Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure
Abstract
1. Introduction
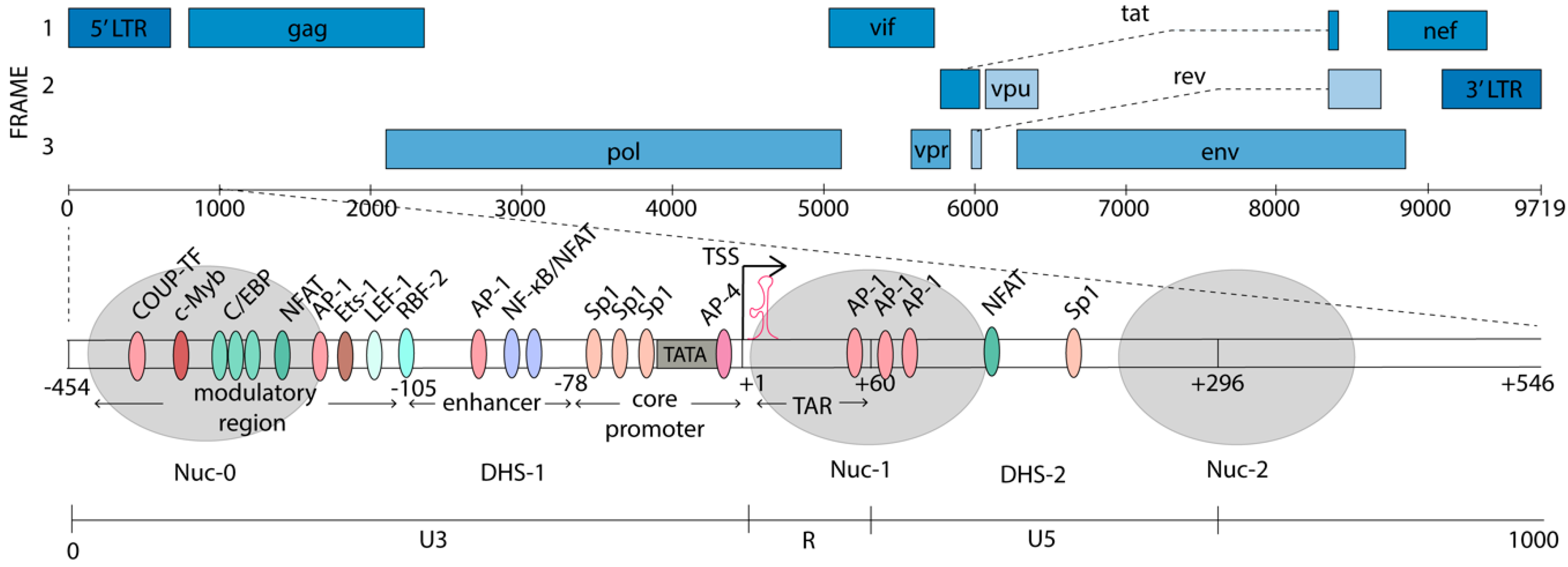
2. HIV-1 Promoter Structure
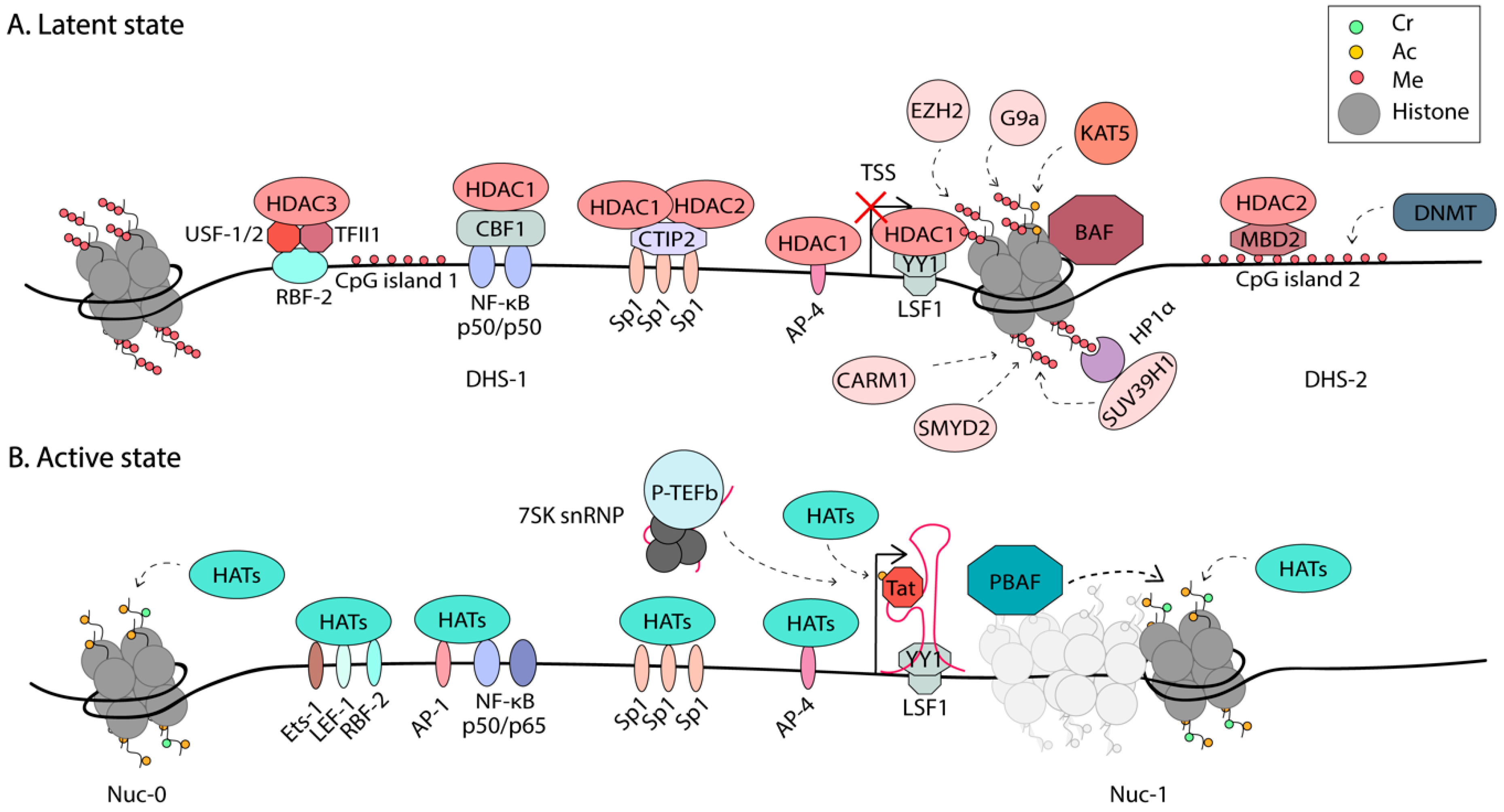
3. Epigenetic Modulation of HIV-1 Latency
3.1. Chromatin Remodeling
3.2. Post-Translational Modifications of Histones
3.3. DNA Methylation of the HIV-1 Genome
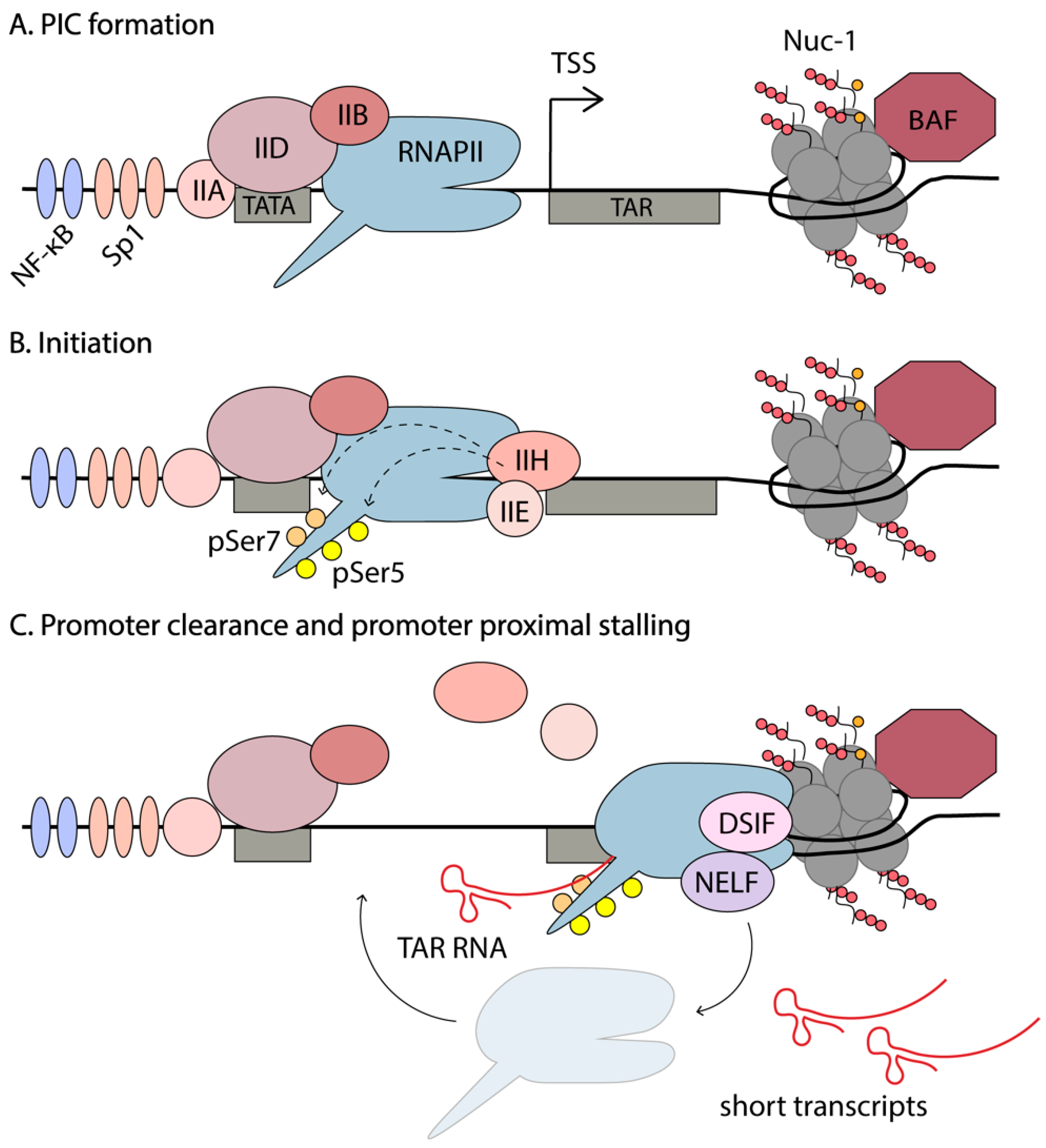
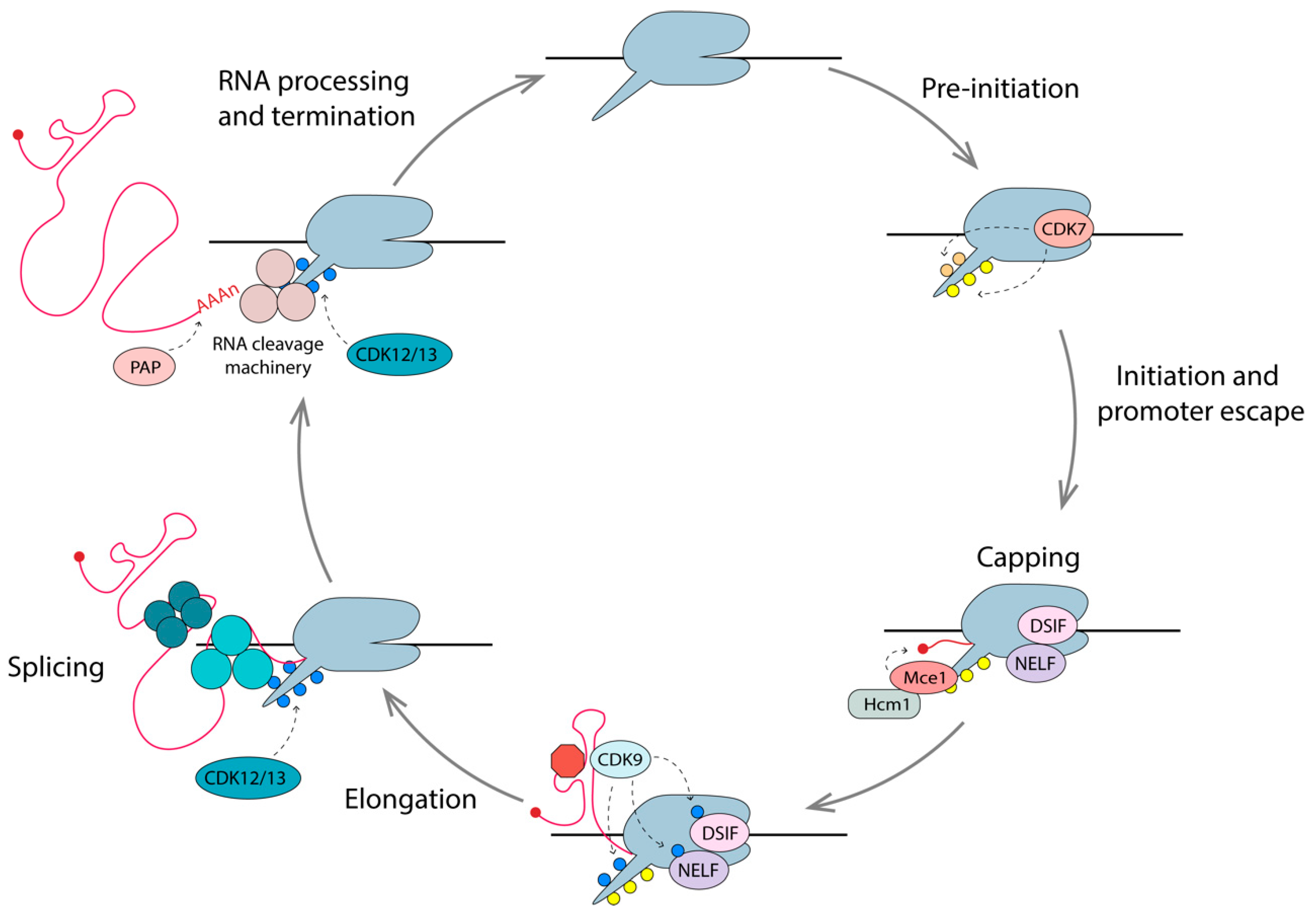
4. Early Stages of Transcription
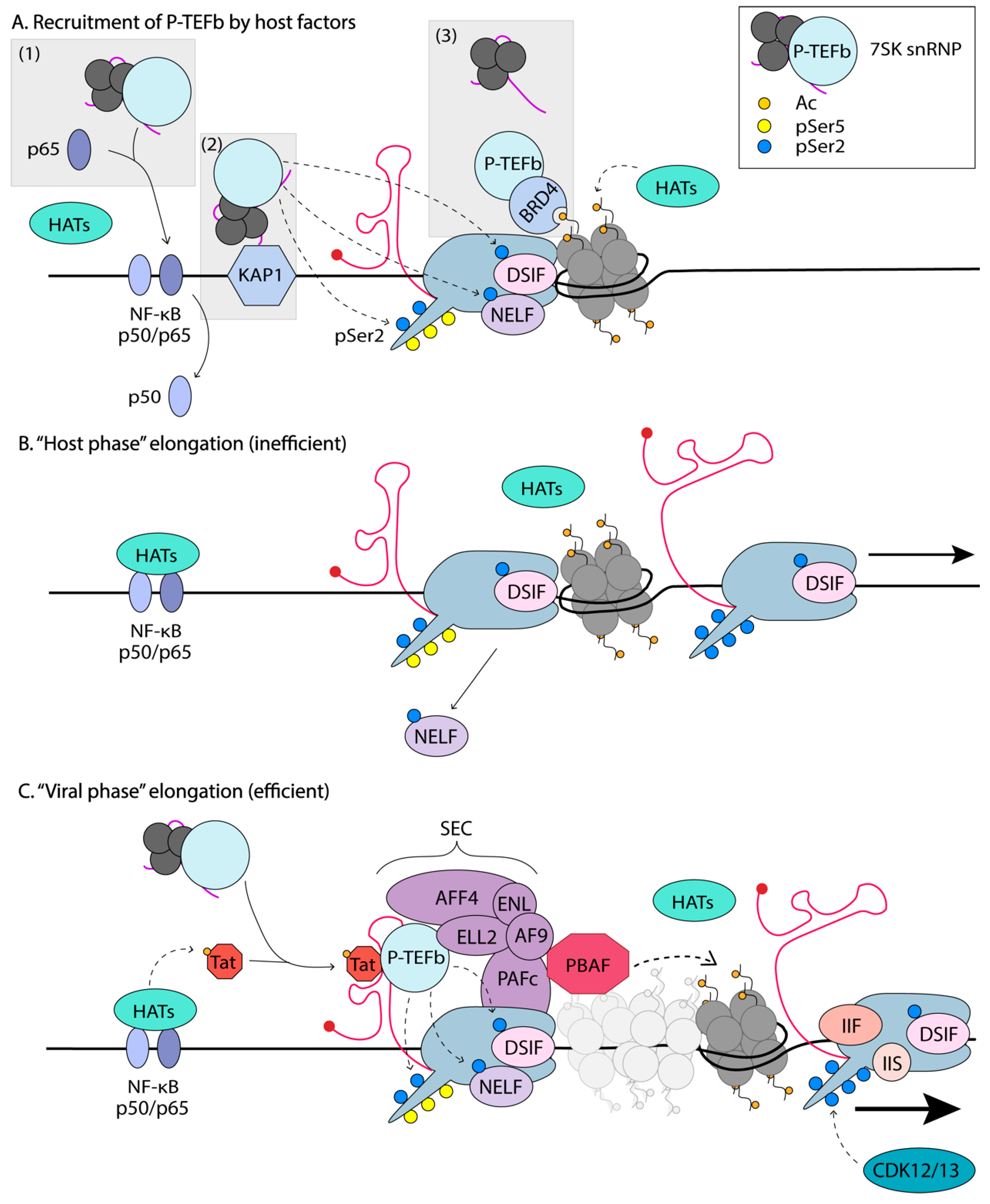
5. Elongation of Transcription
5.1. P-TEFb Regulation
5.2. Delivery of P-TEFb to Promoter-Proximal Paused RNA Polymerase II
6. Events Coupled to Elongation
6.1. Capping
6.2. Splicing
6.3. Pre-mRNA Cleavage and Polyadenylation
7. Therapeutic Strategies and Targets, Recent Advances
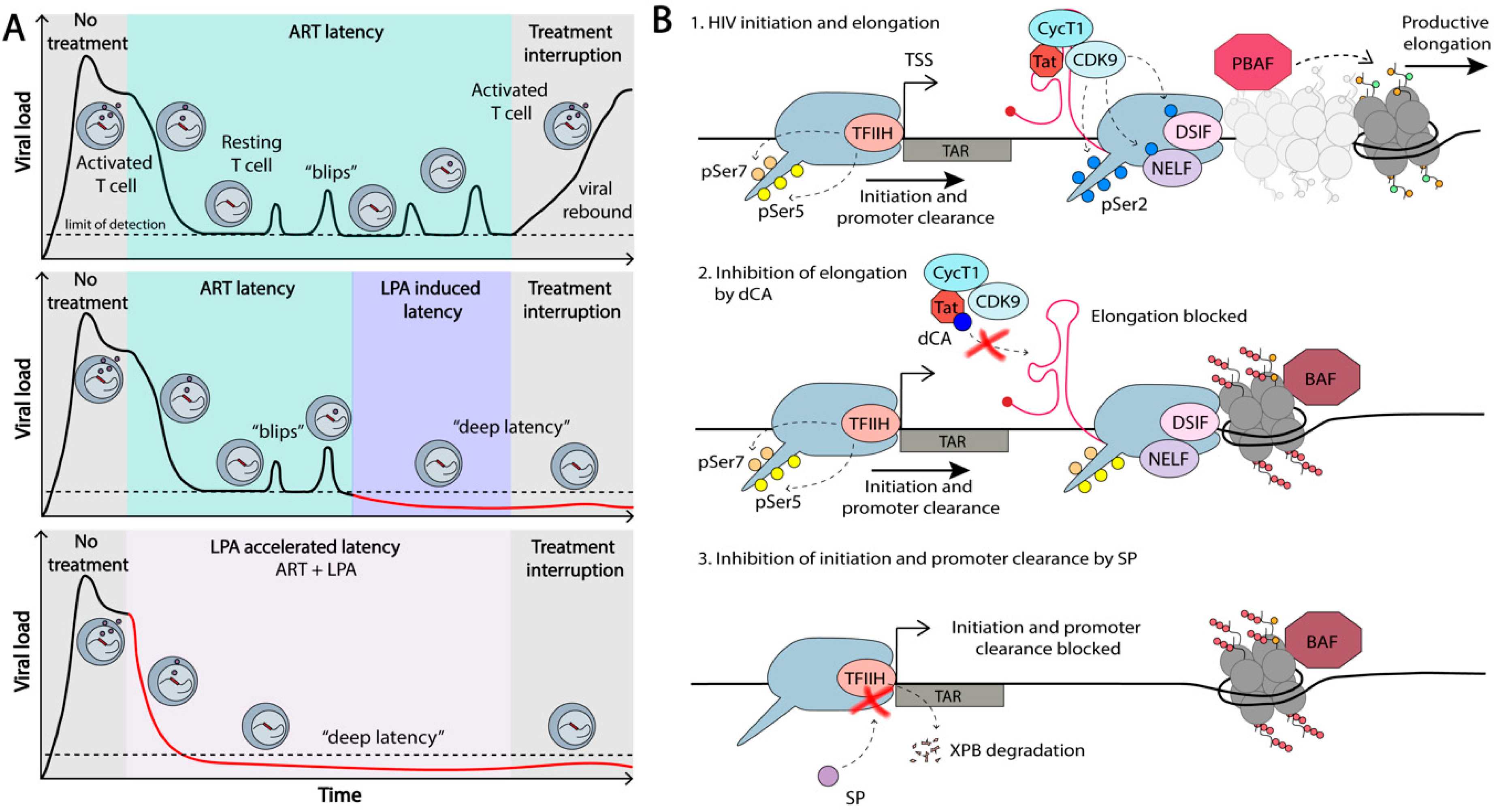
7.1. Shock-and-Kill Approach
7.2. Block-and-Lock Approach
8. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Chun, T.W.; Carruth, L.; Finzi, D.; Shen, X.; DiGiuseppe, J.A.; Taylor, H.; Hermankova, M.; Chadwick, K.; Margolick, J.; Quinn, T.C.; et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997, 387, 183–188. [Google Scholar] [CrossRef]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Wong, J.K.; Hezareh, M.; Günthard, H.F.; Havlir, D.V.; Ignacio, C.C.; Spina, C.A.; Richman, D.D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 1997, 278, 1291–1295. [Google Scholar] [CrossRef] [PubMed]
- Darcis, G.; Van Driessche, B.; Van Lint, C. HIV Latency: Should We Shock or Lock? Trends Immunol. 2017, 38, 217–228. [Google Scholar] [CrossRef]
- Marsden, M.D.; Zack, J.A. HIV cure strategies: A complex approach for a complicated viral reservoir? Future Virol. 2019, 14, 5–8. [Google Scholar] [CrossRef]
- Pitman, M.C.; Lau, J.S.Y.; McMahon, J.H.; Lewin, S.R. Barriers and strategies to achieve a cure for HIV. lancet. HIV 2018, 5, e317–e328. [Google Scholar] [CrossRef]
- Piacenti, F.J. An update and review of antiretroviral therapy. Pharmacotherapy 2006, 26, 1111–1133. [Google Scholar] [CrossRef]
- Schröder, A.R.W.; Shinn, P.; Chen, H.; Berry, C.; Ecker, J.R.; Bushman, F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 2002, 110, 521–529. [Google Scholar] [CrossRef]
