Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Virus
2.3. Plaque Assay
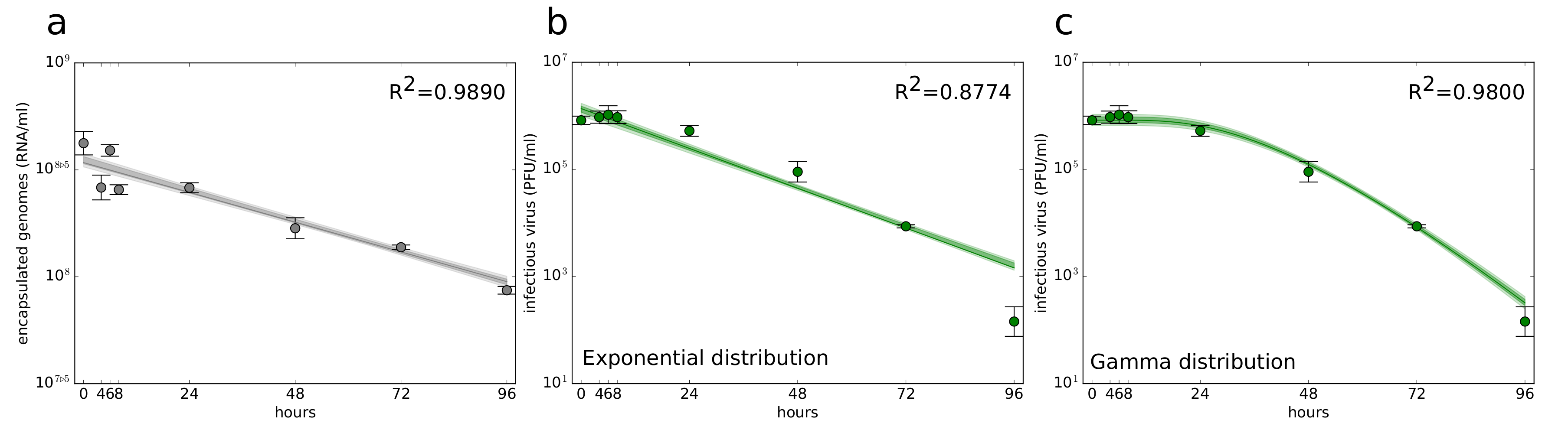
2.4. Decay Curves
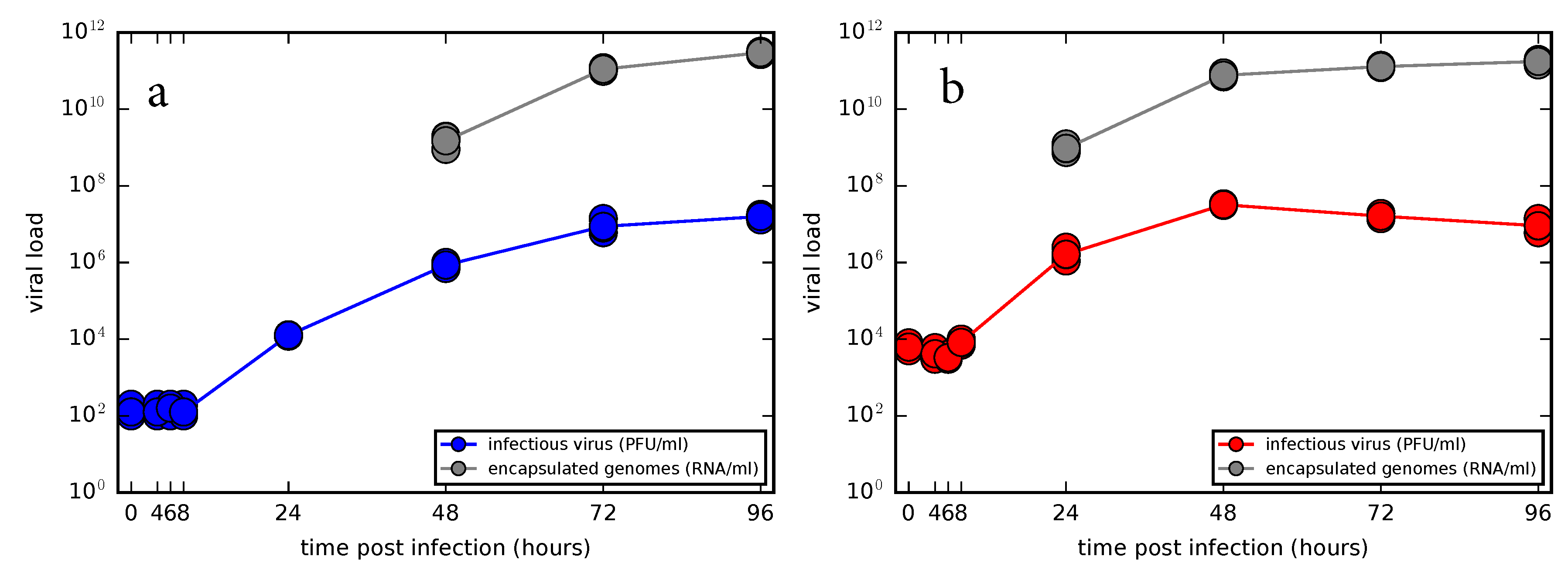
2.5. Growth Curves
2.6. RT-qPCR
2.7. Degradation of Encapsulated Genome and Infectious Virus
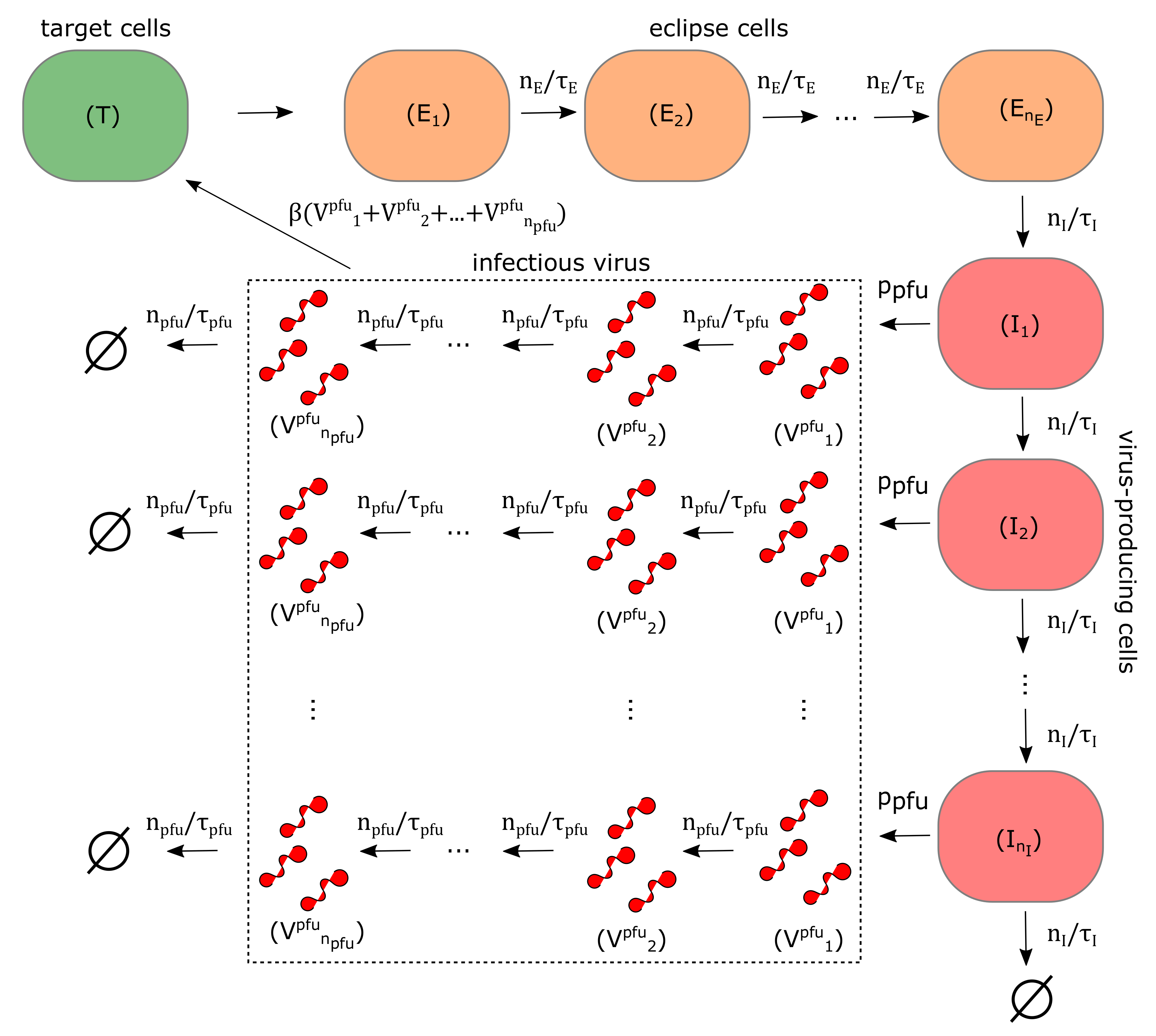
2.8. Mathematical Model of ZIKV In Vitro Kinetics
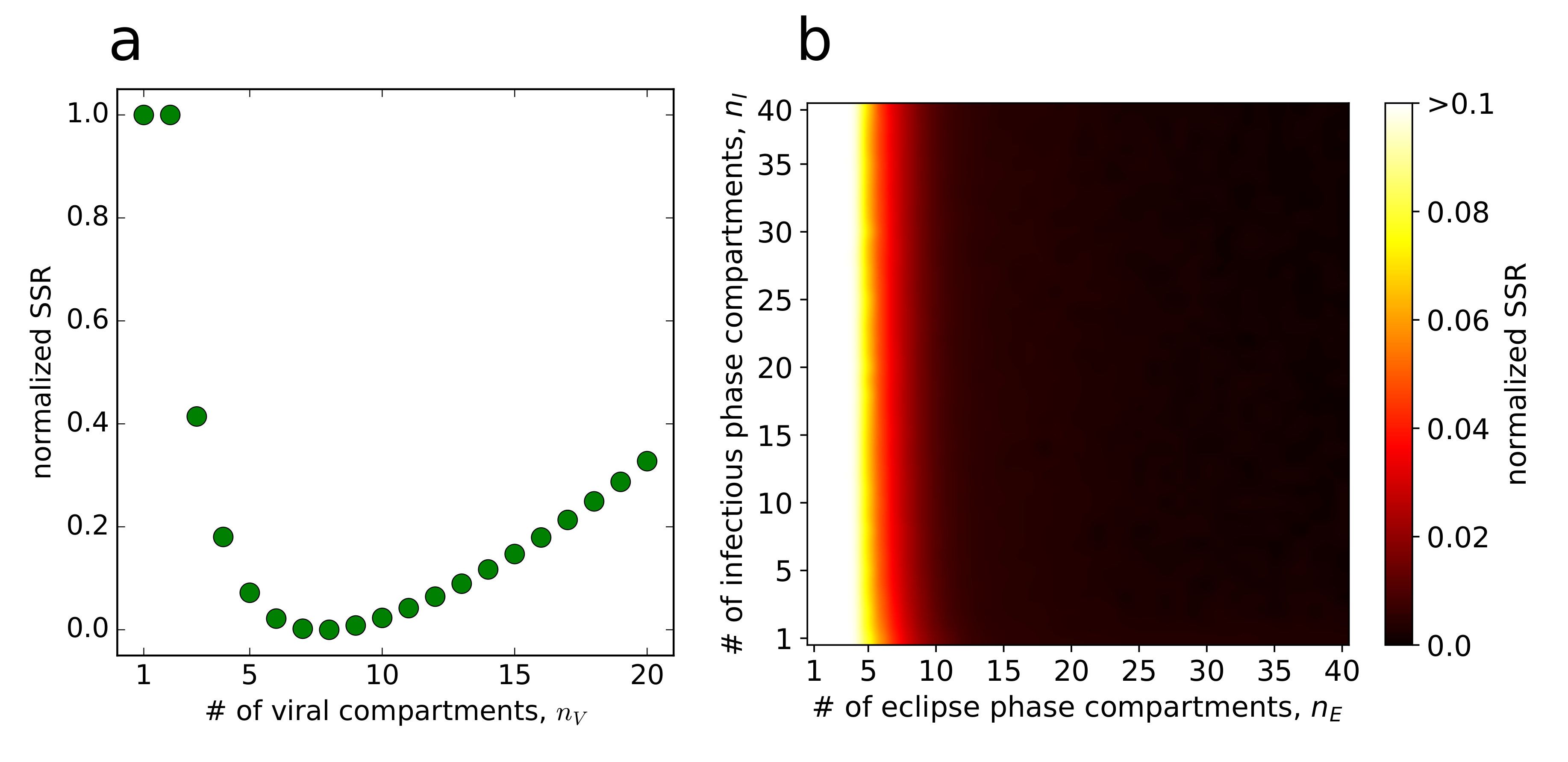
2.9. Selection of Data Points for Parameter Estimation
3. Results
3.1. Quantification of ZIKV Stability Determinants
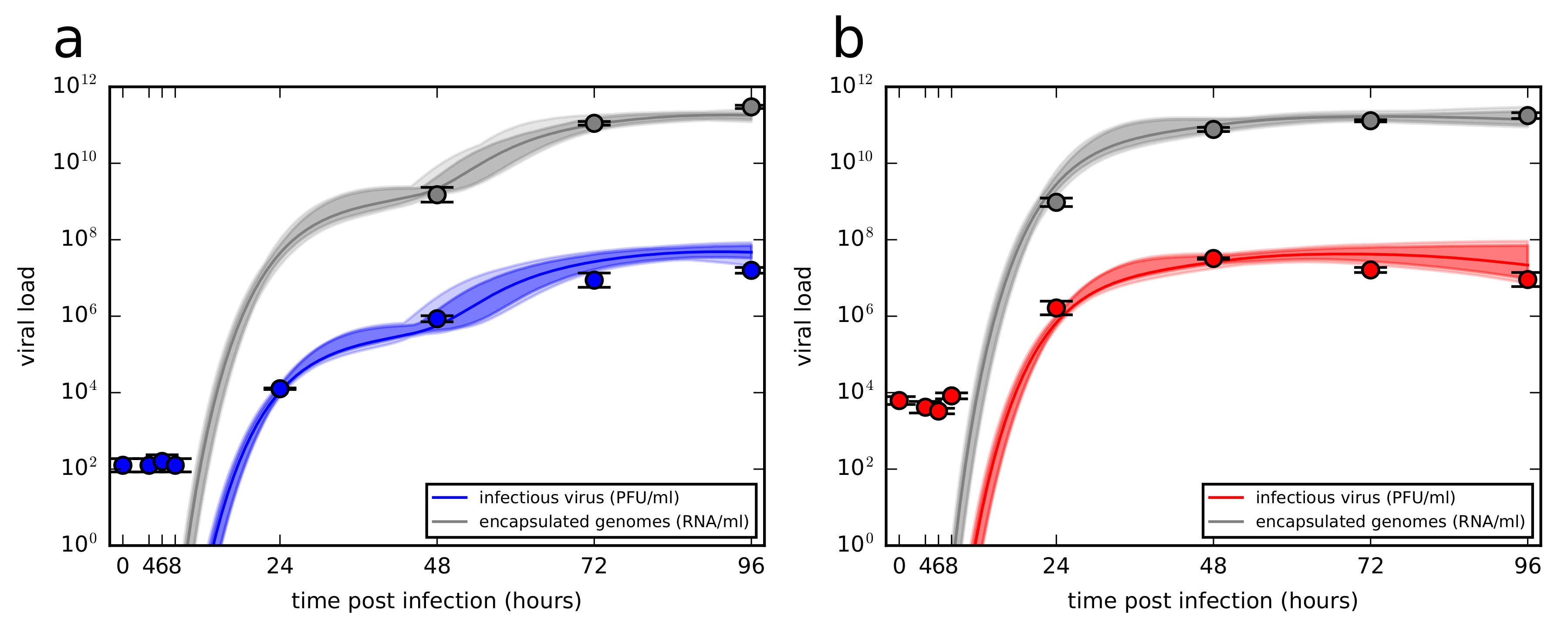
3.2. Experimental Time Course Kinetics of ZIKV Infection In Vitro
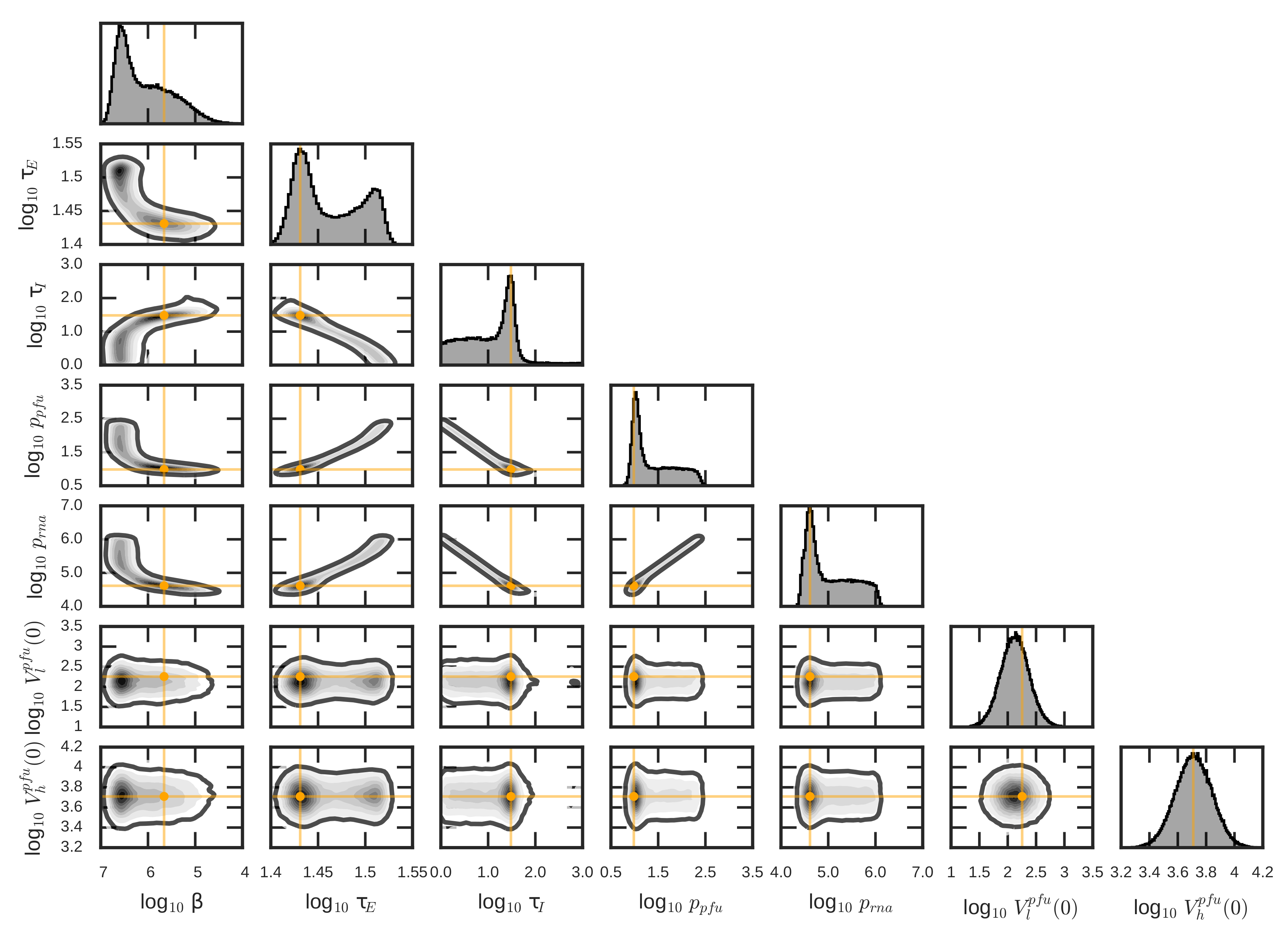
3.3. Quantification of ZIKV Life-Cycle Determinants
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ZIKV | Zika virus |
| MOI | multiplicity of infection |
| RT-qPCR | reverse transcription quantitative polymerase chain reaction |
References
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus (I). Isolations and serological specificity. Trans. R. Soc. Trop. Med. Hyg. 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Lanciotti, R.S.; Kosoy, O.L.; Laven, J.J.; Velez, J.O.; Lambert, A.J.; Johnson, A.J.; Stanfield, S.M.; Duffy, M.R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg. Infect. Dis. 2008, 14, 1232. [Google Scholar] [CrossRef]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.P.; Musso, D.; Ghawche, F. Zika virus infection complicated by Guillain-Barre syndrome—Case report, French Polynesia, December 2013. Eurosurveillance 2014, 19, 20720. [Google Scholar] [CrossRef]
- Dyer, O. Zika virus spreads across Americas as concerns mount over birth defects. BMJ 2015, 351, h6983. [Google Scholar] [CrossRef]
