef]
- Stoler, M.H.; Rhodes, C.R.; Whitbeck, A.; Wolinsky, S.; Chow, L.T.; Broker, T.R. Human papillomavirus type 16 and 18 gene expression in cervical neoplasias. Hum. Pathol. 1992, 23, 117–128. [Google Scholar] [CrossRef]
- Kennedy, I.M.; Haddow, J.K.; Clements, J.B. Analysis of human papillomavirus type 16 late mRNA 3′ processing signals in vitro and in vivo. J. Virol. 1990, 64, 1825–1829. [Google Scholar] [CrossRef]
- Kennedy, I.M.; Haddow, J.K.; Clements, J.B. A negative regulatory element in the human papillomavirus type 16 genome acts at the level of late mRNA stability. J. Virol. 1991, 65, 2093–2097. [Google Scholar] [CrossRef]
- Sokolowski, M.; Zhao, C.; Tan, W.; Schwartz, S. AU-rich mRNA instability elements on human papillomavirus type 1 late mRNAs and c-fos mRNAs interact with the same cellular factors. Oncogene 1997, 15, 2303–2319. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sokolowski, M.; Tan, W.; Schwartz, S. Characterisation and partial purification of cellular factors interacting with a negative element on human papillomavirus type 1 late mRNAs. Virus Res. 1998, 55, 1–13. [Google Scholar] [CrossRef]
- Sokolowski, M.; Furneaux, H.; Schwartz, S. The inhibitory activity of the AU-rich RNA element in the human papillomavirus type 1 late 3′ untranslated region correlates with its affinity for the elav-like HuR protein. J. Virol. 1999, 73, 1080–1091. [Google Scholar] [CrossRef]
- Koffa, M.D.; Graham, S.V.; Takagaki, Y.; Manley, J.L.; Clements, J.B. The human papillomavirus type 16 negative regulatory RNA element interacts with three proteins that act at different posttranscriptional levels. Proc. Natl. Acad. Sci. USA 2000, 97, 4677–4682. [Google Scholar] [CrossRef]
- Cumming, S.; Chuen-Im, T.; Zhang, J.; Graham, S.V. The RNA stability regulator HuR regulates L1 protein expression in vivo in differentiating cervical epithelial cells. Virology 2009, 383, 142–149. [Google Scholar] [CrossRef]
- Jolly, C.E.; Gray, L.J.; Parish, J.L.; Lain, S.; Herrington, C.S. Leptomycin B induces apoptosis in cells containing the whole HPV 16 genome. Int. J. Oncol. 2009, 35, 649–656. [Google Scholar] [CrossRef]
- Steel, J.; Lowen, A.C. Influenza A Virus Reassortment. In Influenza Pathogenesis and Control-Volume I.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 377–401. [Google Scholar]
