Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts
Abstract
1. Introduction
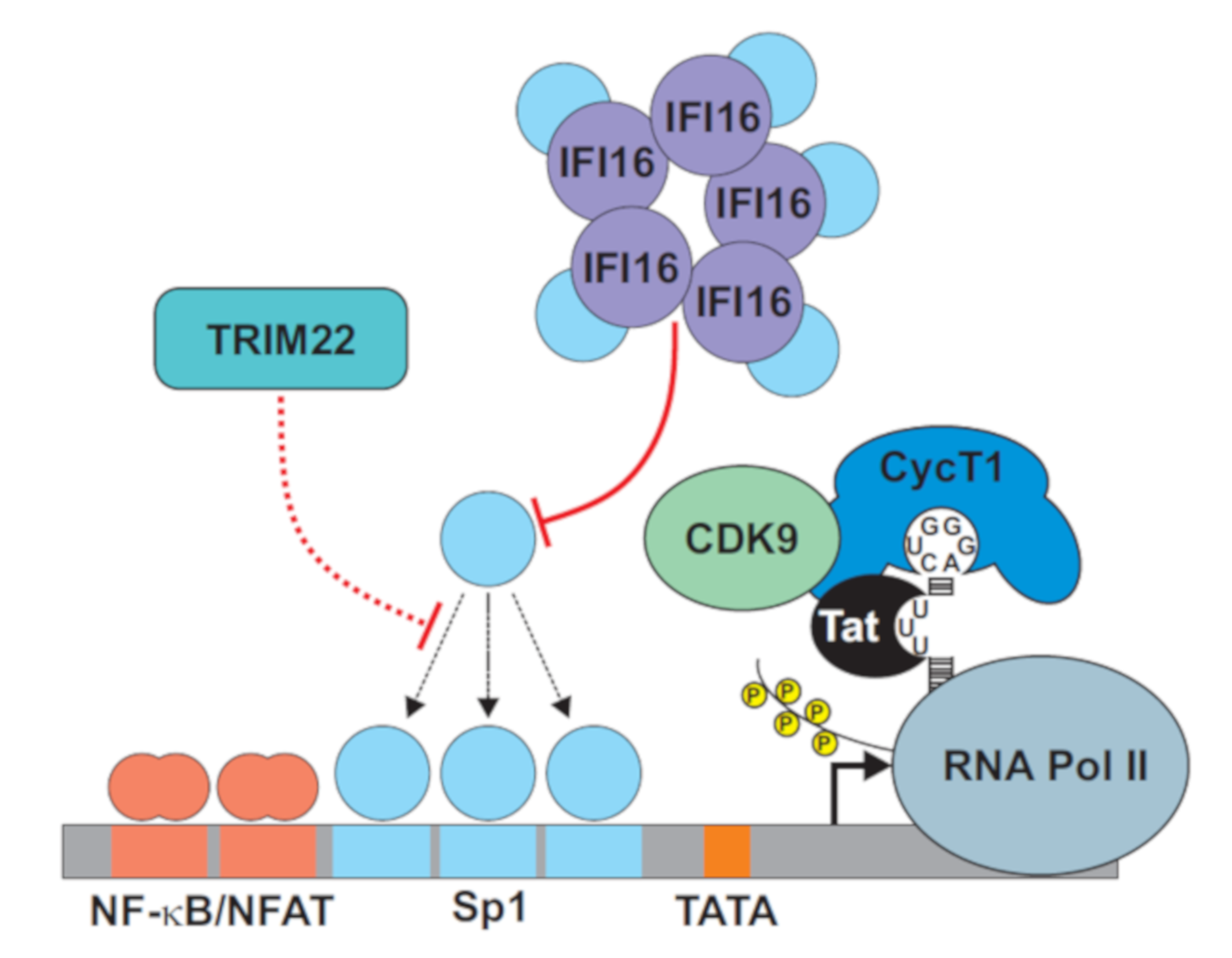
2. Targeting Sp1 for Transcriptional Silencing
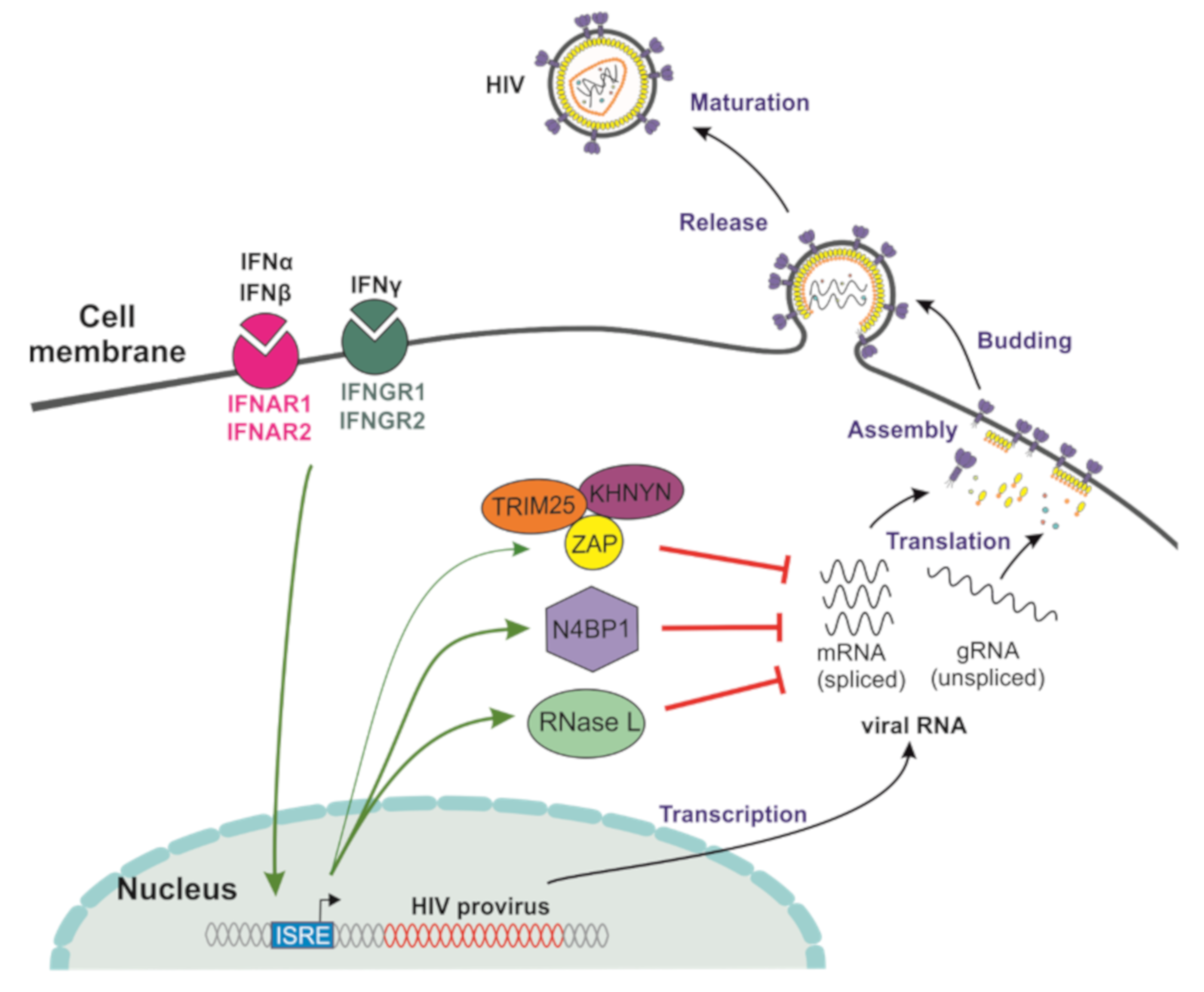
3. Cellular Factors Targeting HIV-1 RNA Transcripts
4. Summary and Perspectives
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Gupta, R.K.; Abdul-Jawad, S.; McCoy, L.E.; Mok, H.P.; Peppa, D.; Salgado, M.; Martinez-Picado, J.; Nijhuis, M.; Wensing, A.M.; Lee, H.; et al. HIV-1 remission following CCR5Δ32/Δ32 haematopoietic stem-cell transplantation. Nature 2019, 568, 244. [Google Scholar] [CrossRef] [PubMed]
- Hutter, G.; Nowak, D.; Mossner, M.; Ganepola, S.; Müßig, A.; Allers, K.; Schneider, T.; Hofmann, J.; Kücherer, C.; Blau, O.; et al. Long-Term Control of HIV by CCR5Δ32/Δ32 Stem-Cell Transplantation. N. Engl. J. Med. 2009, 360, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Ruelas, D.S.; Greene, W.C. An integrated overview of HIV-1 latency. Cell 2013, 155, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Siliciano, R.F.; Greene, W.C. HIV Latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22229121. (accessed on 5 December 2018). [CrossRef] [PubMed]
- Darcis, G.; Van Driessche, B.; Van Lint, C. HIV Latency: Should We Shock or Lock? Trends Immunol. 2017, 38, 217–228. [Google Scholar] [CrossRef] [PubMed]
- Deeks, S.G. Shock and kill. Nature 2012, 487, 439–440. [Google Scholar] [CrossRef]
- Abner, E.; Jordan, A. HIV “shock and kill” therapy: In need of revision. Antivir. Res. 2019, 166, 19–34. [Google Scholar] [CrossRef]
- Vansant, G.; Bruggemans, A.; Janssens, J.; Debyser, Z. Block-and-lock strategies to cure HIV infection. Viruses 2020, 12, 84. [Google Scholar] [CrossRef]
- Marsden, M.D.; Zack, J.A. HIV cure strategies: A complex approach for a complicated viral reservoir? Futur. Virol. 2019, 14, 5–8. [Google Scholar] [CrossRef]
- Chomont, N.; El-Far, M.; Ancuta, P.; Trautmann, L.; A Procopio, F.; Yassine-Diab, B.; Boucher, G.; Boulassel, M.-R.; Ghattas, G.; Brenchley, J.M.; et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009, 15, 893–900. [Google Scholar] [CrossRef]
- Massanella, M.; Fromentin, R.; Chomont, N. Residual inflammation and viral reservoirs: Alliance against an HIV cure. Curr. Opin. HIV AIDS 2016, 11, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Baldauf, H.-M.; Pan, X.; Erikson, E.; Schmidt, S.; Daddacha, W.; Burggraf, M.; Schenková, K.; Ambiel, I.; Wabnitz, G.; Gramberg, T.; et al. SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 2012, 18, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Bukrinsky, M.; Stanwick, T.; Dempsey, M.; Stevenson, M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science 1991, 254, 423–427. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.C.; Zhou, Y.; Kieffer, T.L.; Ruff, C.T.; Buck, C.B.; Siliciano, R.F. Molecular Characterization of Preintegration Latency in Human Immunodeficiency Virus Type 1 Infection. J. Virol. 2002, 76, 8518–8531. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, D.; Srinivasachar, S.; Kirchhoff, F. Potential roles of Nef and Vpu in HIV-1 latency. Futur. Virol. 2019, 14, 227–236. [Google Scholar] [CrossRef]
- Castro, S.; Colomer-Lluch, M.; Serra-Moreno, R. Barriers for HIV Cure: The Latent Reservoir. AIDS Res. Hum. Retroviruses 2018. [Google Scholar] [CrossRef]
- Darcis, G.; Van Driessche, B.; Bouchat, S.; Kirchhoff, F.; Van Lint, C. Molecular Control of HIV and SIV Latency. Curr. Top Microbiol. Immunol. 2017, 417, 1–22. [Google Scholar]
- Khoury, G.; Darcis, G.; Lee, M.Y.; Bouchat, S.; Van Driessche, B.; Purcell, D.F.J.; Van Lint, C. The Molecular Biology of HIV Latency. In Advances in Experimental Medicine and Biology; Springer New York LLC: New York, NY, USA, 2018; pp. 187–212. [Google Scholar]
- Agosto, L.M.; Herring, M.B.; Mothes, W.; Henderson, A. HIV-1-Infected CD4+ T Cells Facilitate Latent Infection of Resting CD4+ T Cells through Cell-Cell Contact. Cell Rep. 2018, 24, 2088–2100. [Google Scholar] [CrossRef]
- Ne, E.; Palstra, R.-J.; Mahmoudi, T. Transcription: Insights From the HIV-1 Promoter. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2018; pp. 191–243. [Google Scholar]
- Ma, X.; Yang, T.; Luo, Y.; Wu, L.; Jiang, Y.; Song, Z.; Pan, T.; Liu, B.; Liu, G.; Liu, J.; et al. TRIM28 promotes HIV-1 latency by SUMOylating CDK9 and inhibiting P-TEFb. eLife 2019, 8. [Google Scholar] [CrossRef]
- Coull, J.J.; Romerio, F.; Sun, J.-M.; Volker, J.L.; Galvin, K.M.; Davie, J.R.; Shi, Y.; Hansen, U.; Margolis, D.M. The Human Factors YY1 and LSF Repress the Human Immunodeficiency Virus Type 1 Long Terminal Repeat via Recruitment of Histone Deacetylase 1. J. Virol. 2000, 74, 6790–6799. [Google Scholar] [CrossRef]
- Barton, K.; Margolis, D.M. Selective Targeting of the Repressive Transcription Factors YY1 and cMyc to Disrupt Quiescent Human Immunodeficiency Viruses. AIDS Res. Hum. Retroviruses 2013, 29, 289–298. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4+T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10, eaap9927. [Google Scholar] [CrossRef]