- Parra, B.; Lizarazo, J.; Jiménez-Arango, J.A.; Zea-Vera, A.F.; González-Manrique, G.; Vargas, J.; Angarita, J.A.; Zuñiga, G.; Lopez-Gonzalez, R.; Beltran, C.L.; et al. Guillain–Barré syndrome associated with Zika virus infection in Colombia. N. Engl. J. Med. 2016, 375, 1513–1523. [Google Scholar] [CrossRef]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika virus associated with microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. Rapid Risk Assessment: Zika Virus Epidemic in the Americas: Potential Association with Microcephaly and Guillain-Barré Syndrome; European Centre for Disease Prevention and Control: Solna, Sweden, 2015. [Google Scholar]
- Ioos, S.; Malletm, H.P.; Goffartm, I.L.; Gauthier, V.; Cardoso, T.; Herida, M. Current Zika virus epidemiology and recent epidemics. Med. Maladies Infect. 2014, 44, 302–307. [Google Scholar] [CrossRef]
- Hiatt, C.W. Kinetics of the inactivation of viruses. Bacteriol. Rev. 1964, 2, 150–163. [Google Scholar] [CrossRef]
- Beauchemin, C.A.A.; Kim, Y.I.; Yu, Q.; Ciaramella, G.; DeVincenzo, J.P. Uncovering critical properties of the human respiratory syncytial virus by combining in vitro assays and in silico analyses. PLoS ONE 2019, 14, e0214708. [Google Scholar] [CrossRef]
- Schmid, B.; Rinas, M.; Ruggieri, A.; Acosta, E.G.; Bartenschlager, M.; Reuter, A.; Fischl, W.; Harder, N.; Bergeest, J.P.; Flossdorf, M.; et al. Live cell analysis and mathematical modeling identify determinants of attenuation of dengue virus 2’-O-methylation mutant. PLoS Pathog. 2015, 11. [Google Scholar] [CrossRef]
- Kostyuchenko, V.A.; Lim, E.X.; Zhang, S.; Fibriansah, G.; Ng, T.S.; Ooi, J.S.; Shi, J.; Lok, S.M. Structure of the thermally stable Zika virus. Nature 2016, 533, 425–428. [Google Scholar] [CrossRef]
- Goo, L.; Dowd, K.A.; Smith, A.R.; Pelc, R.S.; DeMaso, C.R.; Pierson, T.C. Zika virus is not uniquely stable at physiological temperatures compared to other flaviviruses. mBio 2016, 7, e01396-16. [Google Scholar] [CrossRef]
- Ansarah-Sobrinho, C.; Nelson, S.; Jost, C.A.; Whitehead, S.S.; Pierson, T.C. Temperature-dependent production of pseudoinfectious dengue reporter virus particles by complementation. Virology 2008, 381, 67–74. [Google Scholar] [CrossRef]