- Chen, H.C.; Martinez, J.P.; Zorita, E.; Meyerhans, A.; Filion, G.J. Position effects influence HIV latency reversal. Nat. Struct. Mol. Biol. 2017, 24, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Lucic, B.; Chen, H.C.; Kuzman, M.; Zorita, E.; Wegner, J.; Minneker, V.; Wang, W.; Fronza, R.; Laufs, S.; Schmidt, M.; et al. Spatially clustered loci with multiple enhancers are frequent targets of HIV-1 integration. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nicoll, M.P.; Proença, J.T.; Efstathiou, S. The molecular basis of herpes simplex virus latency. FEMS Microbiol. Rev. 2012, 36, 684–705. [Google Scholar] [CrossRef] [PubMed]
- Yukl, S.; Pillai, S.; Li, P.; Chang, K.; Pasutti, W.; Ahlgren, C.; Havlir, D.; Strain, M.; Günthard, H.; Richman, D.; et al. Latently-infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 2009, 387, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Van Lint, C.; Fischle, W.; Paras, P.; Ott, M.; Brady, J.; Verdin, E. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 1996, 93, 6377–6381. [Google Scholar] [CrossRef] [PubMed]
- Emiliani, S.; Fischle, W.; Ott, M.; Van Lint, C.; Amella, C.A.; Verdin, E. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 1998, 72, 1666–1670. [Google Scholar] [CrossRef]
- Ruelas, D.S.; Greene, W.C. An integrated overview of HIV-1 latency. Cell 2013, 155, 519. [Google Scholar] [CrossRef]
- Coiras, M.; López-Huertas, M.R.; Pérez-Olmeda, M.; Alcamí, J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 2009, 7, 798–812. [Google Scholar] [CrossRef]
- Kaczmarek, K.; Morales, A.; Henderson, A.J. T cell transcription factors and their impact on HIV expression. Virol. Res. Treat. 2013, 4, 41–47. [Google Scholar] [CrossRef]
- Fernandez, G.; Zaikos, T.D.; Khan, S.Z.; Jacobi, A.M.; Behlke, M.A.; Zeichner, S.L. Targeting I B Proteins for HIV Latency Activation: The Role of Individual I B and NF- B Proteins. J. Virol. 2013, 87, 3966–3978. [Google Scholar] [CrossRef]
- Van Lint, C.; Bouchat, S.; Marcello, A. HIV-1 transcription and latency: An update. Retrovirology 2013, 10, 67. [Google Scholar] [CrossRef]
- Hakre, S.; Chavez, L.; Shirakawa, K.; Verdin, E. Epigenetic regulation of HIV latency. Curr. Opin. HIV AIDS 2011, 6, 19–24. [Google Scholar] [CrossRef]
- Nguyen, K.; Das, B.; Dobrowolski, C.; Karn, J. Multiple histone lysine methyltransferases are required for the establishment and maintenance of HIV-1 latency. MBio 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ne, E.; Palstra, R.J.; Mahmoudi, T. Transcription: Insights From the HIV-1 Promoter. In International Review of Cell and Molecular Biology; Academic Press: Cambridge, MA, USA, 2018; pp. 191–243. [Google Scholar]