- Asamitsu, K.; Fujinaga, K.; Okamoto, T. HIV Tat/P-TEFb Interaction: A Potential Target for Novel Anti-HIV Therapies. Mol. 2018, 23, 933. [Google Scholar] [CrossRef]
- Jiang, G.; Dandekar, S. Targeting NF-κB Signaling with Protein Kinase C Agonists as an Emerging Strategy for Combating HIV Latency. AIDS Res. Hum. Retroviruses 2015, 31, 4–12. [Google Scholar] [CrossRef]
- Williams, S.; Chen, L.-F.; Kwon, H.; Ruiz-Jarabo, C.M.; Verdin, E.; Greene, W.C. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2005, 25, 139–149. [Google Scholar] [CrossRef]
- Zhou, H.; Xu, M.; Huang, Q.; Gates, A.T.; Zhang, X.; Castle, J.C.; Stec, E.; Ferrer, M.; Strulovici, B.; Hazuda, D.; et al. Genome-Scale RNAi Screen for Host Factors Required for HIV Replication. Cell Host Microbe 2008, 4, 495–504. [Google Scholar] [CrossRef]
- Hotter, D.; Krabbe, T.; Reith, E.; Gawanbacht, A.; Rahm, N.; Ayouba, A.; Van Driessche, B.; Van Lint, C.; Peeters, M.; Kirchhoff, F.; et al. Primate lentiviruses use at least three alternative strategies to suppress NF-κB-mediated immune activation. PLoS Pathog. 2017, 13, e1006598. [Google Scholar] [CrossRef]
- Langer, S.; Hammer, C.; Hopfensperger, K.; Klein, L.; Hotter, D.; De Jesus, P.D.; Herbert, K.M.; Pache, L.; Smith, N.; Van Der Merwe, J.; et al. HIV-1 Vpu is a potent transcriptional suppressor of NF-κB-elicited antiviral immune responses. eLife 2019, 8. [Google Scholar] [CrossRef]
- Sauter, D.; Hotter, D.; Van Driessche, B.; Stürzel, C.M.; Kluge, S.F.; Wildum, S.; Yu, H.; Baumann, B.; Wirth, T.; Plantier, J.-C.; et al. Differential regulation of NF-κB-mediated proviral and antiviral host gene expression by primate lentiviral Nef and Vpu proteins. Cell Rep. 2015, 10, 586–599. [Google Scholar] [CrossRef]
- Giffin, M.J.; Stroud, J.; Bates, D.L.; Von Koenig, K.D.; Hardin, J.; Chen, L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR κB element. Nat. Struct. Mol. Boil. 2003, 10, 800–806. [Google Scholar] [CrossRef]
- Mbonye, U.; Karn, J. Transcriptional control of HIV latency: Cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 2014, 454, 328–339. [Google Scholar] [CrossRef]
- Roux, A.; Leroy, H.; De Muylder, B.; Bracq, L.; Oussous, S.; Dusanter-Fourt, I.; Chougui, G.; Tacine, R.; Randriamampita, C.; Desjardins, D.; et al. FOXO1 transcription factor plays a key role in T cell-HIV-1 interaction. PLoS Pathog. 2019, 15, e1007669. [Google Scholar] [CrossRef]
- Spivak, A.M.; Planelles, V. Novel Latency Reversal Agents for HIV-1 Cure. Annu. Rev. Med. 2018, 69, 421–436. [Google Scholar] [CrossRef]