- Dowd, K.A.; Jost, C.A.; Durbin, A.P.; Whitehead, S.S.; Pierson, T.C. A dynamic landscape for antibody binding modulates antibody-mediated neutralization of West Nile virus. PLoS Pathog. 2011, 7. [Google Scholar] [CrossRef]
- Manning, J.S.; Collins, J.K. Effects of cell culture and laboratory conditions on type 2 dengue virus infectivity. J. Clin. Microbiol. 1979, 10, 235–239. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, H.; Miao, H.; Perelson, A.S. Parameter identifiability and estimation of HIV/AIDS dynamic models. Bull. Math. Biol. 2008, 70, 785–799. [Google Scholar] [CrossRef]
- Verotta, D. Models and estimation methods for clinical HIV-1 data. J. Comput. Appl. Math. 2005, 184, 275–300. [Google Scholar] [CrossRef]
- Miao, H.; Dykes, C.; Demeter, L.M.; Cavenaugh, J.; Park, S.Y.; Perelson, A.S.; Wu, H. Modeling and estimation of kinetic parameters and replicative fitness of HIV-1 from flow-cytometry-based growth competition experiments. Bull. Math. Biol. 2008, 70, 1749–1771. [Google Scholar] [CrossRef]
- Kakizoe, Y.; Nakaoka, S.; Beauchemin, C.A.; Morita, S.; Mori, H.; Igarashi, T.; Aihara, K.; Miura, T.; Iwami, S. A method to determine the duration of the eclipse phase for in vitro infection with a highly pathogenic SHIV strain. Sci. Rep. 2015, 5, 10371. [Google Scholar] [CrossRef]
- Beauchemin, C.A.; Miura, T.; Iwami, S. Duration of SHIV production by infected cells is not exponentially distributed: Implications for estimates of infection parameters and antiviral efficacy. Sci. Rep. 2017, 7, 42765. [Google Scholar] [CrossRef]
- Perelson, A.S.; Ribeiro, R.M. Modeling the within-host dynamics of HIV infection. BMC Biol. 2013, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Guedj, J.; Perelson, A.S. Mathematical modeling of HCV infection: What can it teach us in the era of direct antiviral agents? Antivir. Ther. 2012, 17, 1171. [Google Scholar] [CrossRef] [PubMed]
