Measuring Alphavirus Fidelity Using Non-Infectious Virus Particles
Abstract
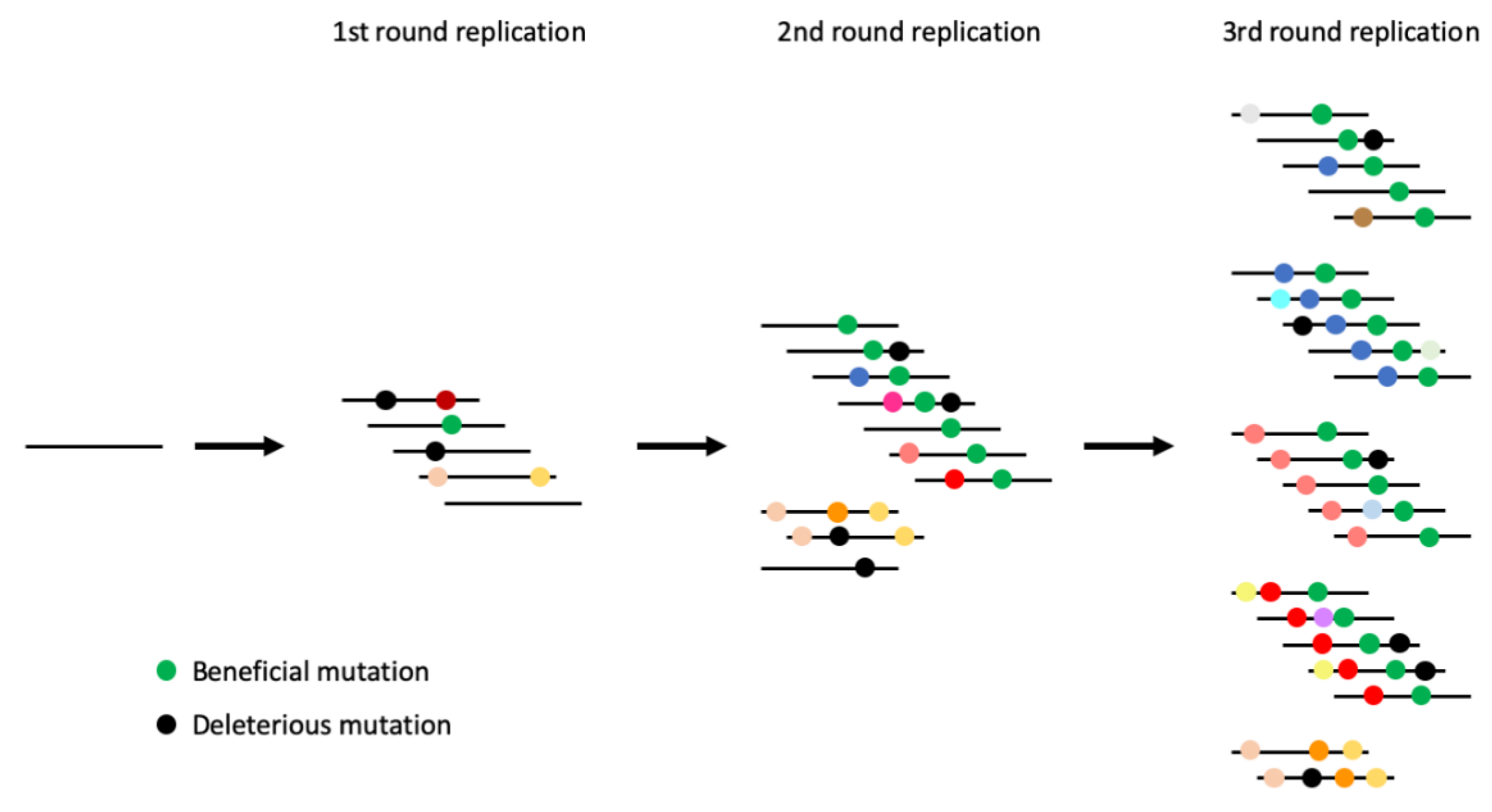
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viruses
2.2. Virus Detection and Replication Assays
2.3. Analysis of Mutation Frequency
2.4. Analysis Counting All Variants (Analysis A)
2.5. Analysis with Downsampled Dataset (Analysis B)
2.6. Statistics
3. Results
3.1. Virus Replication
3.2. Overall Sequence Diversity
3.3. Sequence Diversity across the Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Domingo, E.; Escarmís, C.; Lázaro, E.; Manrubia, S.C. Quasispecies dynamics and RNA virus extinction. Virus. Res. 2005, 107, 129–139. [Google Scholar] [CrossRef]
- Lauring, A.S.; Andino, R. Quasispecies Theory and the Behavior of RNA Viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef]
- Rozen-Gagnon, K.; Stapleford, K.A.; Mongelli, V.; Blanc, H.; Failloux, A.-B.; Saleh, M.-C.; Vignuzzi, M. Alphavirus Mutator Variants Present Host-Specific Defects and Attenuation in Mammalian and Insect Models. PLoS Pathog. 2014, 10, e1003877. [Google Scholar] [CrossRef]
- McDonald, S.; Block, A.; Beaucourt, S.; Moratorio, G.; Vignuzzi, M.; Peersen, O.B. Design of a Genetically Stable High Fidelity Coxsackievirus B3 Polymerase That Attenuates Virus Growth in Vivo. J. Biol. Chem. 2016, 291, 13999–14011. [Google Scholar] [CrossRef]
- Korboukh, V.K.; Lee, C.A.; Acevedo, A.; Vignuzzi, M.; Xiao, Y.; Arnold, J.J.; Hemperly, S.; Graci, J.D.; August, A.; Andino, R.; et al. RNA Virus Population Diversity, an Optimum for Maximal Fitness and Virulence. J. Biol. Chem. 2014, 289, 29531–29544. [Google Scholar] [CrossRef]
- Gnädig, N.F.; Beaucourt, S.; Campagnola, G.; Bordería, A.V.; Sanz-Ramos, M.; Gong, P.; Blanc, H.; Peersen, O.B.; Vignuzzi, M. Coxsackievirus B3 mutator strains are attenuated in vivo. Proc. Natl. Acad. Sci. USA 2012, 109, E2294–E2303. [Google Scholar] [CrossRef]
- Coffey, L.L.; Beeharry, Y.; Bordería, A.V.; Blanc, H.; Vignuzzi, M. Arbovirus high fidelity variant loses fitness in mosquitoes and mice. Proc. Natl. Acad. Sci. USA 2011, 108, 16038–16043. [Google Scholar] [CrossRef]
- Vignuzzi, M.; Wendt, E.; Andino, R. Engineering attenuated virus vaccines by controlling replication fidelity. Nat. Med. 2008, 14, 154–161. [Google Scholar] [CrossRef]
- Pfeiffer, J.K.; Kirkegaard, K. Increased Fidelity Reduces Poliovirus Fitness and Virulence under Selective Pressure in Mice. PLoS Pathog. 2005, 1, e11. [Google Scholar] [CrossRef]
- Meng, T.; Kwang, J. Attenuation of Human Enterovirus 71 High-Replication-Fidelity Variants in AG129 Mice. J. Virol. 2014, 88, 5803–5815. [Google Scholar] [CrossRef]