- Jones, K.; Kadonaga, J.; Luciw, P.; Tjian, R. Activation of the AIDS retrovirus promoter by the cellular transcription factor, Sp1. Science 1986, 232, 755–759. [Google Scholar] [CrossRef]
- Harrich, D.; Garcia, J.; Wu, F.; Mitsuyasu, R.; Gonazalez, J.; Gaynor, R. Role of SP1-binding domains in in vivo transcriptional regulation of the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 1989, 63, 2585–2591. [Google Scholar] [CrossRef]
- Suñé, C.; A García-Blanco, M. Sp1 transcription factor is required for in vitro basal and Tat-activated transcription from the human immunodeficiency virus type 1 long terminal repeat. J. Virol. 1995, 69, 6572–6576. [Google Scholar] [CrossRef]
- Di Pietro, A.; Kajaste-Rudnitski, A.; Oteiza, A.; Nicora, L.; Towers, G.J.; Mechti, N.; Vicenzi, E. TRIM22 Inhibits Influenza A Virus Infection by Targeting the Viral Nucleoprotein for Degradation. J. Virol. 2013, 87, 4523–4533. [Google Scholar] [CrossRef]
- Turrini, F.; Marelli, S.; Kajaste-Rudnitski, A.; Lusic, M.; Van Lint, C.; Das, A.T.; Harwig, A.; Berkhout, B.; Vicenzi, E. HIV-1 transcriptional silencing caused by TRIM22 inhibition of Sp1 binding to the viral promoter. Retrovirology 2015, 12, 104. [Google Scholar] [CrossRef]
- Kajaste-Rudnitski, A.; Marelli, S.S.; Pultrone, C.; Pertel, T.; Uchil, P.D.; Mechti, N.; Mothes, W.; Poli, G.; Luban, J.; Vicenzi, E. TRIM22 Inhibits HIV-1 Transcription Independently of Its E3 Ubiquitin Ligase Activity, Tat, and NF-κB-Responsive Long Terminal Repeat Elements. J. Virol. 2011, 85, 5183–5196. [Google Scholar] [CrossRef]
- Gao, B.; Duan, Z.; Xu, W.; Xiong, S. Tripartite motif?containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear?located RING domain†. Hepatology 2009, 50, 424–433. [Google Scholar] [CrossRef]
- Yang, C.; Zhao, X.; Sun, D.; Yang, L.; Chong, C.; Pan, Y.; Chi, X.; Gao, Y.; Wang, M.; Shi, X.; et al. Interferon alpha (IFNα)-induced TRIM22 interrupts HCV replication by ubiquitinating NS5A. Cell. Mol. Immunol. 2015, 13, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Eldin, P.; Papon, L.; Oteiza, A.; Brocchi, E.; Lawson, T.G.; Mechti, N. TRIM22 E3 ubiquitin ligase activity is required to mediate antiviral activity against encephalomyocarditis virus. J. Gen. Virol. 2009, 90, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Turrini, F.; Saliu, F.; Forlani, G.; Das, A.T.; Van Lint, C.; Accolla, R.S.; Berkhout, B.; Poli, G.; Vicenzi, E. Interferon-inducible TRIM22 contributes to maintenance of HIV-1 proviral latency in T cell lines. Virus Res. 2019, 269, 197631. [Google Scholar] [CrossRef]