- Aston, P. A new model for the dynamics of hepatitis C infection: Derivation, analysis and implications. Viruses 2018, 10, 195. [Google Scholar] [CrossRef]
- Rihan, F.A.; Sheek-Hussein, M.; Tridane, A.; Yafia, R. Dynamics of hepatitis C virus infection: Mathematical modeling and parameter estimation. Math. Model. Nat. Phenom. 2017, 12, 33–47. [Google Scholar] [CrossRef]
- Arthur, J.G.; Tran, H.T.; Aston, P. Feasibility of parameter estimation in hepatitis C viral dynamics models. J. Inverse Ill-Posed Probl. 2017, 25, 69–80. [Google Scholar] [CrossRef]
- Perelson, A.S.; Ribeiro, R.M. Estimating drug efficacy and viral dynamic parameters: HIV and HCV. Stat. Med. 2008, 27, 4647–4657. [Google Scholar] [CrossRef]
- Krakauer, D.C.; Komarova, N.L. Levels of selection in positive-strand virus dynamics. J. Evol. Biol. 2003, 16, 64–73. [Google Scholar] [CrossRef]
- Regoes, R.R.; Crotty, S.; Antia, R.; Tanaka, M.M. Optimal replication of poliovirus within cells. Am. Nat. 2005, 165, 364–373. [Google Scholar] [CrossRef]
- Schulte, M.B.; Draghi, J.A.; Plotkin, J.B.; Andino, R. Experimentally guided models reveal replication principles that shape the mutation distribution of RNA viruses. Elife 2015, 4, e03753. [Google Scholar] [CrossRef]
- Boianelli, A.; Nguyen, V.; Ebensen, T.; Schulze, K.; Wilk, E.; Sharma, N.; Stegemann-Koniszewski, S.; Bruder, D.; Toapanta, F.; Guzmán, C.; et al. Modeling influenza virus infection: A roadmap for influenza research. Viruses 2015, 7, 5274–5304. [Google Scholar] [CrossRef]
- Pinilla, L.T.; Holder, B.P.; Abed, Y.; Boivin, G.; Beauchemin, C.A. The H275Y neuraminidase mutation of the pandemic A/H1N1 influenza virus lengthens the eclipse phase and reduces viral output of infected cells, potentially compromising fitness in ferrets. J. Virol. 2012, 86, 10651–10660. [Google Scholar] [CrossRef] [PubMed]
- Holder, B.P.; Beauchemin, C.A. Exploring the effect of biological delays in kinetic models of influenza within a host or cell culture. BMC Public Health 2011, 11, S10. [Google Scholar] [CrossRef] [PubMed]