- Smith, G.J.D.; Bahl, J.; Vijaykrishna, D.; Zhang, A.J.X.; Poon, L.L.M.; Chen, H.; Webster, R.; Peiris, J.S.M.; Guan, Y. Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. USA 2009, 106, 11709–11712. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.T.-Y.; Zhu, H.; Wang, J.; Smith, D.K.; Holmes, E.C.; Webster, R.G.; Webby, R.; Peiris, J.M.; Guan, Y. Reassortment Events among Swine Influenza A Viruses in China: Implications for the Origin of the 2009 Influenza Pandemic. J. Virol. 2011, 85, 10279–10285. [Google Scholar] [CrossRef] [PubMed]
- Fang, R.; Jou, W.M.; Huylebroeck, D.; Devos, R.; Fiers, W. Complete structure of A/duck/Ukraine/63 influenza hemagglutinin gene: Animal virus as progenitor of human H3 Hong Kong 1968 influenza hemagglutinin. Cell 1981, 25, 315–323. [Google Scholar] [CrossRef]
- Saunders-Hastings, P.; Krewski, D. Reviewing the History of Pandemic Influenza: Understanding Patterns of Emergence and Transmission. Pathogens 2016, 5, 66. [Google Scholar] [CrossRef]
- Nickol, M.E.; Kindrachuk, J. A year of terror and a century of reflection: Perspectives on the great influenza pandemic of 1918–1919. BMC Infect. Dis. 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Krug, R.M.; Broni, B.A.; Bouloy, M. Are the 5′ ends of influenza viral mrnas synthesized in vivo donated by host mRNAs? Cell 1979, 18, 329–334. [Google Scholar] [CrossRef]
- Fodor, E.; Pritlove, D.C.; Brownlee, G.G. The influenza virus panhandle is involved in the initiation of transcription. J. Virol. 1994, 68, 4092–4096. [Google Scholar] [CrossRef]
- Engelhardt, O.G.; Smith, M.; Fodor, E. Association of the Influenza A Virus RNA-Dependent RNA Polymerase with Cellular RNA Polymerase II. J. Virol. 2005, 79, 5812–5818. [Google Scholar] [CrossRef]
- De Vlugt, C.; Sikora, D.; Pelchat, M. Insight into Influenza: A Virus Cap-Snatching. Viruses 2018, 10, 641. [Google Scholar] [CrossRef]
- Inglis, S.C.; Brown, C.M. Spliced and unspliced RNAs encoded by virion RNA segment 7 of influenza virus. Nucleic Acids Res. 1981, 9, 2727–2740. [Google Scholar] [CrossRef]
- Lamb, R.A.; Lai, C.-J. Spliced and unspliced messenger RNAs synthesized from cloned influenza virus M DNA in an SV40 vector: Expression of the influenza virus membrane protein (M1). Virology 1982, 123, 237–256. [Google Scholar] [CrossRef]