- Xie, X.; Wang, H.; Zeng, J.; Li, C.; Zhou, G.; Yang, D.; Yu, L. Foot-and-mouth disease virus low-fidelity polymerase mutants are attenuated. Arch. Virol. 2014, 159, 2641–2650. [Google Scholar] [CrossRef] [PubMed]
- Kautz, T.F.; Guerbois, M.; Khanipov, K.; Patterson, E.I.; Langsjoen, R.M.; Yun, R.; Warmbrod, K.L.; Fofanov, Y.; Weaver, S.C.; Forrester, N.L. Low-fidelity Venezuelan equine encephalitis virus polymerase mutants to improve live-attenuated vaccine safety and efficacy. Virus. Evol. 2018, 4, vey004. [Google Scholar] [CrossRef] [PubMed]
- Pringle, C. Genetic characteristics of conditional lethal mutants of vesicular stomatitis virus induced by 5-fluorouracil, 5-azacytidine, and ethyl methane sulfonate. J. Virol. 1970, 5, 559–567. [Google Scholar] [CrossRef]
- Pathak, V.; Temin, H. 5-Azacytidine and RNA secondary structure increase the retrovirus mutation rate. J. Virol. 1992, 66, 3093–3100. [Google Scholar] [CrossRef] [PubMed]
- Crotty, S.; Cameron, C.E.; Andino, R. RNA virus error catastrophe: Direct molecular test by using ribavirin. Proc. Natl. Acad. Sci. USA 2001, 98, 6895–6900. [Google Scholar] [CrossRef]
- Crotty, S.; Maag, D.; Arnold, J.J.; Zhong, W.; Lau, J.Y.; Hong, Z.; Andino, R.; Cameron, C.E. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 2000, 6, 1375–1379. [Google Scholar] [CrossRef]
- Warmbrod, L.K.; Patterson, E.I.; Kautz, T.F.; Stanton, A.; Rockx-Brouwer, D.; Kalveram, B.K.; Khanipov, K.; Thangamani, S.; Fofanov, Y.; Forrester, N.L. Viral RNA-dependent RNA polymerase mutants display an altered mutation spectrum resulting in attenuation in both mosquito and vertebrate hosts. Plos Pathog. 2019, 15, e1007610. [Google Scholar] [CrossRef]
- Pfeiffer, J.K.; Kirkegaard, K. A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc. Natl. Acad. Sci. USA 2003, 100, 7289–7294. [Google Scholar] [CrossRef]
- Arias, A.; Arnold, J.J.; Sierra, M.; Smidansky, E.D.; Domingo, E.; Cameron, C.E. Determinants of RNA-Dependent RNA Polymerase (In)fidelity Revealed by Kinetic Analysis of the Polymerase Encoded by a Foot-and-Mouth Disease Virus Mutant with Reduced Sensitivity to Ribavirin▿. J. Virol. 2008, 82, 12346–12355. [Google Scholar] [CrossRef]
- Slyke, G.A.; Arnold, J.J.; Lugo, A.J.; Griesemer, S.B.; Moustafa, I.M.; Kramer, L.D.; Cameron, C.E.; Ciota, A.T. Sequence-Specific Fidelity Alterations Associated with West Nile Virus Attenuation in Mosquitoes. PLoS Pathog. 2015, 11, e1005009. [Google Scholar]