- Holder, B.P.; Liao, L.E.; Simon, P.; Boivin, G.; Beauchemin, C.A. Design considerations in building in silico equivalents of common experimental influenza virus assays. Autoimmunity 2011, 44, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Simon, P.F.; de La Vega, M.A.; Paradis, É.; Mendoza, E.; Coombs, K.M.; Kobasa, D.; Beauchemin, C.A. Avian influenza viruses that cause highly virulent infections in humans exhibit distinct replicative properties in contrast to human H1N1 viruses. Sci. Rep. 2016, 6, 24154. [Google Scholar] [CrossRef]
- Paradis, E.G.; Pinilla, L.T.; Holder, B.P.; Abed, Y.; Boivin, G.; Beauchemin, C.A. Impact of the H275Y and I223V mutations in the neuraminidase of the 2009 pandemic influenza virus in vitro and evaluating experimental reproducibility. PLoS ONE 2015, 10, e0126115. [Google Scholar] [CrossRef]
- Iwami, S.; Holder, B.P.; Beauchemin, C.A.A.; Morita, S.; Tada, T.; Sato, K.; Igarashi, T.; Miura, T. Quantification system for the viral dynamics of a highly pathogenic simian/human immunodeficiency virus based on an in vitro experiment and a mathematical model. Retrovirology 2012, 9, 18. [Google Scholar] [CrossRef]
- Banerjee, S.; Guedj, J.; Ribeiro, R.M.; Moses, M.; Perelson, A.S. Estimating biologically relevant parameters under uncertainty for experimental within-host murine West Nile virus infection. J. R. Soc. Interface 2016, 13, 20160130. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Binder, S.C.; Boianelli, A.; Meyer-Hermann, M.; Hernandez-Vargas, E.A. Ebola virus infection modeling and identifiability problems. Front. Microbiol. 2015, 6, 257. [Google Scholar] [CrossRef]
- Nguyen, V.K.; Hernandez-Vargas, E.A. Windows of opportunity for Ebola virus infection treatment and vaccination. Sci. Rep. 2017, 7, 8975. [Google Scholar] [CrossRef]
- Desmyter, J.; Melnick, J.L.; Rawls, W.E. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 1968, 2, 955–961. [Google Scholar] [CrossRef]