- Duverger, A.; Wolschendorf, F.; Zhang, M.; Wagner, F.; Hatcher, B.; Jones, J.; Cron, R.Q.; van der Sluis, R.M.; Jeeninga, R.E.; Berkhout, B.; et al. An AP-1 Binding Site in the Enhancer/Core Element of the HIV-1 Promoter Controls the Ability of HIV-1 To Establish Latent Infection. J. Virol. 2013, 87, 2264–2277. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, P.L.; Sheline, C.T.; Cannon, K.; Voz, M.L.; Pazin, M.J.; Kadonaga, J.T.; Jones, K.A. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome- assembled DNA in vitro. Genes Dev. 1995, 9, 2090–2104. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, W.; Barreto, K.; Raithatha, S.; Sadowski, I. An Upstream YY1 Binding Site on the HIV-1 LTR Contributes to Latent Infection. PLoS ONE 2013, 8, e77052. [Google Scholar] [CrossRef] [PubMed]
- Karn, J. Tackling Tat. J. Mol. Biol. 1999, 293, 235–254. [Google Scholar] [CrossRef] [PubMed]
- Sodroski, J.; Rosen, C.; Wong-Staal, F.; Zaki Salahuddin, S.; Popovic, M.; Arya, S.; Gallo, R.C.; Haseltine, W.A. Trans-acting transcriptional regulation of human T-cell leukemia virus type III long terminal repeat. Science (80-) 1985, 227, 171–173. [Google Scholar] [CrossRef]
- Sodroski, J.; Patarca, R.; Rosen, C.; Wong-Staal, F.; Haseltine, W. Location of the trans-activating region on the genome of human T-cell lymphotropic virus type III. Science (80-) 1985, 229, 74–77. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. The Molecular Basis for Human Immunodeficiency Virus Latency. Annu. Rev. Virol. 2017, 4, 261–285. [Google Scholar] [CrossRef]
- Kim, J.B.; Sharp, P.A. Positive Transcription Elongation Factor b Phosphorylates hSPT5 and RNA Polymerase II Carboxyl-terminal Domain Independently of Cyclin-dependent Kinase-activating Kinase. J. Biol. Chem. 2001, 276, 12317–12323. [Google Scholar] [CrossRef]
- Fong, Y.W.; Zhou, Q. Relief of Two Built-In Autoinhibitory Mechanisms in P-TEFb Is Required for Assembly of a Multicomponent Transcription Elongation Complex at the Human Immunodeficiency Virus Type 1 Promoter. Mol. Cell. Biol. 2000, 20, 5897–5907. [Google Scholar] [CrossRef]
- Kuzmina, A.; Krasnopolsky, S.; Taube, R. Super elongation complex promotes early HIV transcription and its function is modulated by P-TEFb. Transcription 2017, 8, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Karn, J. Tat, a novel regulator of HIV transcription and latency The Tat/TAR RNA interaction. Available online: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.540.5110&rep=rep1&type=pdf (accessed on 12 April 2020).