- Griesemer, S.B.; Kramer, L.D.; Slyke, G.A.; Pata, J.D.; Gohara, D.W.; Cameron, C.E.; Ciota, A.T. Mutagen resistance and mutation restriction of St. Louis encephalitis virus. J. Gen. Virol. 2017, 98, 201–211. [Google Scholar] [CrossRef]
- Pauly, M.D.; Lyons, D.M.; Fitzsimmons, W.J.; Lauring, A.S. Epistatic Interactions within the Influenza A Virus Polymerase Complex Mediate Mutagen Resistance and Replication Fidelity. Msphere 2017, 2, e00323-17. [Google Scholar] [CrossRef]
- Kautz, T.F.; Forrester, N.L. RNA Virus Fidelity Mutants: A Useful Tool for Evolutionary Biology or a Complex Challenge? Viruses 2018, 10, 600. [Google Scholar] [CrossRef]
- Poirier, E.Z.; Mounce, B.C.; Rozen-Gagnon, K.; Hooikaas, P.J.; Stapleford, K.A.; Moratorio, G.; Vignuzzi, M. Low-fidelity polymerases of alphaviruses recombine at higher rates to overproduce defective interfering particles. J. Virol. 2015, 90, 2446–2454. [Google Scholar] [CrossRef]
- Salminen, A.; Wahlberg, J.; Lobigs, M.; Liljeström, P.; Garoff, H. Membrane fusion process of Semliki Forest virus. II: Cleavage-dependent reorganization of the spike protein complex controls virus entry. J. Cell Biol. 1992, 116, 349–357. [Google Scholar] [CrossRef]
- Heidner, H.; McKnight, K.; Davis, N.; Johnston, R. Lethality of PE2 incorporation into Sindbis virus can be suppressed by second-site mutations in E3 and E2. J. Virol. 1994, 68, 2683–2692. [Google Scholar] [CrossRef]
- Davis, N.L.; Brown, K.W.; Greenwald, G.F.; Zajac, A.J.; Zacny, V.L.; Smith, J.F.; Johnston, R.E. Attenuated Mutants of Venezuelan Equine Encephalitis Virus Containing Lethal Mutations in the PE2 Cleavage Signal Combined with a Second-Site Suppressor Mutation in E1. Virology 1995, 212, 102–110. [Google Scholar] [CrossRef]
- Smith, D.R.; Adams, P.A.; Kenney, J.L.; Wang, E.; Weaver, S.C. Venezuelan equine encephalitis virus in the mosquito vector Aedes taeniorhynchus: Infection initiated by a small number of susceptible epithelial cells and a population bottleneck. Virology 2008, 372, 176–186. [Google Scholar] [CrossRef]
- Duffus, W.; Levy-Mintz, P.; Klimjack, M.; Kielian, M. Mutations in the putative fusion peptide of Semliki Forest virus affect spike protein oligomerization and virus assembly. J. Virol. 1995, 69, 2471–2479. [Google Scholar] [CrossRef]
- Kielian, M.; Klimjack, M.; Ghosh, S.; Duffus, W. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J. Cell Biol. 1996, 134, 863–872. [Google Scholar] [CrossRef]
- Patterson, E.I.; Warmbrod, K.L.; Bouyer, D.H.; Forrester, N.L. Evaluation of the inactivation of Venezuelan equine encephalitis virus by several common methods. J. Virol. Methods 2018, 254, 31–34. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 May 2020).