- Osada, N.; Kohara, A.; Yamaji, T.; Hirayama, N.; Kasai, F.; Sekizuka, T.; Kuroda, M.; Hanada, K. The genome landscape of the African green monkey kidney-derived Vero cell line. DNA Res. 2014, 21, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Pastorino, B.; Bessaud, M.; Grandadam, M.; Murri, S.; Tolou, H.J.; Peyrefitte, C.N. Development of a TaqMan® RT-PCR assay without RNA extraction step for the detection and quantification of African Chikungunya viruses. J. Virol. Methods 2005, 124, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, T.B.; Bangham, C.R. Cycles, chaos, and evolution in virus cultures: A model of defective interfering particles. Proc. Natl. Acad. Sci. USA 1994, 91, 8685–8689. [Google Scholar] [CrossRef] [PubMed]
- Liao, L.E.; Iwami, S.; Beauchemin, C.A.A. (In)validating experimentally derived knowledge about influenza A defective interfering particles. J. R. Soc. Interface 2016, 13, 20160412. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Mathematical Ideas in Biology; Cambridge University Press: Cambridge, UK, 1968; Volume 550. [Google Scholar]
- Macdonald, R.D.; Yamamoto, T.; Fedorak, P. Statistical prediction of the number of cells surviving infection by an autointerfering virus using the Poisson distribution. J. Theor. Biol. 1977, 68, 355–363. [Google Scholar] [CrossRef]
- Foreman-Mackey, D.; Hogg, D.W.; Lang, D.; Goodman, J. emcee: The MCMC hammer. Publ. Astron. Soc. Pac. 2013, 125, 306–312. [Google Scholar] [CrossRef]
- Goodman, J.; Weare, J. Ensemble samplers with affine invariance. Commun. Appl. Math. Comput. Sci. 2010, 5, 65–80. [Google Scholar] [CrossRef]
- Lloyd, A.L. The dependence of viral parameter estimates on the assumed viral life cycle: Limitations of studies of viral load data. Proc. R. Soc. B 2001, 268, 847–854. [Google Scholar] [CrossRef]