- Brady, J.; Kashanchi, F. Tat gets the “Green” light on transcription initiation. Retrovirology 2005, 2, 69. [Google Scholar] [CrossRef] [PubMed]
- Kao, S.Y.; Calman, A.F.; Luciw, P.A.; Peterlin, B.M. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 1987, 330, 489–493. [Google Scholar] [CrossRef]
- Selby, M.J.; Bain, E.S.; Luciw, P.A.; Peterlin, B.M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989, 3, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, M.B.; Baltimore, D.; Frankel, A.D. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 1991, 88, 4045–4049. [Google Scholar] [CrossRef]
- Laspia, M.F.; Rice, A.P.; Mathews, M.B. HIV-1 Tat protein increases transcriptional initiation and stabilizes elongation. Cell 1989, 59, 283–292. [Google Scholar] [CrossRef]
- Raha, T.; Cheng, S.W.G.; Green, M.R. HIV-1 Tat stimulates transcription complex assembly through recruitment of TBP in the absence of TAFs. PLoS Biol. 2005, 3, 0221–0230. [Google Scholar] [CrossRef]
- Ping, Y.H.; Rana, T.M. Tat-associated kinase (P-TEFb): A component of transcription preinitiation and elongation complexes. J. Biol. Chem. 1999, 274, 7399–7404. [Google Scholar] [CrossRef]
- Marzio, G.; Tyagi, M.; Gutierrez, M.I.; Giacca, M. HIV-1 Tat transactivator recruits p300 and CREB-binding protein histone acetyltransferases to the viral promoter. Proc. Natl. Acad. Sci. USA 1998, 95, 13519–13524. [Google Scholar] [CrossRef]
- Lusic, M.; Marcello, A.; Cereseto, A.; Giacca, M. Regulation of HIV-1 gene expression by histone acetylation and factor recruitment at the LTR promoter. EMBO J. 2003, 22, 6550–6561. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Reeder, J.E.; Kwak, Y.T.; McNamara, R.P.; Forst, C.V.; D’Orso, I. HIV Tat controls RNA Polymerase II and the epigenetic landscape to transcriptionally reprogram target immune cells. Elife 2015, 4, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, L.S.; Burnett, J.C.; Toettcher, J.E.; Arkin, A.P.; Schaffer, D.V. Stochastic gene expression in a lentiviral positive-feedback loop: HIV-1 Tat fluctuations drive phenotypic diversity. Cell 2005, 122, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Razooky, B.S.; Pai, A.; Aull, K.; Rouzine, I.M.; Weinberger, L.S. A Hardwired HIV Latency Program. Cell 2015, 160, 990–1001. [Google Scholar] [CrossRef] [PubMed]
- Jadlowsky, J.K.; Wong, J.Y.; Graham, A.C.; Dobrowolski, C.; Devor, R.L.; Adams, M.D.; Fujinaga, K.; Karn, J. Negative Elongation Factor Is Required for the Maintenance of Proviral Latency but Does Not Induce Promoter-Proximal Pausing of RNA Polymerase II on the HIV Long Terminal Repeat. Mol. Cell. Biol. 2014, 34, 1911–1928. [Google Scholar] [CrossRef]
- Klaver, B.; Berkhout, B. Comparison of 5’ and 3’ long terminal repeat promoter function in human immunodeficiency virus. J. Virol. 1994, 68, 3830–3840. [Google Scholar] [CrossRef]
- Zapata, J.C.; Campilongo, F.; Barclay, R.A.; DeMarino, C.; Iglesias-Ussel, M.D.; Kashanchi, F.; Romerio, F. The Human Immunodeficiency Virus 1 ASP RNA promotes viral latency by recruiting the Polycomb Repressor Complex 2 and promoting nucleosome assembly. Virology 2017, 506, 34–44. [Google Scholar] [CrossRef]
- Kobayashi-Ishihara, M.; Yamagishi, M.; Hara, T.; Matsuda, Y.; Takahashi, R.; Miyake, A.; Nakano, K.; Yamochi, T.; Ishida, T.; Watanabe, T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology 2012, 9, 38. [Google Scholar] [CrossRef]
- Pereira, L.A. SURVEY AND SUMMARY A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000, 28, 663–668. [Google Scholar] [CrossRef]
- Verdin, E.; Paras, P.; Van Lint, C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993, 12, 3249–3259. [Google Scholar] [CrossRef]
- Rafati, H.; Parra, M.; Hakre, S.; Moshkin, Y.; Verdin, E.; Mahmoudi, T. Repressive LTR Nucleosome Positioning by the BAF Complex Is Required for HIV Latency. PLoS Biol. 2011, 9, e1001206. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, T. The BAF complex and HIV latency. Transcription 2012, 3, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.J.; Zou, X.; Calame, K.L. C/EBP proteins activate transcription from the human immunodeficiency virus type 1 long terminal repeat in macrophages/monocytes. J. Virol. 1995, 69, 5337–5344. [Google Scholar] [CrossRef] [PubMed]
- Giacca, M.; Gutierrez, M.I.; Menzo, S.; D’Adda Di Fagagna, F.; Flaschi, A. A human binding site for transcription factor USF/MLTF mimics the negative regulatory element of human immunodeficiency virus type 1. Virology 1992, 186, 133–147. [Google Scholar] [CrossRef]
- Rosen, C.A.; Sodroski, J.G.; Haseltine, W.A. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell 1985, 41, 813–823. [Google Scholar] [CrossRef]
- Pi-Yun, C.; Stellrecht, K.; Melana, S.; Pogo, B.G.T. Elements in the long terminal repeat of HIV-1 that interact with nuclear extracts from Jurkat cells persistently infected with vaccinia virus. Virus Res. 1994, 34, 127–138. [Google Scholar] [CrossRef]
- Beg, A.A.; Ruben, S.M.; Scheinman, R.I.; Haskill, S.; Rosen, C.A.; Baldwin, A.S. IκB interacts with the nuclear localization sequences of the subunits of NF-κB: A mechanism for cytoplasmic retention. Genes Dev. 1992, 6, 1899–1913. [Google Scholar] [CrossRef]
- Ghosh, S.; Baltimore, D. Activation in vitro of NF-κB by phosphorylation of its inhibitor IκB. Nature 1990, 344, 678–682. [Google Scholar] [CrossRef]
- Kim, Y.K.; Bourgeois, C.F.; Pearson, R.; Tyagi, M.; West, M.J.; Wong, J.; Wu, S.Y.; Chiang, C.M.; Karn, J. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 2006, 25, 3596–3604. [Google Scholar] [CrossRef]
- Giffin, M.J.; Stroud, J.C.; Bates, D.L.; Von Koenig, K.D.; Hardin, J.; Chen, L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR κB element. Nat. Struct. Biol. 2003, 10, 800–806. [Google Scholar] [CrossRef]
- Bosque, A.; Planelles, V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009, 113, 58–65. [Google Scholar] [CrossRef]
- Cron, R.Q.; Bartz, S.R.; Clausell, A.; Bort, S.J.; Klebanoff, S.J.; Lewis, D.B. NFAT1 enhances HIV-1 gene expression in primary human CD4 T cells. Clin. Immunol. 2000, 94, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Vaeth, M. NFAT control of immune function: New Frontiers for an Abiding Trooper. F1000Research 2018, 7, 260. [Google Scholar]
- Pessler, F.; Cron, R.Q. Reciprocal regulation of the nuclear factor of activated T cells and HIV-1. Genes Immun. 2004, 5, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Khan, S.Z.; Rawat, P.; Augustine, T.; Raynes, D.A.; Guerriero, V.; Mitra, D. HSP70 binding protein 1 (HspBP1) suppresses HIV-1 replication by inhibiting NF-B mediated activation of viral gene expression. Nucleic Acids Res. 2015, 44, 1613–1629. [Google Scholar] [CrossRef] [PubMed]
- Montano, M.A.; Kripke, K.; Norina, C.D.; Achacoso, P.; Herzenberg, L.A.; Roy, A.L.; Nolan, G.P. NF-κB homodimer binding within the HIV-1 initiator region and interactions with TFII-I. Proc. Natl. Acad. Sci. USA 1996, 93, 12376–12381. [Google Scholar] [CrossRef]
- Garcia, J.A.; Harrich, D.; Soultanakis, E.; Wu, F.; Mitsuyasu, R.; Gaynor, R.B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989, 8, 765–778. [Google Scholar] [CrossRef]
- Zenzie-Gregory, B.; Sheridan, P.; Jones, K.A.; Smale, S.T. HIV-1 core promoter lacks a simple initiator element but contains a bipartite activator at the transcription start site. J. Biol. Chem. 1993, 268, 15823–15832. [Google Scholar]
- Jones, K.A.; Kadonaga, J.T.; Luciw, P.A.; Tjian, R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science (80-) 1986, 232, 755–759. [Google Scholar] [CrossRef]
- Nilson, K.A.; Price, D.H. The Role of RNA Polymerase II Elongation Control in HIV-1 Gene Expression, Replication, and Latency. Genet. Res. Int. 2011, 2011, 1–9. [Google Scholar] [CrossRef][Green Version]
- Sodroski, J.G.; Rosen, C.A.; Haseltine, W.A. Trans-acting transcriptional activation of the long terminal repeat of human T lymphotropic viruses in infected cells. Science (80-) 1984, 225, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; Silverman, R.; Cell, K.J. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 1989, 59, 273–282. [Google Scholar] [CrossRef]
- Muesing, M.; Smith, D.; Cell, D.C. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell 1987, 48, 691–701. [Google Scholar] [CrossRef]
- Mariño-Ramírez, L.; Kann, M.G.; Shoemaker, B.A.; Landsman, D. Histone structure and nucleosome stability. Expert Rev. Proteomics 2005, 2, 719–729. [Google Scholar] [CrossRef] [PubMed]
- Schones, D.E.; Cui, K.; Cuddapah, S.; Roh, T.Y.; Barski, A.; Wang, Z.; Wei, G.; Zhao, K. Dynamic Regulation of Nucleosome Positioning in the Human Genome. Cell 2008, 132, 887–898. [Google Scholar] [CrossRef]
- Clapier, C.R.; Iwasa, J.; Cairns, B.R.; Peterson, C.L. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 2017, 18, 407–422. [Google Scholar] [CrossRef]
- Stasevich, T.J.; Hayashi-Takanaka, Y.; Sato, Y.; Maehara, K.; Ohkawa, Y.; Sakata-Sogawa, K.; Tokunaga, M.; Nagase, T.; Nozaki, N.; McNally, J.G.; et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature 2014, 516, 272–275. [Google Scholar] [CrossRef]
- Olsen, C.A. Expansion of the lysine acylation landscape. Angew. Chemie Int. Ed. 2012, 51, 3755–3756. [Google Scholar] [CrossRef]
- Bowman, G.D.; Poirier, M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2012, 38, 23–38. [Google Scholar] [CrossRef]