- Rohr, O.; Aunis, D.; Schaeffer, E. COUP-TF and Sp1 Interact and Cooperate in the Transcriptional Activation of the Human Immunodeficiency Virus Type 1 Long Terminal Repeat in Human Microglial Cells. J. Boil. Chem. 1997, 272, 31149–31155. [Google Scholar] [CrossRef]
- Hotter, D.; Bosso, M.; Jønsson, K.L.; Krapp, C.; Stürzel, C.M.; Das, A.; Littwitz-Salomon, E.; Berkhout, B.; Russ, A.; Wittmann, S.; et al. IFI16 Targets the Transcription Factor Sp1 to Suppress HIV-1 Transcription and Latency Reactivation. Cell Host Microbe 2019, 25, 858–872. [Google Scholar] [CrossRef]
- Jakobsen, M.R.; Bak, R.; Andersen, A.; Berg, R.K.; Jensen, S.B.; Jin, T.; Laustsen, A.; Hansen, K.; Østergaard, L.; Fitzgerald, K.A.; et al. PNAS Plus: From the Cover: IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. Proc. Natl. Acad. Sci USA 2013, 110, E4571–E4580. [Google Scholar] [CrossRef]
- Jønsson, K.L.; Laustsen, A.; Krapp, C.; Skipper, K.; Thavachelvam, K.; Hotter, D.; Egedal, J.H.; Kjolby, M.; Mohammadi, P.; Prabakaran, T.; et al. IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat. Commun. 2017, 8, 14391. [Google Scholar] [CrossRef]
- Monroe, K.M.; Yang, Z.; Johnson, J.R.; Geng, X.; Doitsh, G.; Krogan, N.J.; Greene, W.C. IFI16 DNA Sensor Is Required for Death of Lymphoid CD4 T Cells Abortively Infected with HIV. Science 2013, 343, 428–432. [Google Scholar] [CrossRef]
- Kerur, N.; Veettil, M.V.; Sharma-Walia, N.; Bottero, V.; Sadagopan, S.; Otageri, P.; Chandran, B. IFI16 Acts as a Nuclear Pathogen Sensor to Induce the Inflammasome in Response to Kaposi Sarcoma-Associated Herpesvirus Infection. Cell Host Microbe 2011, 9, 363–375. [Google Scholar] [CrossRef]
- Orzalli, M.H.; DeLuca, N.A.; Knipe, D.M. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc. Natl. Acad. Sci USA 2012, 109, E3008–E3017. [Google Scholar] [CrossRef]
- Gariano, G.R.; Dell’Oste, V.; Bronzini, M.; Gatti, D.; Luganini, A.; De Andrea, M.; Gribaudo, G.; Gariglio, M.; Landolfo, S. The Intracellular DNA Sensor IFI16 Gene Acts as Restriction Factor for Human Cytomegalovirus Replication. PLoS Pathog. 2012, 8, e1002498. Available online: http://www.ncbi.nlm.nih.gov/pubmed/22291595 (accessed on 16 July 2018). [CrossRef]
- Diner, B.A.; Lum, K.K.; Toettcher, J.E.; Cristea, I.M. Viral DNA Sensors IFI16 and Cyclic GMP-AMP Synthase Possess Distinct Functions in Regulating Viral Gene Expression, Immune Defenses, and Apoptotic Responses during Herpesvirus Infection. mBio 2016, 7. [Google Scholar] [CrossRef]