- Lamb, R.A.; Choppin, P.W.; Chanock, R.M.; Lai, C.J. Mapping of the two overlapping genes for polypeptides NS1 and NS2 on RNA segment 8 of influenza virus genome. Proc. Natl. Acad. Sci. USA 1980, 77, 1857–1861. [Google Scholar] [CrossRef]
- Jackson, D.A.; Caton, A.J.; McCready, S.J.; Cook, P.R. Influenza virus RNA is synthesized at fixed sites in the nucleus. Nature 1982, 296, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Turrell, L.; Lyall, J.W.; Tiley, L.S.; Fodor, E.; Vreede, F.T. The role and assembly mechanism of nucleoprotein in influenza A virus ribonucleoprotein complexes. Nat. Commun. 2013, 4, 1591. [Google Scholar] [CrossRef]
- Resa-Infante, P.; Recuero-Checa, M.A.; Zamarreño, N.; Llorca, O.; Ortín, J. Structural and Functional Characterization of an Influenza Virus RNA Polymerase-Genomic RNA Complex. J. Virol. 2010, 84, 10477–10487. [Google Scholar] [CrossRef] [PubMed]
- Arranz, R.; Coloma, R.; Chichon, F.J.; Conesa, J.J.; Carrascosa, J.L.; Valpuesta, J.M.; Ortín, J.; Martín-Benito, J.; Ruiz-Garcia, D. The Structure of Native Influenza Virion Ribonucleoproteins. Science 2012, 338, 1634–1637. [Google Scholar] [CrossRef]
- Read, E.K.C.; Digard, P. Individual influenza a virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J. Gen. Virol. 2010, 91, 1290–1301. [Google Scholar] [CrossRef]
- Larsen, S.; Bui, S.; Pérez, V.; Mohammad, A.; Medina-Ramirez, H.; Newcomb, L.L. Influenza polymerase encoding mRNAs utilize atypical mRNA nuclear export. Virol. J. 2014, 11, 1–11. [Google Scholar] [CrossRef]
- Bier, K.; York, A.; Fodor, E. Cellular cap-binding proteins associate with influenza virus mRNAs. J. Gen. Virol. 2011, 92, 1627–1634. [Google Scholar] [CrossRef]
- Poon, L.L.M.; Pritlove, D.C.; Fodor, E.; Brownlee, G.G. Direct Evidence that the Poly(A) Tail of Influenza A Virus mRNA Is Synthesized by Reiterative Copying of a U Track in the Virion RNA Template. J. Virol. 1999, 73, 3473–3476. [Google Scholar] [CrossRef] [PubMed]
- Amorim, M.J.; Read, E.K.; Dalton, R.M.; Medcalf, L.; Digard, P. Nuclear Export of Influenza a Virus mRNAs Requires Ongoing RNA Polymerase II Activity. Traffic 2007, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mor, A.; White, A.; Zhang, K.; Thompson, M.; Esparza, M.; Muñoz-Moreno, R.; Koide, K.; Lynch, K.W.; García-Sastre, A.; Fontoura, B.M.A. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 2016, 1, 16069. [Google Scholar] [CrossRef] [PubMed]