- Adelman, K.; Lis, J.T. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 2012, 13, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Easley, R.; Carpio, L.; Dannenberg, L.; Choi, S.; Alani, D.; Van Duyne, R.; Guendel, I.; Klase, Z.; Agbottah, E.; Kehn-Hall, K.; et al. Transcription through the HIV-1 nucleosomes: Effects of the PBAF complex in Tat activated transcription. Virology 2010, 405, 322–333. [Google Scholar] [CrossRef]
- Col, E.; Caron, C.; Seigneurin-Berny, D.; Gracia, J.; Favier, A.; Khochbin, S. The Histone Acetyltransferase, hGCN5, Interacts with and Acetylates the HIV Transactivator, Tat. J. Biol. Chem. 2001, 276, 28179–28184. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Mousseau, G.; Valente, S.T. Tat inhibition by didehydro-Cortistatin A promotes heterochromatin formation at the HIV-1 long terminal repeat. Epigenetics Chromatin 2019, 12, 23. [Google Scholar] [CrossRef]
- Belotserkovskaya, R.; Oh, S.; Bondarenko, V.A.; Orphanides, G.; Studitsky, V.M.; Reinberg, D. FACT facilitates transcription-dependent nucleosome alteration. Science (80-) 2003, 301, 1090–1093. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Santoso, N.; Power, D.; Simpson, S.; Dieringer, M.; Miao, H.; Gurova, K.; Giam, C.Z.; Elledge, S.J.; Zhu, J. FACT proteins, SUPT16H and SSRP1, are transcriptional suppressors of HIV-1 and HTLV-1 that facilitate viral latency. J. Biol. Chem. 2015, 290, 27297–27310. [Google Scholar] [CrossRef] [PubMed]
- Jean, M.J.; Hayashi, T.; Huang, H.; Brennan, J.; Simpson, S.; Purmal, A.; Gurova, K.; Keefer, M.C.; Kobie, J.J.; Santoso, N.G.; et al. Curaxin CBL0100 blocks HIV-1 replication and reactivation through inhibition of viral transcriptional elongation. Front. Microbiol. 2017, 8, 2007. [Google Scholar] [CrossRef] [PubMed]
- Conrad, R.J.; Fozouni, P.; Thomas, S.; Sy, H.; Zhang, Q.; Zhou, M.M.; Ott, M. The Short Isoform of BRD4 Promotes HIV-1 Latency by Engaging Repressive SWI/SNF Chromatin-Remodeling Complexes. Mol. Cell 2017, 67, 1001–1012.e6. [Google Scholar] [CrossRef] [PubMed]
- Marian, C.A.; Stoszko, M.; Wang, L.; Leighty, M.W.; de Crignis, E.; Maschinot, C.A.; Gatchalian, J.; Carter, B.C.; Chowdhury, B.; Hargreaves, D.C.; et al. Small Molecule Targeting of Specific BAF (mSWI/SNF) Complexes for HIV Latency Reversal. Cell Chem. Biol. 2018, 25, 1443–1455.e14. [Google Scholar] [CrossRef]
- Zhu, J.; Gaiha, G.D.; John, S.P.; Pertel, T.; Chin, C.R.; Gao, G.; Qu, H.; Walker, B.D.; Elledge, S.J.; Brass, A.L. Reactivation of Latent HIV-1 by Inhibition of BRD4. Cell Rep. 2012, 2, 807–816. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. Transcriptional control of HIV latency: Cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 2014, 454–455, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Moon, K.J.; Mochizuki, K.; Zhou, M.; Jeong, H.S.; Brady, J.N.; Ozato, K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell 2005, 19, 523–534. [Google Scholar]
- Mbonye, U.; Wang, B.; Gokulrangan, G.; Shi, W.; Yang, S.; Karn, J. Cyclin-dependent kinase 7 (CDK7)-mediated phosphorylation of the CDK9 activation loop promotes P-TEFb assembly with Tat and proviral HIV reactivation. J. Biol. Chem. 2018, 293, 10009–10025. [Google Scholar] [CrossRef] [PubMed]
- Bisgrove, D.A.; Mahmoudi, T.; Henklein, P.; Verdin, E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 13690–13695. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef]
- Yang, Z.; Yik, J.H.N.; Chen, R.; He, N.; Moon, K.J.; Ozato, K.; Zhou, Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell 2005, 19, 535–545. [Google Scholar] [CrossRef]
- D’Orso, I.; Frankel, A.D. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat. Struct. Mol. Biol. 2010, 17, 815–821. [Google Scholar] [CrossRef]
- Laird, G.M.; Bullen, C.K.; Rosenbloom, D.I.S.; Martin, A.R.; Hill, A.L.; Durand, C.M.; Siliciano, J.D.; Siliciano, R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015, 125, 1901–1912. [Google Scholar] [CrossRef]
- Alghamdi, S.; Khan, I.; Beeravolu, N.; McKee, C.; Thibodeau, B.; Wilson, G.; Chaudhry, G.R. BET protein inhibitor JQ1 inhibits growth and modulates WNT signaling in mesenchymal stem cells. Stem Cell Res. Ther. 2016, 7, 22. [Google Scholar] [CrossRef]
- Lee, D.U.; Katavolos, P.; Palanisamy, G.; Katewa, A.; Sioson, C.; Corpuz, J.; Pang, J.; DeMent, K.; Choo, E.; Ghilardi, N.; et al. Nonselective inhibition of the epigenetic transcriptional regulator BET induces marked lymphoid and hematopoietic toxicity in mice. Toxicol. Appl. Pharmacol. 2016, 300, 47–54. [Google Scholar] [CrossRef]
- Korb, E.; Herre, M.; Zucker-Scharff, I.; Darnell, R.B.; Allis, C.D. BET protein Brd4 activates transcription in neurons and BET inhibitor Jq1 blocks memory in mice. Nat. Neurosci. 2015, 18, 1464–1473. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar] [CrossRef]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Raisner, R.; Kharbanda, S.; Jin, L.; Romero, F.A.; Magnuson, S.; Gascoigne Correspondence, K.E. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell 2018, 24, 1722–1729. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Jiang, D.; Tian, J.; Liu, L.; Lu, T.; Huang, X.; Sun, H. Acetylation of H3K4, H3K9, and H3K27 mediated by p300 regulates the expression of GATA4 in cardiocytes. Genes Dis. 2019, 6, 318–325. [Google Scholar] [CrossRef]
- Zhang, Z.; Nikolai, B.C.; Gates, L.A.; Jung, S.Y.; Siwak, E.B.; He, B.; Rice, A.P.; O’Malley, B.W.; Feng, Q. Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency. Nucleic Acids Res. 2017, 45, 9348–9360. [Google Scholar] [CrossRef]
- Narlikar, G.J.; Fan, H.Y.; Kingston, R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell 2002, 108, 475–487. [Google Scholar] [CrossRef]
- He, G.; Margolis, D.M. Counterregulation of Chromatin Deacetylation and Histone Deacetylase Occupancy at the Integrated Promoter of Human Immunodeficiency Virus Type 1 (HIV-1) by the HIV-1 Repressor YY1 and HIV-1 Activator Tat. Mol. Cell. Biol. 2002, 22, 2965–2973. [Google Scholar] [CrossRef]
- Rasmussen, T.A.; Tolstrup, M.; Brinkmann, C.R.; Olesen, R.; Erikstrup, C.; Solomon, A.; Winckelmann, A.; Palmer, S.; Dinarello, C.; Buzon, M.; et al. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: A phase 1/2, single group, clinical trial. Lancet HIV 2014, 1, e13–e21. [Google Scholar] [CrossRef]
- Elliott, J.H.; Wightman, F.; Solomon, A.; Ghneim, K.; Ahlers, J.; Cameron, M.J.; Smith, M.Z.; Spelman, T.; McMahon, J.; Velayudham, P.; et al. Activation of HIV Transcription with Short-Course Vorinostat in HIV-Infected Patients on Suppressive Antiretroviral Therapy. PLoS Pathog. 2014, 10, e1004473. [Google Scholar] [CrossRef]
- Søgaard, O.S.; Graversen, M.E.; Leth, S.; Olesen, R.; Brinkmann, C.R.; Nissen, S.K.; Kjaer, A.S.; Schleimann, M.H.; Denton, P.W.; Hey-Cunningham, W.J.; et al. The Depsipeptide Romidepsin Reverses HIV-1 Latency In Vivo. PLOS Pathog. 2015, 11, e1005142. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.G.; Chiang, V.; Fyne, E.; Balakrishnan, M.; Barnes, T.; Graupe, M.; Hesselgesser, J.; Irrinki, A.; Murry, J.P.; Stepan, G.; et al. Histone Deacetylase Inhibitor Romidepsin Induces HIV Expression in CD4 T Cells from Patients on Suppressive Antiretroviral Therapy at Concentrations Achieved by Clinical Dosing. PLoS Pathog. 2014, 10, e1004071. [Google Scholar] [CrossRef] [PubMed]
- Leth, S.; Schleimann, M.H.; Nissen, S.K.; Højen, J.F.; Olesen, R.; Graversen, M.E.; Jørgensen, S.; Kjær, A.S.; Denton, P.W.; Mørk, A. Combined effect of Vacc-4x, recombinant human granulocyte macrophage colony-stimulating factor vaccination, and romidepsin on the HIV-1 reservoir (REDUC): A single-arm, phase 1B/2A trial. Lancet HIV 2016, 3, e463–e472. [Google Scholar] [CrossRef]
- Filippakopoulos, P.; Picaud, S.; Mangos, M.; Keates, T.; Lambert, J.P.; Barsyte-Lovejoy, D.; Felletar, I.; Volkmer, R.; Müller, S.; Pawson, T.; et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012, 149, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Horikoshi, M. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes to Cells 1998, 3, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Mbonye, U.; Feng, Z.; Wang, X.; Gao, X.; Karn, J.; Zhou, Q. The KAT5-Acetyl-Histone4-Brd4 axis silences HIV-1 transcription and promotes viral latency. PLoS Pathog. 2018, 14, e1007012. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Cho, W.K.; Chu, C.K.; Keedy, K.S.; Archin, N.M.; Margolis, D.M.; Karn, J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J. Virol. 2011, 85, 9078–9089. [Google Scholar] [CrossRef]
- Boehm, D.; Jeng, M.; Camus, G.; Gramatica, A.; Schwarzer, R.; Johnson, J.R.; Hull, P.A.; Montano, M.; Sakane, N.; Pagans, S.; et al. SMYD2-Mediated Histone Methylation Contributes to HIV-1 Latency. Cell Host Microbe 2017, 21, 569–579.e6. [Google Scholar] [CrossRef] [PubMed]
- Congdon, L.M.; Houston, S.I.; Veerappan, C.S.; Spektor, T.M.; Rice, J.C. PR-Set7-mediated monomethylation of histone H4 lysine 20 at specific genomic regions induces transcriptional repression. J. Cell. Biochem. 2010, 110, 609–619. [Google Scholar] [CrossRef]
- Trojer, P.; Li, G.; Sims, R.J.; Vaquero, A.; Kalakonda, N.; Boccuni, P.; Lee, D.; Erdjument-Bromage, H.; Tempst, P.; Nimer, S.D.; et al. L3MBTL1, a Histone-Methylation-Dependent Chromatin Lock. Cell 2007, 129, 915–928. [Google Scholar] [CrossRef]