- Kinney, R.M.; Johnson, B.J.; Welch, J.B.; Tsuchiya, K.R.; Trent, D.W. The full-length nucleotide sequences of the virulent Trinidad donkey strain of Venezuelan equine encephalitis virus and its attenuated vaccine derivative, strain TC-83. Virology 1989, 170, 19–30. [Google Scholar] [CrossRef]
- Albayrak, L.; Khanipov, K.; Pimenova, M.; Golovko, G.; Rojas, M.; Pavlidis, I.; Chumakov, S.; Aguilar, G.; Chávez, A.; Widger, W.R.; et al. The ability of human nuclear DNA to cause false positive low-abundance heteroplasmy calls varies across the mitochondrial genome. BMC Genom. 2016, 17, 1017. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Moratorio, G.; Henningsson, R.; Barbezange, C.; Carrau, L.; Bordería, A.V.; Blanc, H.; Beaucourt, S.; Poirier, E.Z.; Vallet, T.; Boussier, J.; et al. Attenuation of RNA viruses by redirecting their evolution in sequence space. Nat. Microbiol. 2017, 2, 17088. [Google Scholar] [CrossRef]
- Vignuzzi, M.; Stone, J.K.; Arnold, J.J.; Cameron, C.E.; Andino, R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 2006, 439, 344. [Google Scholar] [CrossRef]
- Riemersma, K.K.; Steiner, C.; Singapuri, A.; Coffey, L.L. Chikungunya Virus Fidelity Variants Exhibit Differential Attenuation and Population Diversity in Cell Culture and Adult Mice. J. Virol. 2018, 93, e01606-18. [Google Scholar] [CrossRef]
- Dubuisson, J.; Rice, C. Sindbis virus attachment: Isolation and characterization of mutants with impaired binding to vertebrate cells. J. Virol. 1993, 67, 3363–3374. [Google Scholar] [CrossRef]
- Sebastian-Martin, A.; Barrioluengo, V.; Menendez-Arias, L. Transcriptional inaccuracy threshold attenuates differences in RNA-dependent DNA synthesis fidelity between retroviral reverse transciptases. Sci. Rep. 2018, 8, 627. [Google Scholar] [CrossRef]
- Peng, Q.; Satya, R.V.; Lewis, M.; Randad, P.; Wang, Y. Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genom. 2015, 15, 589. [Google Scholar] [CrossRef]
- Patterson, E.I.; Khanipov, K.; Rojas, M.M.; Kautz, T.F.; Rockx-Brouwer, D.; Golovko, G.; Albayrak, L.; Fofanov, Y.; Forrester, N.L. Mosquito bottlenecks alter viral mutant swarm in a tissue and time-dependent manner with contraction and expansion of variant positions and diversity. Virus. Evol. 2018, 4, vey001. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Beane, J.E.; Ebel, G.D. RNAi Targeting of West Nile Virus in Mosquito Midguts Promotes Virus Diversification. PLoS Pathog. 2009, 5, e1000502. [Google Scholar] [CrossRef] [PubMed]
- Myles, K.M.; Morazzani, E.M.; Adelman, Z.N. Origins of alphavirus-derived small RNAs in mosquitoes. RNA Biol. 2009, 6, 387–391. [Google Scholar] [CrossRef]