- Johnson, K.E.; Bottero, V.; Flaherty, S.; Dutta, S.; Singh, V.V.; Chandran, B. IFI16 Restricts HSV-1 Replication by Accumulating on the HSV-1 Genome, Repressing HSV-1 Gene Expression, and Directly or Indirectly Modulating Histone Modifications. PLoS Pathog. 2014, 10, e1004503. [Google Scholar] [CrossRef]
- Li, T.; Diner, B.A.; Chen, J.; Cristea, I.M. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. Proc. Natl. Acad. Sci USA 2012, 109, 10558–10563. [Google Scholar] [CrossRef]
- Sohn, J.; Morrone, S.; Wang, T.; Hooy, R. The Cooperative Assembly of IFI16 Filaments on dsDNA Provides Insights into Host Defense Strategy. Biophys. J. 2015, 108, 40a. [Google Scholar] [CrossRef][Green Version]
- Haronikova, L.; Coufal, J.; Kejnovská, I.; Jagelská, E.B.; Fojta, M.; Dvořáková, P.; Muller, P.; Vojtesek, B.; Brázda, V. IFI16 Preferentially Binds to DNA with Quadruplex Structure and Enhances DNA Quadruplex Formation. PLoS ONE 2016, 11, e0157156. [Google Scholar] [CrossRef]
- Perrone, R.; Lavezzo, E.; Palù, G.; Richter, S.N. Conserved presence of G-quadruplex forming sequences in the Long Terminal Repeat Promoter of Lentiviruses. Sci. Rep. 2017, 7, 2018. [Google Scholar] [CrossRef]
- Cigno, I.L.; De Andrea, M.; Borgogna, C.; Albertini, S.; Landini, M.M.; Peretti, A.; Johnson, K.E.; Chandran, B.; Landolfo, S.; Gariglio, M. The Nuclear DNA Sensor IFI16 Acts as a Restriction Factor for Human Papillomavirus Replication through Epigenetic Modifications of the Viral Promoters. J. Virol. 2015, 89, 7506–7520. [Google Scholar] [CrossRef]
- Orzalli, M.H.; Conwell, S.E.; Berrios, C.; DeCaprio, J.A.; Knipe, D.M. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc. Natl. Acad. Sci USA 2013, 110, E4492–E4501. [Google Scholar] [CrossRef]
- Safe, S.; Imanirad, P.; Sreevalsan, S.; Nair, V.; Jutooru, I. Transcription factor Sp1, also known as specificity protein 1 as a therapeutic target. Expert Opin. Ther. Targets 2014, 18, 759–769. [Google Scholar] [CrossRef]
- Bachu, M.; Yalla, S.; Asokan, M.; Verma, A.; Neogi, U.; Sharma, S.; Murali, R.V.; Mukthey, A.B.; Bhatt, R.; Chatterjee, S.; et al. Multiple NF-κB Sites in HIV-1 Subtype C Long Terminal Repeat Confer Superior Magnitude of Transcription and Thereby the Enhanced Viral Predominance*. J. Boil. Chem. 2012, 287, 44714–44735. [Google Scholar] [CrossRef] [PubMed]
- Kluge, S.F.; Sauter, D.; Kirchhoff, F. SnapShot: Antiviral Restriction Factors. Cell 2015, 163, 774. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Bieniasz, P.D. HIV Restriction Factors and Mechanisms of Evasion. Cold Spring Harb. Perspect. Med. 2012, 2, a006940. [Google Scholar] [CrossRef]