- Jiang, G.; Nguyen, D.; Archin, N.M.; Yukl, S.A.; Méndez-Lagares, G.; Tang, Y.; Elsheikh, M.M.; Thompson, G.R.; Hartigan-O’Connor, D.J.; Margolis, D.M.; et al. HIV latency is reversed by ACSS2-driven histone crotonylation. J. Clin. Investig. 2018, 128, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Sabari, B.R.; Tang, Z.; Huang, H.; Yong-Gonzalez, V.; Molina, H.; Kong, H.E.; Dai, L.; Shimada, M.; Cross, J.R.; Zhao, Y.; et al. Intracellular Crotonyl-CoA Stimulates Transcription through p300-Catalyzed Histone Crotonylation. Mol. Cell 2015, 58, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sabari, B.R.; Panchenko, T.; Wen, H.; Zhao, D.; Guan, H.; Wan, L.; Huang, H.; Tang, Z.; Zhao, Y.; et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol. Cell 2016, 62, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Lerner, P.; Guadalupe, M.; Donovan, R.; Hung, J.; Flamm, J.; Prindiville, T.; Sankaran-Walters, S.; Syvanen, M.; Wong, J.K.; George, M.D.; et al. The Gut Mucosal Viral Reservoir in HIV-Infected Patients Is Not the Major Source of Rebound Plasma Viremia following Interruption of Highly Active Antiretroviral Therapy. J. Virol. 2011, 85, 4772–4782. [Google Scholar] [CrossRef][Green Version]
- Sabari, B.R.; Zhang, D.; Allis, C.D.; Zhao, Y. Metabolic regulation of gene expression through histone acylations. Nat. Rev. Mol. Cell Biol. 2017, 18, 90–101. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Villa, M.; Sanin, D.E.; Buck, M.D.; O’Sullivan, D.; Ching, R.; Matsushita, M.; Grzes, K.M.; Winkler, F.; Chang, C.H.; et al. Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep. 2019, 27, 2063–2074.e5. [Google Scholar] [CrossRef]
- Wu, J.Q.; Dwyer, D.E.; Dyer, W.B.; Yang, Y.H.; Wang, B.; Saksena, N.K. Genome-wide analysis of primary CD4+ and CD8+ T cell transcriptomes shows evidence for a network of enriched pathways associated with HIV disease. Retrovirology 2011, 8, 18. [Google Scholar] [CrossRef]
- Ishida, T.; Hamano, A.; Koiwa, T.; Watanabe, T. 5’ long terminal repeat (LTR)-selective methylation of latently infected HIV-1 provirus that is demethylated by reactivation signals. Retrovirology 2006, 3, 69. [Google Scholar] [CrossRef]
- Kauder, S.E.; Bosque, A.; Lindqvist, A.; Planelles, V.; Verdin, E. Epigenetic Regulation of HIV-1 Latency by Cytosine Methylation. PLoS Pathog. 2009, 5, e1000495. [Google Scholar] [CrossRef]
- Blazkova, J.; Trejbalova, K.; Gondois-Rey, F.; Halfon, P.; Philibert, P.; Guiguen, A.; Verdin, E.; Olive, D.; Van Lint, C.; Hejnar, J.; et al. CpG Methylation Controls Reactivation of HIV from Latency. PLoS Pathog. 2009, 5, e1000554. [Google Scholar] [CrossRef]
- Kint, S.; Trypsteen, W.; De Spiegelaere, W.; Malatinkova, E.; Kinloch-De Loes, S.; De Meyer, T.; Van Criekinge, W.; Vandekerckhove, L. Underestimated effect of intragenic HIV-1 DNA methylation on viral transcription in infected individuals. Clin. Epigenetics 2020, 12, 36. [Google Scholar] [CrossRef] [PubMed]
- Compe, E.; Genes, C.M.; Braun, C.; Coin, F.; Egly, J.M. TFIIE orchestrates the recruitment of the TFIIH kinase module at promoter before release during transcription. Nat. Commun. 2019, 10, 2084. [Google Scholar] [CrossRef]
- Greber, B.J.; Toso, D.B.; Fang, J.; Nogales, E. The complete structure of the human TFIIH core complex. Elife 2019, 8, e44771. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Roeder, R.G. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010, 11, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, A.; Pauls, E.; Badia, R.; Riveira-Muñoz, E.; Bailana, B.C.E.; Esté, J.A. Characterization of the influence of mediator complex in HIV-1 transcription. J. Biol. Chem. 2014, 289, 27665–27676. [Google Scholar] [CrossRef] [PubMed]
- Cary, D.C.; Rheinberger, M.; Rojc, A.; Peterlin, B.M. HIV Transcription Is Independent of Mediator Kinases. AIDS Res. Hum. Retroviruses 2019, 35, 710–717. [Google Scholar] [CrossRef] [PubMed]
- Mediouni, S.; Chinthalapudi, K.; Ekka, M.K.; Usui, I.; Jablonski, J.A.; Clementz, M.A.; Mousseau, G.; Nowak, J.; Macherla, V.R.; Beverage, J.N.; et al. Didehydro-Cortistatin A Inhibits HIV-1 by Specifically Binding to the Unstructured Basic Region of Tat. MBio 2019, 10, e02662-18. [Google Scholar] [CrossRef]
- Compe, E.; Egly, J.M. TFIIH: When transcription met DNA repair. Nat. Rev. Mol. Cell Biol. 2012, 13, 343–354. [Google Scholar] [CrossRef]
- Yan, C.; Dodd, T.; He, Y.; Tainer, J.A.; Tsutakawa, S.E.; Ivanov, I. Transcription preinitiation complex structure and dynamics provide insight into genetic diseases. Nat. Struct. Mol. Biol. 2019, 26, 397–406. [Google Scholar] [CrossRef]
- Tirode, F.; Busso, D.; Coin, F.; Egly, J.M. Reconstitution of the transcription factor TFIIH: Assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol. Cell 1999, 3, 87–95. [Google Scholar] [CrossRef]
- Greifenberg, A.K.; Hönig, D.; Pilarova, K.; Düster, R.; Bartholomeeusen, K.; Bösken, C.A.; Anand, K.; Blazek, D.; Geyer, M. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell Rep. 2016, 14, 320–331. [Google Scholar] [CrossRef]
- Nemec, C.M.; Yang, F.; Gilmore, J.M.; Hintermair, C.; Ho, Y.H.; Tseng, S.C.; Heidemann, M.; Zhang, Y.; Florens, L.; Gasch, A.P.; et al. Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proc. Natl. Acad. Sci. USA 2017, 114, E3944–E3953. [Google Scholar] [CrossRef]
- Rice, A.P. Roles of CDKs in RNA polymerase II transcription of the HIV-1 genome. Transcription 2019, 10, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Glover-Cutter, K.; Larochelle, S.; Erickson, B.; Zhang, C.; Shokat, K.; Fisher, R.P.; Bentley, D.L. TFIIH-Associated Cdk7 Kinase Functions in Phosphorylation of C-Terminal Domain Ser7 Residues, Promoter-Proximal Pausing, and Termination by RNA Polymerase II. Mol. Cell. Biol. 2009, 29, 5455–5464. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Takagi, T.; Wada, T.; Yano, K.; Furuya, A.; Sugimoto, S.; Hasegawa, J.; Handa, H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 1999, 97, 41–51. [Google Scholar] [CrossRef]
- Wada, T.; Takagi, T.; Yamaguchi, Y.; Watanabe, D.; Handa, H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998, 17, 7395–7403. [Google Scholar] [CrossRef]
- Garber, M.E.; Jones, K.A. HIV-1 Tat: Coping with negative elongation factors. Curr. Opin. Immunol. 1999, 11, 460–465. [Google Scholar] [CrossRef]
- Galbraith, M.D.; Donner, A.J.; Espinosa, J.M. CDK8: A positive regulator of transcription. Transcription 2010, 1, 4–12. [Google Scholar] [CrossRef]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Liang, K.; Gao, X.; Gilmore, J.M.; Florens, L.; Washburn, M.P.; Smith, E.; Shilatifard, A. Characterization of Human Cyclin-Dependent Kinase 12 (CDK12) and CDK13 Complexes in C-Terminal Domain Phosphorylation, Gene Transcription, and RNA Processing. Mol. Cell. Biol. 2015, 35, 928–938. [Google Scholar] [CrossRef]
- Berro, R.; Pedati, C.; Kehn-Hall, K.; Wu, W.; Klase, Z.; Even, Y.; Geneviere, A.M.; Ammosova, T.; Nekhai, S.; Kashanchi, F. CDK13, a New Potential Human Immunodeficiency Virus Type 1 Inhibitory Factor Regulating Viral mRNA Splicing. J. Virol. 2008, 82, 7155–7166. [Google Scholar] [CrossRef]
- Barboric, M.; Nissen, R.M.; Kanazawa, S.; Jabrane-Ferrat, N.; Peterlin, B.M. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 2001, 8, 327–337. [Google Scholar] [CrossRef]
- Devaiah, B.N.; Lewis, B.A.; Cherman, N.; Hewitt, M.C.; Albrecht, B.K.; Robey, P.G.; Ozato, K.; Sims, R.J.; Singer, D.S. BRD4 is an atypical kinase that phosphorylates Serine2 of the RNA Polymerase II carboxy-terminal domain. Proc. Natl. Acad. Sci. USA 2012, 109, 6927–6932. [Google Scholar] [CrossRef] [PubMed]
- Morton, E.L.; Forst, C.V.; Zheng, Y.; Depaula-Silva, A.B.; Ramirez, N.G.P. Transcriptional Circuit Fragility Influences HIV Proviral Fate. Cell 2019, 27, 154–171. [Google Scholar] [CrossRef] [PubMed]
- Lagha, M.; Bothma, J.P.; Esposito, E.; Ng, S.; Stefanik, L.; Tsui, C.; Johnston, J.; Chen, K.; Gilmour, D.S.; Zeitlinger, J.; et al. Paused Pol II coordinates tissue morphogenesis in the drosophila embryo. Cell 2013, 153, 976. [Google Scholar] [CrossRef]
- Boettiger, A.N.; Levine, M. Synchronous and stochastic patterns of gene activation in the drosophila embryo. Science (80-) 2009, 325, 471–473. [Google Scholar] [CrossRef] [PubMed]
- Shao, W.; Zeitlinger, J. Paused RNA polymerase II inhibits new transcriptional initiation. Nat. Genet. 2017, 49, 1045–1051. [Google Scholar] [CrossRef] [PubMed]
- Seila, A.C.; Core, L.J.; Lis, J.T.; Sharp, P.A. Divergent transcription: A new feature of active promoters. Cell Cycle 2009, 8, 2557–2564. [Google Scholar] [CrossRef]
- Kim, Y.K.; Mbonye, U.; Hokello, J.; Karn, J. T-Cell receptor signaling enhances transcriptional elongation from latent HIV proviruses by activating P-TEFb through an ERK-dependent pathway. J. Mol. Biol. 2011, 410, 896–916. [Google Scholar] [CrossRef]
- Wei, P.; Garber, M.E.; Fang, S.M.; Fischer, W.H.; Jones, K.A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 1998, 92, 451–462. [Google Scholar] [CrossRef]
- Rahl, P.B.; Lin, C.Y.; Seila, A.C.; Flynn, R.A.; Mccuine, S.; Burge, C.B.; Sharp, P.A.; Young, R.A. c-Myc Regulates Transcriptional Pause Release. Cell 2010, 141, 432–445. [Google Scholar] [CrossRef] [PubMed]
- Wada, T.; Takagi, T.; Yamaguchi, Y.; Ferdous, A.; Imai, T.; Hirose, S.; Sugimoto, S.; Yano, K.; Hartzog, G.A.; Winston, F.; et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998, 12, 343–356. [Google Scholar] [CrossRef] [PubMed]