- Pereira, C.F.; Read, E.K.C.; Wise, H.M.; Amorim, M.J.; Digard, P. Influenza A Virus NS1 Protein Promotes Efficient Nuclear Export of Unspliced Viral M1 mRNA. J. Virol. 2017, 91, e00528-17. [Google Scholar] [CrossRef]
- Pereira, C.F.; Wise, H.M.; Kurian, D.; Pinto, R.M.; Amorim, M.J.; Gill, A.C.; Digard, P. Effects of mutations in the effector domain of influenza A virus NS1 protein. BMC Res. Notes 2018, 11, 673. [Google Scholar] [CrossRef]
- Manzoor, R.; Igarashi, M.; Takada, A. Influenza a Virus M2 Protein: Roles from Ingress to Egress. Int. J. Mol. Sci. 2017, 18, 2649. [Google Scholar] [CrossRef]
- Chiba, S.; Hill-Batorski, L.; Neumann, G.; Kawaoka, Y. The Cellular DExD/H-Box RNA Helicase UAP56 Co-localizes With the Influenza A Virus NS1 Protein. Front. Microbiol. 2018, 9, 2192. [Google Scholar] [CrossRef]
- Esparza, M.; Mor, A.; Niederstrasser, H.; White, K.M.; White, A.; Zhang, K.; Gao, S.; Wang, J.; Liang, J.; Sho, S.; et al. Chemical intervention of influenza virus mRNA nuclear export. PLoS Pathog. 2020, 16, e1008407. [Google Scholar] [CrossRef]
- Robb, N.C.; Jackson, D.; Vreede, F.T.; Fodor, E. Splicing of influenza A virus NS1 mRNA is independent of the viral NS1 protein. J. Gen. Virol. 2010, 91, 2331–2340. [Google Scholar] [CrossRef]
- Ren, X.; Yu, Y.; Li, H.; Huang, J.; Zhou, A.; Liu, S.; Hu, P.; Li, B.; Qi, W.; Liao, M. Avian Influenza A Virus Polymerase Recruits Cellular RNA Helicase eIF4A3 to Promote Viral mRNA Splicing and Spliced mRNA Nuclear Export. Front. Microbiol. 2019, 10, 1625. [Google Scholar] [CrossRef]
- Diot, C.; Fournier, G.; Dos Santos, M.; Magnus, J.; Komarova, A.; Van Der Werf, S.; Munier, S.; Naffakh, N. Influenza A Virus Polymerase Recruits the RNA Helicase DDX19 to Promote the Nuclear Export of Viral mRNAs. Sci. Rep. 2016, 6, 33763. [Google Scholar] [CrossRef] [PubMed]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, R.E.; Talon, J.; Palese, P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998, 17, 288–296. [Google Scholar] [CrossRef]
- Bui, M.; Wills, E.G.; Helenius, A.; Whittaker, G.R. Role of the Influenza Virus M1 Protein in Nuclear Export of Viral Ribonucleoproteins. J. Virol. 2000, 74, 1781–1786. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Liu, X.; Yu, M.; Li, J.; Jia, X.; Bi, Y.; Sun, L.; Gao, G.F.; Liu, W. A Nuclear Export Signal in the Matrix Protein of Influenza A Virus Is Required for Efficient Virus Replication. J. Virol. 2012, 86, 4883–4891. [Google Scholar] [CrossRef]