- Mittler, J.E.; Sulzer, B.; Neumann, A.U.; Perelson, A.S. Influence of delayed viral production on viral dynamics in HIV-1 infected patients. Math. Biosci. 1998, 152, 143–163. [Google Scholar] [CrossRef]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika virus infection in human skin cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef]
- Turpin, J.; Frumence, E.; Desprès, P.; Viranaicken, W.; Krejbich-Trotot, P. The Zika virus delays cell death through the anti-apoptotic Bcl-2 family proteins. Cells 2019, 8, 1338. [Google Scholar] [CrossRef] [PubMed]
- Dixit, N.M.; Perelson, A.S. HIV dynamics with multiple infections of target cells. Proc. Natl. Acad. Sci. USA 2005, 102, 8198–8203. [Google Scholar] [CrossRef] [PubMed]
- Cummings, K.W.; Levy, D.N.; Wodarz, D. Increased burst size in multiply infected cells can alter basic virus dynamics. Biol. Direct 2012, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Heldt, F.S.; Frensing, T.; Pflugmacher, A.; Gröpler, R.; Peschel, B.; Reichl, U. Multiscale modeling of influenza A virus infection supports the development of direct-acting antivirals. PLoS Comput. Biol. 2013, 9, e1003372. [Google Scholar] [CrossRef]
- Moser, L.A.; Boylan, B.T.; Moreira, F.R.; Myers, L.J.; Svenson, E.L.; Fedorova, N.B.; Pickett, B.E.; Bernard, K.A. Growth and adaptation of Zika virus in mammalian and mosquito cells. PLoS Negl. Trop. Dis. 2018, 12, e0006880. [Google Scholar] [CrossRef]
- Willard, K.A.; Demakovsky, L.; Tesla, B.; Goodfellow, F.T.; Stice, S.L.; Murdock, C.C.; Brindley, M.A. Zika virus exhibits lineage-specific phenotypes in cell culture, in Aedes aegypti mosquitoes, and in an embryo model. Viruses 2017, 9, 383. [Google Scholar] [CrossRef]
- Smith, D.R.; Sprague, T.R.; Hollidge, B.S.; Valdez, S.M.; Padilla, S.L.; Bellanca, S.A.; Golden, J.W.; Coyne, S.R.; Kulesh, D.A.; Miller, L.J.; et al. African and Asian Zika virus isolates display phenotypic differences both in vitro and in vivo. Am. J. Trop. Med. Hyg. 2018, 98, 432–444. [Google Scholar] [CrossRef]
| Forward primer (5 to 3) | TCGTTGCCCAACACAAG |
| Reverse primer (5 to 3) | CCACTAATGTTCTTTTGCAGACAT |