- Vos, S.M.; Farnung, L.; Urlaub, H.; Cramer, P. Structure of paused transcription complex Pol II–DSIF–NELF. Nature 2018, 560, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Price, D.H. Controlling the Elongation Phase of Transcription with P-TEFb. Mol. Cell 2006, 23, 297–305. [Google Scholar] [CrossRef]
- Fujinaga, K.; Irwin, D.; Huang, Y.; Taube, R.; Kurosu, T.; Peterlin, B.M. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004, 24, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Couturier, J.; Orozco, A.F.; Liu, H.; Budhiraja, S.; Siwak, E.B.; Nehete, P.N.; Sastry, K.J.; Rice, A.P.; Lewis, D.E. Regulation of cyclin T1 during HIV replication and latency establishment in human memory CD4 T cells. Virol. J. 2019, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Budhiraja, S.; Famiglietti, M.; Bosque, A.; Planelles, V.; Rice, A.P. Cyclin T1 and CDK9 T-Loop Phosphorylation Are Downregulated during Establishment of HIV-1 Latency in Primary Resting Memory CD4+ T Cells. J. Virol. 2013, 87, 1211–1220. [Google Scholar] [CrossRef]
- Chiang, K.; Sung, T.L.; Rice, A.P. Regulation of Cyclin T1 and HIV-1 Replication by MicroRNAs in Resting CD4+ T Lymphocytes. J. Virol. 2012, 86, 3244–3252. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, T.; Price, D.H. RNA Polymerase II Elongation Control. Annu. Rev. Biochem. 2012, 81, 119–143. [Google Scholar] [CrossRef]
- Sung, T.L.; Rice, A.P. Effects of prostratin on Cyclin TI/P-TEFb function and the gene expression profile in primary resting CD4+ T cells. Retrovirology 2006, 3, 66. [Google Scholar] [CrossRef]
- Garber, M.E.; Mayall, T.P.; Suess, E.M.; Meisenhelder, J.; Thompson, N.E.; Jones, K.A. CDK9 Autophosphorylation Regulates High-Affinity Binding of the Human Immunodeficiency Virus Type 1 Tat-P-TEFb Complex to TAR RNA. Mol. Cell. Biol. 2000, 20, 6958–6969. [Google Scholar] [CrossRef]
- Larochelle, S.; Amat, R.; Glover-Cutter, K.; Sansó, M.; Zhang, C.; Allen, J.J.; Shokat, K.M.; Bentley, D.L.; Fisher, R.P. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat. Struct. Mol. Biol. 2012, 19, 1108–1115. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Price, J.P.; Byers, S.A.; Cheng, D.; Peng, J.; Price, D.H. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005, 280, 28819–28826. [Google Scholar] [CrossRef] [PubMed]
- Krueger, B.J.; Varzavand, K.; Cooper, J.J.; Price, D.H. The Mechanism of Release of P-TEFb and HEXIM1 from the 7SK snRNP by Viral and Cellular Activators Includes a Conformational Change in 7SK. PLoS ONE 2010, 5, e12335. [Google Scholar] [CrossRef]
- Cho, S.; Schroeder, S.; Kaehlcke, K.; Kwon, H.S.; Pedal, A.; Herker, E.; Schnoelzer, M.; Ott, M. Acetylation of cyclin T1 regulates the equilibrium between active and inactive P-TEFb in cells. EMBO J. 2009, 28, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Faust, T.B.; Li, Y.; Bacon, C.W.; Jang, G.M.; Weiss, A.; Jayaraman, B.; Newton, B.W.; Krogan, N.J.; D’orso, I.; Frankel, A.D. The HIV-1 tat protein recruits a ubiquitin ligase to reorganize the 7SK snRNP for transcriptional activation. Elife 2018, 7, e31879. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, D.; Pierstorff, E.; Luo, K. Transcription elongation factor P-TEFb mediates Tat activation of HIV-1 transcription at multiple stages. EMBO J. 1998, 17, 3681–3691. [Google Scholar] [CrossRef]
- Luo, Z.; Lin, C.; Guest, E.; Garrett, A.S.; Mohaghegh, N.; Swanson, S.; Marshall, S.; Florens, L.; Washburn, M.P.; Shilatifard, A. The Super Elongation Complex Family of RNA Polymerase II Elongation Factors: Gene Target Specificity and Transcriptional Output. Mol. Cell. Biol. 2012, 32, 2608–2617. [Google Scholar] [CrossRef] [PubMed]
- He, N.; Chan, C.K.; Sobhian, B.; Chou, S.; Xue, Y.; Liu, M.; Alber, T.; Benkirane, M.; Zhou, Q. Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc. Natl. Acad. Sci. USA 2011, 108, E636–E645. [Google Scholar] [CrossRef]
- Takahashi, H.; Parmely, T.J.; Sato, S.; Tomomori-Sato, C.; Banks, C.A.S.; Kong, S.E.; Szutorisz, H.; Swanson, S.K.; Martin-Brown, S.; Washburn, M.P.; et al. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 2011, 146, 92–104. [Google Scholar] [CrossRef]
- McNamara, R.P.; Reeder, J.E.; McMillan, E.A.; Bacon, C.W.; McCann, J.L.; D’Orso, I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol. Cell 2016, 61, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Spina, C.A.; Anderson, J.; Archin, N.M.; Bosque, A.; Chan, J.; Famiglietti, M.; Greene, W.C.; Kashuba, A.; Lewin, S.R.; Margolis, D.M.; et al. An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients. PLoS Pathog. 2013, 9, 1–15. [Google Scholar] [CrossRef]
- Boireau, S.; Maiuri, P.; Basyuk, E.; De La Mata, M.; Knezevich, A.; Pradet-Balade, B.; Bäcker, V.; Kornblihtt, A.; Marcello, A.; Bertrand, E. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 2007, 179, 291–304. [Google Scholar] [CrossRef]
- Singh, J.; Padgett, R.A. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009, 16, 1128–1133. [Google Scholar] [CrossRef] [PubMed]
- Fish, R.N.; Kane, C.M. Promoting elongation with transcript cleavage stimulatory factors. Biochim. Biophys. Acta - Gene Struct. Expr. 2002, 1577, 287–307. [Google Scholar] [CrossRef]
- Price, D.H.; Sluder, A.E.; Greenleaf, A.L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol. Cell. Biol. 1989, 9, 1465–1475. [Google Scholar] [CrossRef]
- Custódio, N.; Carmo-Fonseca, M. Co-transcriptional splicing and the CTD code. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 395–411. [Google Scholar] [CrossRef]
- Moteki, S.; Price, D. Functional coupling of capping and transcription of mRNA. Mol. Cell 2002, 10, 599–609. [Google Scholar] [CrossRef]
- Wen, Y.; Shatkin, A.J. Transcription elongation factor hSPT5 stimulates mRNA capping. Genes Dev. 1999, 13, 1774–1779. [Google Scholar] [CrossRef] [PubMed]
- Sims, R.J.; Belotserkovskaya, R.; Reinberg, D. Elongation by RNA polymerase II: The short and long of it. Genes Dev. 2004, 18, 2437–2468. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Kiong Ho, C.; Saha, N.; Schwer, B.; Shuman, S.; Rana, T.M. Tat Stimulates Cotranscriptional Capping of HIV mRNA. Mol. Cell. 2002, 10, 585–597. [Google Scholar] [CrossRef]
- Komarnitsky, P.; Cho, E.J.; Buratowski, S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000, 14, 2452–2460. [Google Scholar] [CrossRef] [PubMed]
- Bowman, E.A.; Kelly, W.G. RNA Polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation: A tail of two kinases. Nucl. 2014, 5, 224–236. [Google Scholar] [CrossRef] [PubMed]
- Emery, A.; Zhou, S.; Pollom, E.; Swanstrom, R. Characterizing HIV-1 Splicing by Using Next-Generation Sequencing. J. Virol. 2017, 91, e02515-16. [Google Scholar] [CrossRef]
- Das, R.; Yu, J.; Zhang, Z.; Gygi, M.P.; Krainer, A.R.; Gygi, S.P.; Reed, R. SR Proteins Function in Coupling RNAP II Transcription to Pre-mRNA Splicing. Mol. Cell 2007, 26, 867–881. [Google Scholar] [CrossRef]
- Yoh, S.M.; Cho, H.; Pickle, L.; Evans, R.M.; Jones, K.A. The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export. Genes Dev. 2007, 21, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Berro, R.; Kehn, K.; de la Fuente, C.; Pumfery, A.; Adair, R.; Wade, J.; Colberg-Poley, A.M.; Hiscott, J.; Kashanchi, F. Acetylated Tat Regulates Human Immunodeficiency Virus Type 1 Splicing through Its Interaction with the Splicing Regulator p32. J. Virol. 2006, 80, 3189–3204. [Google Scholar] [CrossRef] [PubMed]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25, 2158–2172. [Google Scholar] [CrossRef] [PubMed]
- Davidson, L.; Muniz, L.; West, S. 3’ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014, 28, 342–356. [Google Scholar] [CrossRef]
- Wahle, E.; Rüegsegger, U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999, 23, 277–295. [Google Scholar] [CrossRef]
- Colgan, D.F.; Manley, J.L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997, 11, 2755–2766. [Google Scholar] [CrossRef] [PubMed]
- Descours, B.; Petitjean, G.; López-Zaragoza, J.L.; Bruel, T.; Raffel, R.; Psomas, C.; Reynes, J.; Lacabaratz, C.; Levy, Y.; Schwartz, O.; et al. CD32a is a marker of a CD4 T-cell HIV reservoir harbouring replication-competent proviruses. Nature 2017, 543, 564–567. [Google Scholar] [CrossRef] [PubMed]
- Fromentin, R.; Bakeman, W.; Lawani, M.B.; Khoury, G.; Hartogensis, W.; DaFonseca, S.; Killian, M.; Epling, L.; Hoh, R.; Sinclair, E.; et al. CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence during ART. PLOS Pathog. 2016, 12, e1005761. [Google Scholar] [CrossRef] [PubMed]
- Hogan, L.E.; Vasquez, J.; Hobbs, K.S.; Hanhauser, E.; Aguilar-Rodriguez, B.; Hussien, R.; Thanh, C.; Gibson, E.A.; Carvidi, A.B.; Smith, L.C.B.; et al. Increased HIV-1 transcriptional activity and infectious burden in peripheral blood and gut-associated CD4+ T cells expressing CD30. PLOS Pathog. 2018, 14, e1006856. [Google Scholar] [CrossRef]
- Darcis, G.; Kootstra, N.A.; Hooibrink, B.; van Montfort, T.; Maurer, I.; Groen, K.; Jurriaans, S.; Bakker, M.; van Lint, C.; Berkhout, B.; et al. CD32+CD4+ T Cells Are Highly Enriched for HIV DNA and Can Support Transcriptional Latency. Cell Rep. 2020, 30, 2284–2296.e3. [Google Scholar] [CrossRef] [PubMed]