- Harris, R.S.; Hultquist, J.F.; Evans, D.T. The Restriction Factors of Human Immunodeficiency Virus*. J. Boil. Chem. 2012, 287, 40875–40883. [Google Scholar] [CrossRef]
- Ezelle, H.J.; Malathi, K.; Hassel, B.A. The roles of RNase-L in antimicrobial immunity and the cytoskeleton-associated innate response. Int. J. Mol. Sci. 2016, 17, 74. Available online: http://www.ncbi.nlm.nih.gov/pubmed/26760998 (accessed on 13 March 2020). [CrossRef]
- Chakrabarti, A.; Banerjee, S.; Franchi, L.; Loo, Y.-M.; Gale, M.; Núñez, G.; Silverman, R.H. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 2015, 17, 466–477. [Google Scholar] [CrossRef]
- Malathi, K.; Dong, B.; Gale, M.; Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef]
- Li, Y.; Banerjee, S.; Wang, Y.; Goldstein, S.A.; Dong, B.; Gaughan, C.; Silverman, R.H.; Weiss, S.R. Activation of RNase L is dependent on OAS3 expression during infection with diverse human viruses. Proc. Natl. Acad. Sci USA 2016, 113, 2241–2246. [Google Scholar] [CrossRef]
- Brennan-Laun, S.E.; Ezelle, H.J.; Li, X.-L.; Hassel, B.A. RNase-L Control of Cellular mRNAs: Roles in Biologic Functions and Mechanisms of Substrate Targeting. J. Interf. Cytokine Res. 2014, 34, 275–288. [Google Scholar] [CrossRef]
- Martinand, C.; Montavon, C.; Salehzada, T.; Silhol, M.; LeBleu, B.; Bisbal, C. RNase L Inhibitor Is Induced during Human Immunodeficiency Virus Type 1 Infection and Down Regulates the 2-5A/RNase L Pathway in Human T Cells. J. Virol. 1999, 73, 290–296. [Google Scholar] [CrossRef]
- García, M.Á.; Gil, J.; Ventoso, I.; Guerra, S.; Domingo, E.; Rivas, C.; Esteban, M. Impact of Protein Kinase PKR in Cell Biology: From Antiviral to Antiproliferative Action. Microbiol. Mol. Boil. Rev. 2006, 70, 1032–1060. [Google Scholar] [CrossRef]
- Dauber, B.; Wolff, T. Activation of the Antiviral Kinase PKR and Viral Countermeasures. Viruses 2009, 1, 523–544. [Google Scholar] [CrossRef]
- Dias Junior, A.G.; Sampaio, N.G.; Rehwinkel, J. A Balancing Act: MDA5 in Antiviral Immunity and Autoinflammation. In Trends in Microbiology; Elsevier Ltd.: Amsterdam, The Netherlands, 2019; Volume 27, pp. 75–85. [Google Scholar]
- Ireland, D.; Stohlman, S.A.; Hinton, D.R.; Kapil, P.; Silverman, R.H.; Atkinson, R.A.; Bergmann, C. RNase L Mediated Protection from Virus Induced Demyelination. PLoS Pathog. 2009, 5, e1000602. [Google Scholar] [CrossRef] [PubMed]
- Andersen, J.B.; Mazan-Mamczarz, K.; Zhan, M.; Gorospe, M.; Hassel, B.A. Ribosomal protein mRNAs are primary targets of regulation in RNase-L-induced senescence. RNA Boil. 2009, 6, 305–315. [Google Scholar] [CrossRef]