- Sim, S.; Aw, P.P.; Wilm, A.; Teoh, G.; Hue, K.; Nguyen, N.; Nagarajan, N.; Simmons, C.P.; Hibberd, M.L. Tracking Dengue Virus Intra-host Genetic Diversity during Human-to-Mosquito Transmission. PLoS Neglect. Trop. D 2015, 9, e0004052. [Google Scholar] [CrossRef]
- Stapleford, K.A.; Moratorio, G.; Henningsson, R.; Chen, R.; Matheus, S.; Enfissi, A.; Weissglas-Volkov, D.; Isakov, O.; Blanc, H.; Mounce, B.C.; et al. Whole-Genome Sequencing Analysis from the Chikungunya Virus Caribbean Outbreak Reveals Novel Evolutionary Genomic Elements. PLoS Neglect. Trop. D 2016, 10, e0004402. [Google Scholar] [CrossRef]
- Gotte, B.; Liu, L.; McInerney, G.M. The enigmatic Alphavirus non-structural protein 3 (nsP3) revealing its secrets at last. Viruses 2018, 10, 105. [Google Scholar] [CrossRef]
- Collins, N.D.; Beck, A.S.; Widen, S.G.; Wood, T.G.; Higgs, S.; Barrett, A.D. Structural and nonstructural genes contribute to the genetic diversity of RNA viruses. mBio 2018, 9, e01871-18. [Google Scholar] [CrossRef]


| Mutation | Gene | TC-83 E3Δ56-59 | TC-83 E3Δ56-59 nsP4-G14R | TC-83 E3Δ56-59 nsP4-A96T | TC-83 E3Δ56-59 nsP4-C488Y |
|---|---|---|---|---|---|
| C5540A | nsP3 | 0.0778 (0.1348) | 0.1316 (0.2111) | 0.1273 (0.2013) | 0.1371 (0.2025) |
| C5555A | nsP3 | 0.0253 (0.1621) | 0.0287 (0.2084) | 0.0355 (0.1937) | 0.0326 (0.1987) |
| A7653C | capsid | 0.0426 (0.0549) | 0.0277 (0.0350) | 0.0270 (0.0371) | 0.0328 (0.0367) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patterson, E.I.; Khanipov, K.; Swetnam, D.M.; Walsdorf, S.; Kautz, T.F.; Thangamani, S.; Fofanov, Y.; Forrester, N.L. Measuring Alphavirus Fidelity Using Non-Infectious Virus Particles. Viruses 2020, 12, 546. https://doi.org/10.3390/v12050546
Patterson EI, Khanipov K, Swetnam DM, Walsdorf S, Kautz TF, Thangamani S, Fofanov Y, Forrester NL. Measuring Alphavirus Fidelity Using Non-Infectious Virus Particles. Viruses. 2020; 12(5):546. https://doi.org/10.3390/v12050546
Chicago/Turabian StylePatterson, Edward I., Kamil Khanipov, Daniele M. Swetnam, Samantha Walsdorf, Tiffany F. Kautz, Saravanan Thangamani, Yuriy Fofanov, and Naomi L. Forrester. 2020. "Measuring Alphavirus Fidelity Using Non-Infectious Virus Particles" Viruses 12, no. 5: 546. https://doi.org/10.3390/v12050546
APA StylePatterson, E. I., Khanipov, K., Swetnam, D. M., Walsdorf, S., Kautz, T. F., Thangamani, S., Fofanov, Y., & Forrester, N. L. (2020). Measuring Alphavirus Fidelity Using Non-Infectious Virus Particles. Viruses, 12(5), 546. https://doi.org/10.3390/v12050546