| Probe (5 [6-FAM] to 3) | GCCTACCTTGACAAGCAATCAGACACTCA |
| Parameter | Description | Units | Value | 95% CrR |
|---|---|---|---|---|
| decay time of encapsulated genomes | h | 74.86 | [70.31, 76.43] | |
| initial concentration of encapsulated genomes | RNA/mL | 3.42 | [3.38, 3.65] |
| Parameter | Description | Units | Value | 95% CrR | ||
|---|---|---|---|---|---|---|
| decay time of infectious virus | h | 14.02 | 39.55 | [13.87, 14.73] | [38.93, 40.22] | |
| initial concentration of infectious virus | PFU/mL | 13.64 | 8.30 | [11.64, 15.17] | [7.46, 9.55] | |
| Parameter | Description | Units | Value | 95% CrR |
|---|---|---|---|---|
| rate of infection by infectious virus | mL/(PFU × h) | 2.19 | [0.165, 15.15] | |
| length of eclipse phase | h | 27 | [25.94, 33.13] | |
| length of infectious phase | h | 30.41 | [1.171, 191.07] | |
| infectious virus production rate | PFU/(cell × mL × h) | 9.65 | [8.05, 214.12] | |
| encapsulated genome production rate | RNA/(cell × mL × h) | 4.11 | [2.752, 99.37] | |
| residual infectious virus in low MOI infection | PFU/mL | 1.80 | [0.44, 4.17] | |
| residual infectious virus in high MOI infection | PFU/mL | 5.12 | [2.88, 9.13] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernhauerová, V.; Rezelj, V.V.; Vignuzzi, M. Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection. Viruses 2020, 12, 547. https://doi.org/10.3390/v12050547
Bernhauerová V, Rezelj VV, Vignuzzi M. Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection. Viruses. 2020; 12(5):547. https://doi.org/10.3390/v12050547
Chicago/Turabian StyleBernhauerová, Veronika, Veronica V. Rezelj, and Marco Vignuzzi. 2020. "Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection" Viruses 12, no. 5: 547. https://doi.org/10.3390/v12050547
APA StyleBernhauerová, V., Rezelj, V. V., & Vignuzzi, M. (2020). Modelling Degradation and Replication Kinetics of the Zika Virus In Vitro Infection. Viruses, 12(5), 547. https://doi.org/10.3390/v12050547