- Yasuda, J.; Nakada, S.; Kato, A.; Toyoda, T.; Ishihama, A. Molecular Assembly of Influenza Virus: Association of the NS2 Protein with Virion Matrix. Virology 1993, 196, 249–255. [Google Scholar] [CrossRef]
- Richardson, J.C.; Akkina, R.K. NS2 protein of influenza virus is found in purified virus and phosphorylated in infected cells. Arch. Virol. 1991, 116, 69–80. [Google Scholar] [CrossRef]
- Neumann, G.; Hughes, M.T.; Kawaoka, Y. Influenza A virus NS2 protein mediates vRNP nuclear export through NES-independent interaction with hCRM1. EMBO J. 2000, 19, 6751–6758. [Google Scholar] [CrossRef]
- Iwatsuki-Horimoto, K.; Horimoto, T.; Fujii, Y.; Kawaoka, Y. Generation of Influenza A Virus NS2 (NEP) Mutants with an Altered Nuclear Export Signal Sequence. J. Virol. 2004, 78, 10149–10155. [Google Scholar] [CrossRef]
- Yu, M.; Liu, X.; Cao, S.; Zhao, Z.; Zhang, K.; Xie, Q.; Chen, C.; Gao, S.; Bi, Y.; Sun, L.; et al. Identification and Characterization of Three Novel Nuclear Export Signals in the Influenza A Virus Nucleoprotein. J. Virol. 2012, 86, 4970–4980. [Google Scholar] [CrossRef]
- Chen, J.; Huang, S.; Chen, Z. Human cellular protein nucleoporin hNup98 interacts with influenza A virus NS2/nuclear export protein and overexpression of its GLFG repeat domain can inhibit virus propagation. J. Gen. Virol. 2010, 91, 2474–2484. [Google Scholar] [CrossRef] [PubMed]
- Akyazi, B.Ş.; PİRİNÇAL, A.; Kawaguchi, A.; Nagata, K.; Turan, K. Interaction of influenza A virus NS2/NEP protein with the amino-terminal part of Nup214. Turk. J. Biol. 2020, 44, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Chen, J.; Chen, Q.; Wang, H.; Yao, Y.; Chen, Z.; Chen, J.; Chen, J. A Second CRM1-Dependent Nuclear Export Signal in the Influenza A Virus NS2 Protein Contributes to the Nuclear Export of Viral Ribonucleoproteins. J. Virol. 2013, 87, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, M.; Zheng, W.; Liu, W. Nucleocytoplasmic Shuttling of Influenza A Virus Proteins. Viruses 2015, 7, 2668–2682. [Google Scholar] [CrossRef]
- Chutiwitoonchai, N.; Aida, Y. NXT1, a Novel Influenza A NP Binding Protein, Promotes the Nuclear Export of NP via a CRM1-Dependent Pathway. Viruses 2016, 8, 209. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Fang, M.; Chen, X.; Zeng, X. A human cell polarity protein Lgl2 regulates influenza A virus nucleoprotein exportation from nucleus in MDCK cells. J. Biosci. 2020, 45, 1–9. [Google Scholar] [CrossRef]