- Bruel, T.; Schwartz, O. Markers of the HIV-1 reservoir: Facts and controversies. Curr. Opin. HIV AIDS 2018, 13, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Darcis, G.; Berkhout, B.; Pasternak, A.O. The quest for cellular markers of HIV reservoirs: Any color you like. Front. Immunol. 2019, 10, 2251. [Google Scholar] [CrossRef]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Laskey, S.B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Hoh, R.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.S.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540. [Google Scholar] [CrossRef] [PubMed]
- Norton, N.J.; Fun, A.; Bandara, M.; Wills, M.R.; Mok, H.P.; Lever, A.M.L. Innovations in the quantitative virus outgrowth assay and its use in clinical trials. Retrovirology 2017, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Falcinelli, S.D.; Ceriani, C.; Margolis, D.M.; Archin, N.M. New Frontiers in Measuring and Characterizing the HIV Reservoir. Front. Microbiol. 2019, 10, 2878. [Google Scholar] [CrossRef] [PubMed]
- Battivelli, E.; Dahabieh, M.S.; Abdel-Mohsen, M.; Svensson, J.P.; Da Silva, I.T.; Cohn, L.B.; Gramatica, A.; Deeks, S.; Greene, W.C.; Pillai, S.K.; et al. Distinct chromatin functional states correlate with HIV latency reactivation in infected primary CD4+ T cells. Elife 2018, 7, e34655. [Google Scholar] [CrossRef]
- Rasmussen, T.A.; Lewin, S.R. Shocking HIV out of hiding: Where are we with clinical trials of latency reversing agents? Curr. Opin. HIV AIDS 2016, 11, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Day, C.L.; Kaufmann, D.E.; Kiepiela, P.; Brown, J.A.; Moodley, E.S.; Reddy, S.; Mackey, E.W.; Miller, J.D.; Leslie, A.J.; DePierres, C.; et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 2006, 443, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Cillo, A.R.; Sobolewski, M.D.; Bosch, R.J.; Fyne, E.; Piatak, M.; Coffin, J.M.; Mellors, J.W. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc. Natl. Acad. Sci. USA 2014, 111, 7078–7083. [Google Scholar] [CrossRef] [PubMed]
- Grau-Expósito, J.; Luque-Ballesteros, L.; Navarro, J.; Curran, A.; Burgos, J.; Ribera, E.; Torrella, A.; Planas, B.; Badía, R.; Martin-Castillo, M.; et al. Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS Pathog. 2019, 15, e1007991. [Google Scholar] [CrossRef]
- Zhao, M.; De Crignis, E.; Rokx, C.; Verbon, A.; van Gelder, T.; Mahmoudi, T.; Katsikis, P.D.; Mueller, Y.M. T cell toxicity of HIV latency reversing agents. Pharmacol. Res. 2019, 139, 524–534. [Google Scholar] [CrossRef]
- Clutton, G.; Xu, Y.; Baldoni, P.L.; Mollan, K.R.; Kirchherr, J.; Newhard, W.; Cox, K.; Kuruc, J.D.; Kashuba, A.; Barnard, R.; et al. The differential short- and long-term effects of HIV-1 latency-reversing agents on T cell function. Sci. Rep. 2016, 6, 1–17. [Google Scholar]
- Dar, R.D.; Hosmane, N.N.; Arkin, M.R.; Siliciano, R.F.; Weinberger, L.S. Screening for noise in gene expression identifies drug synergies. Science (80-) 2014, 344, 1392–1396. [Google Scholar] [CrossRef]
- Campbell, G.R.; Bruckman, R.S.; Chu, Y.L.; Trout, R.N.; Spector, S.A. SMAC Mimetics Induce Autophagy-Dependent Apoptosis of HIV-1-Infected Resting Memory CD4+ T Cells. Cell Host Microbe 2018, 24, 689–702.e7. [Google Scholar] [CrossRef]
- Kessing, C.F.; Nixon, C.C.; Li, C.; Tsai, P.; Takata, H.; Mousseau, G.; Ho, P.T.; Honeycutt, J.B.; Fallahi, M.; Trautmann, L.; et al. In Vivo Suppression of HIV Rebound by Didehydro-Cortistatin A, a “Block-and-Lock” Strategy for HIV-1 Treatment. Cell Rep. 2017, 21, 600–611. [Google Scholar] [CrossRef]
- Vansant, G.; Bruggemans, A.; Janssens, J.; Debyser, Z. Block-and-lock strategies to cure HIV infection. Viruses 2020, 12, 84. [Google Scholar] [CrossRef] [PubMed]
- Ramsköld, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, M.M.; Tang, Y.; Li, D.; Jiang, G. Deep latency: A new insight into a functional HIV cure. EBioMedicine 2019, 45, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Mousseau, G.; Clementz, M.A.; Bakeman, W.N.; Nagarsheth, N.; Cameron, M.; Shi, J.; Baran, P.; Fromentin, R.; Chomont, N.; Valente, S.T. An Analog of the Natural Steroidal Alkaloid Cortistatin A Potently Suppresses Tat-Dependent HIV Transcription. Cell Host Microbe 2012, 12, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Mousseau, G.; Kessing, C.F.; Fromentin, R.; Trautmann, L.; Chomont, N.; Valente, S.T. The tat inhibitor didehydro-cortistatin a prevents HIV-1 reactivation from latency. MBio 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Dornadula, G.; Zhang, H.; VanUitert, B.; Stern, J.; Lawrence Livornese, J.; Ingerman, M.J.; Witek, J.; Kedanis, R.J.; Natkin, J.; DeSimone, J.; et al. Residual HIV-1 RNA in Blood Plasma of Patients Taking Suppressive Highly Active Antiretroviral Therapy. JAMA 1999, 282, 1627. [Google Scholar] [CrossRef] [PubMed]
- Klatt, N.R.; Chomont, N.; Douek, D.C.; Deeks, S.G. Immune activation and HIV persistence: Implications for curative approaches to HIV infection. Immunol. Rev. 2013, 254, 326–342. [Google Scholar] [CrossRef]
- Chao, S.H.; Fujinaga, K.; Marion, J.E.; Taube, R.; Sausville, E.A.; Senderowicz, A.M.; Peterlin, B.M.; Price, D.H. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 2000, 275, 28345–28348. [Google Scholar] [CrossRef] [PubMed]
- Blachly, J.S.; Byrd, J.C. Emerging drug profile: Cyclin-dependent kinase inhibitors. Leuk. Lymphoma 2013, 54, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Anderson, I.; Low, J.S.; Weston, S.; Weinberger, M.; Zhyvoloup, A.; Labokha, A.A.; Corazza, G.; Kitson, R.A.; Moody, C.J.; Marcello, A.; et al. Heat shock protein 90 controls HIV-1 reactivation from latency. Proc. Natl. Acad. Sci. USA 2014, 111, E1528–E1537. [Google Scholar] [CrossRef]
- Joshi, P.; Maidji, E.; Stoddart, C.A. Inhibition of heat shock protein 90 prevents HIV rebound. J. Biol. Chem. 2016, 291, 10332–10346. [Google Scholar] [CrossRef]
- Alekseev, S.; Ayadi, M.; Brino, L.; Egly, J.M.; Larsen, A.K.; Coin, F. A small molecule screen identifies an inhibitor of DNA repair inducing the degradation of TFIIH and the Chemosensitization of tumor cells to platinum. Chem. Biol. 2014, 21, 398–407. [Google Scholar] [CrossRef] [PubMed]
- Alekseev, S.; Nagy, Z.; Sandoz, J.; Weiss, A.; Egly, J.M.; Le May, N.; Coin, F. Transcription without XPB Establishes a Unified Helicase-Independent Mechanism of Promoter Opening in Eukaryotic Gene Expression. Mol. Cell 2017, 65, 504–514.e4. [Google Scholar] [CrossRef]
- Elinoff, J.M.; Chen, L.Y.; Dougherty, E.J.; Awad, K.S.; Wang, S.; Biancotto, A.; Siddiqui, A.H.; Weir, N.A.; Cai, R.; Sun, J.; et al. Spironolactone-induced degradation of the TFIIH core complex XPB subunit suppresses NF-κB and AP-1 signalling. Cardiovasc. Res. 2018, 114, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Lacombe, B.; Morel, M.; Margottin-Goguet, F.; Ramirez, B.C. Specific Inhibition of HIV Infection by the Action of Spironolactone in T Cells. J. Virol. 2016, 90, 10972–10980. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Verma, D.; Thompson, J.; Swaminathan, S. Spironolactone blocks Epstein–Barr virus production by inhibiting EBV SM protein function. Proc. Natl. Acad. Sci. USA 2016, 113, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Szalat, R.; Samur, M.; Fulciniti, M.; Lopez, M.; Nanjappa, P.; Cleynen, A.; Wen, K.; Kumar, S.; Perini, T.; Calkins, A.; et al. Nucleotide excision repair is a potential therapeutic target in multiple myeloma. Leukemia 2018, 32, 111–119. [Google Scholar] [CrossRef]
- Compe, E.; Egly, J.M. Nucleotide Excision Repair and Transcriptional Regulation: TFIIH and Beyond. Annu. Rev. Biochem. 2016, 85, 265–290. [Google Scholar] [CrossRef]
- Besnard, E.; Hakre, S.; Kampmann, M.; Lim, H.W.; Hosmane, N.N.; Martin, A.; Bassik, M.C.; Verschueren, E.; Battivelli, E.; Chan, J.; et al. The mTOR Complex Controls HIV Latency. Cell Host Microbe 2016, 20, 785–797. [Google Scholar] [CrossRef]
- Winter, G.E.; Mayer, A.; Buckley, D.L.; Erb, M.A.; Roderick, J.E.; Vittori, S.; Reyes, J.M.; di Iulio, J.; Souza, A.; Ott, C.J.; et al. BET Bromodomain Proteins Function as Master Transcription Elongation Factors Independent of CDK9 Recruitment. Mol. Cell 2017, 67, 5–18.e19. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.; Liu, Z.; Alamer, E.; Fan, X.; Chen, H.; Endsley, J.; Gelman, B.B.; Tian, B.; Kim, J.H.; Michael, N.L.; et al. Structure-guided drug design identifies a BRD4-selective small molecule that suppresses HIV. J. Clin. Investig. 2019, 129, 3361–3373. [Google Scholar] [CrossRef] [PubMed]
- Das, B.; Dobrowolski, C.; Luttge, B.; Valadkhan, S.; Chomont, N.; Johnston, R.; Bacchetti, P.; Hoh, R.; Gandhi, M.; Deeks, S.G.; et al. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proc. Natl. Acad. Sci. USA 2018, 115, E7795–E7804. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mori, L.; Valente, S.T. Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure. Viruses 2020, 12, 529. https://doi.org/10.3390/v12050529
Mori L, Valente ST. Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure. Viruses. 2020; 12(5):529. https://doi.org/10.3390/v12050529
Chicago/Turabian StyleMori, Luisa, and Susana T. Valente. 2020. "Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure" Viruses 12, no. 5: 529. https://doi.org/10.3390/v12050529
APA StyleMori, L., & Valente, S. T. (2020). Key Players in HIV-1 Transcriptional Regulation: Targets for a Functional Cure. Viruses, 12(5), 529. https://doi.org/10.3390/v12050529