- Kumar, S.; Yeo, D.; Muralidharan, N.H.; Lai, S.K.; Tong, C.; Tan, B.H.; Sugrue, R. Impaired Nuclear Export of the Ribonucleoprotein Complex and Virus-Induced Cytotoxicity Combine to Restrict Propagation of the A/Duck/Malaysia/02/2001 (H9N2) Virus in Human Airway Cells. Cells 2020, 9, 355. [Google Scholar] [CrossRef]
- Sun, Q.; Chen, X.; Zhou, Q.; Burstein, E.; Yang, S.; Jia, D. Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduct. Target. Ther. 2016, 1, 16010. [Google Scholar] [CrossRef]
- Chari, A.; Vogl, D.T.; Gavriatopoulou, M.; Nooka, A.K.; Yee, A.J.; Huff, C.A.; Moreau, P.; Dingli, D.; Cole, C.; Lonial, S.; et al. Oral Selinexor-Dexamethasone for Triple-Class Refractory Multiple Myeloma. N. Engl. J. Med. 2019, 381, 727–738. [Google Scholar] [CrossRef]
- Perwitasari, O.; Johnson, S.; Yan, X.; Howerth, E.; Shacham, S.; Landesman, Y.; Baloglu, E.; McCauley, D.; Tamir, S.; Tompkins, S.M.; et al. Verdinexor, a Novel Selective Inhibitor of Nuclear Export, Reduces Influenza A Virus Replication In Vitro and In Vivo. J. Virol. 2014, 88, 10228–10243. [Google Scholar] [CrossRef]
- Perwitasari, O.; Johnson, S.; Yan, X.; Register, E.; Crabtree, J.; Gabbard, J.; Howerth, E.; Shacham, S.; Carlson, R.; Tamir, S.; et al. Antiviral Efficacy of Verdinexor In Vivo in Two Animal Models of Influenza A Virus Infection. PLoS ONE 2016, 11, e0167221. [Google Scholar] [CrossRef] [PubMed]
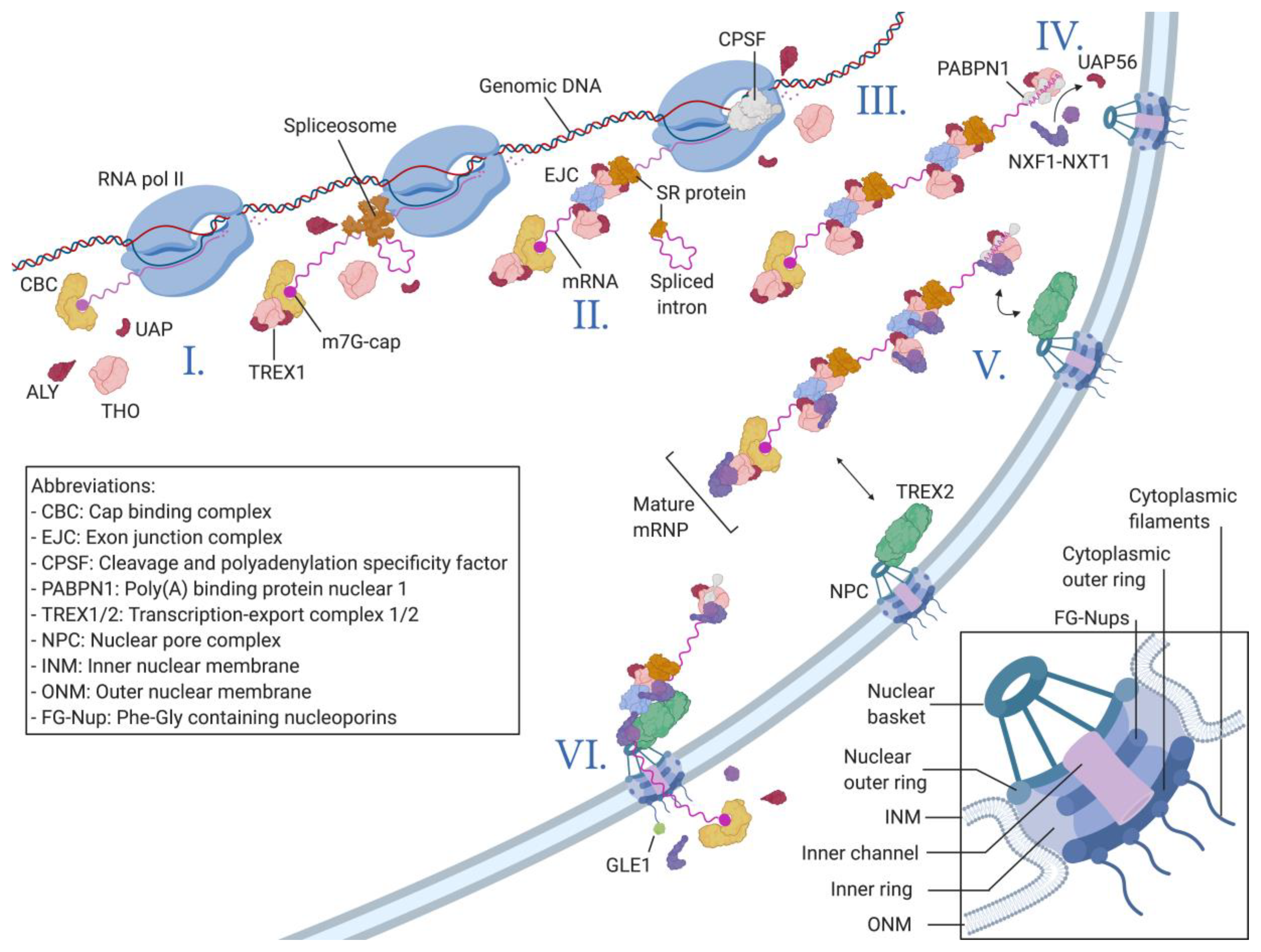
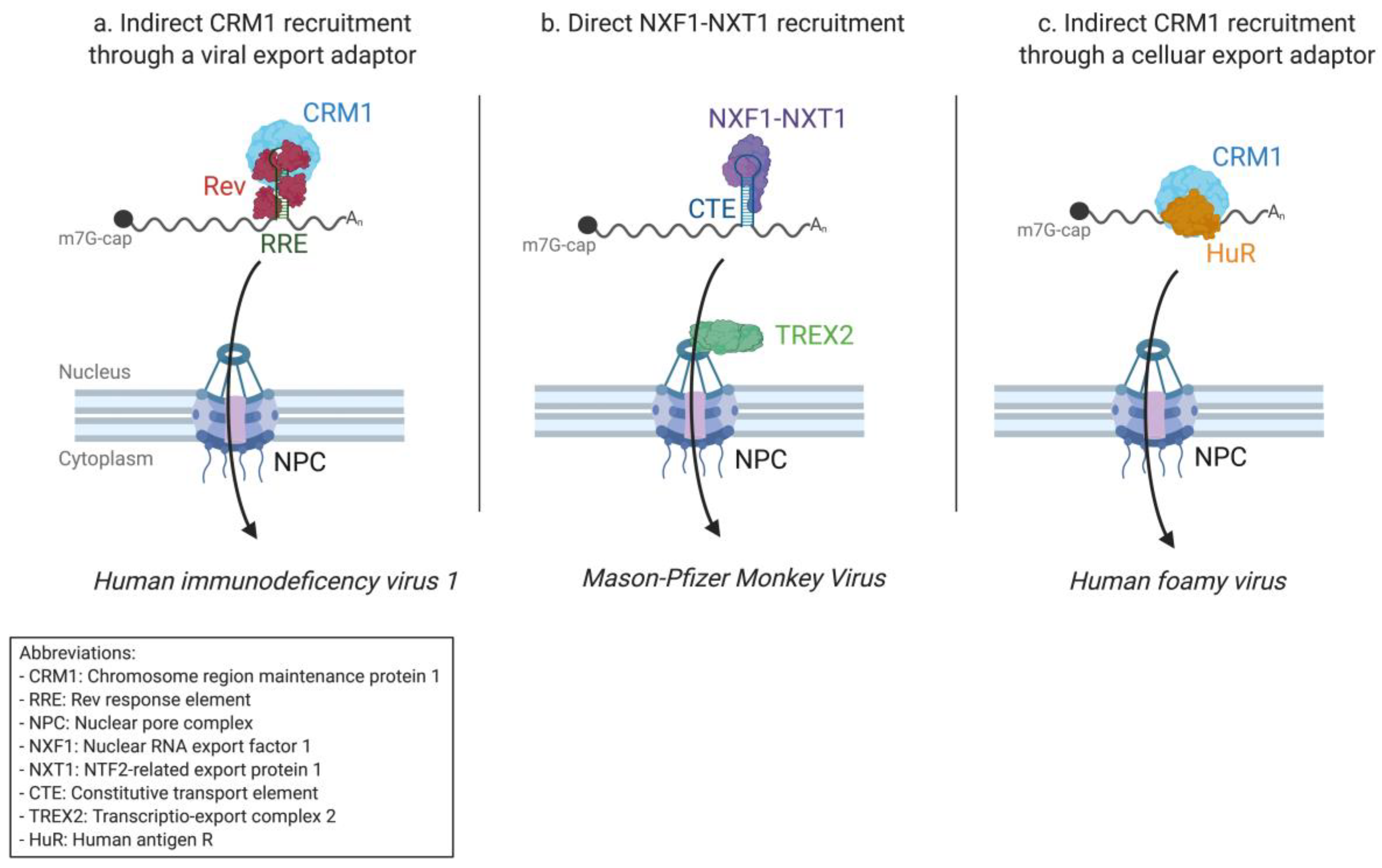


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gales, J.P.; Kubina, J.; Geldreich, A.; Dimitrova, M. Strength in Diversity: Nuclear Export of Viral RNAs. Viruses 2020, 12, 1014. https://doi.org/10.3390/v12091014
Gales JP, Kubina J, Geldreich A, Dimitrova M. Strength in Diversity: Nuclear Export of Viral RNAs. Viruses. 2020; 12(9):1014. https://doi.org/10.3390/v12091014
Chicago/Turabian StyleGales, Jón Pol, Julie Kubina, Angèle Geldreich, and Maria Dimitrova. 2020. "Strength in Diversity: Nuclear Export of Viral RNAs" Viruses 12, no. 9: 1014. https://doi.org/10.3390/v12091014
APA StyleGales, J. P., Kubina, J., Geldreich, A., & Dimitrova, M. (2020). Strength in Diversity: Nuclear Export of Viral RNAs. Viruses, 12(9), 1014. https://doi.org/10.3390/v12091014