- Hassel, B.; Zhou, A.; Sotomayor, C.; Maran, A.; Silverman, R. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 1993, 12, 3297–3304. [Google Scholar] [CrossRef]
- Chakrabarti, A.; Jha, B.K.; Silverman, R.H. New insights into the role of RNase L in innate immunity. J. Interferon Cytokine Res. 2011, 37, 49–57. [Google Scholar] [CrossRef]
- Ghimire, D.; Rai, M.; Gaur, R. Novel host restriction factors implicated in HIV-1 replication. J. Gen. Virol. Microbiol. Soc. 2018, 99, 435–446. [Google Scholar] [CrossRef]
- Odon, V.; Fros, J.J.; Goonawardane, N.; Dietrich, I.; Ibrahim, A.; Alshaikhahmed, K.; Nguyen, D.; Simmonds, P. The role of ZAP and OAS3/RNAseL pathways in the attenuation of an RNA virus with elevated frequencies of CpG and UpA dinucleotides. Nucleic Acids Res. 2019, 47, 8061–8083. [Google Scholar] [CrossRef]
- Gupte, R.; Liu, Z.; Kraus, W.L. PARPs and ADP-ribosylation: Recent advances linking molecular functions to biological outcomes. Genes Dev. 2017, 31, 101–126. [Google Scholar] [CrossRef]
- Kerns, J.A.; Emerman, M.; Malik, H.S. Positive Selection and Increased Antiviral Activity Associated with the PARP-Containing Isoform of Human Zinc-Finger Antiviral Protein. PLoS Genet. 2008, 4, e21. [Google Scholar] [CrossRef]
- Pascal, J.M. The comings and goings of PARP-1 in response to DNA damage. In DNA Repair; Elsevier B.V.: Amsterdam, The Netherlands, 2018; Volume 71, pp. 177–182. [Google Scholar]
- Posavec Marjanović, M.; Crawford, K.; Ahel, I. PARP, transcription and chromatin modeling. In Seminars in Cell and Developmental Biology; Academic Press: Cambridge, MA, USA, 2017; Volume 63, pp. 102–113. [Google Scholar]
- Gao, G.; Guo, X.; Goff, S.P. Inhibition of Retroviral RNA Production by ZAP, a CCCH-Type Zinc Finger Protein. Science 2002, 297, 1703–1706. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Witteveldt, J.; Evans, D.J.; Simmonds, P. The influence of CpG and UpA dinucleotide frequencies on RNA virus replication and characterization of the innate cellular pathways underlying virus attenuation and enhanced replication. Nucleic Acids Res. 2014, 42, 4527–4545. [Google Scholar] [CrossRef]
- Karlin, S.; Doerfler, W.; Cardon, L.R. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J. Virol. 1994, 68, 2889–2897. [Google Scholar] [CrossRef]
- Antzin-Anduetza, I.; Mahiet, C.; Granger, L.A.; Odendall, C.; Swanson, C.M. Increasing the CpG dinucleotide abundance in the HIV-1 genomic RNA inhibits viral replication. Retrovirology 2017, 14, 49. [Google Scholar] [CrossRef]
- Burns, C.C.; Campagnoli, R.; Shaw, J.; Vincent, A.; Jorba, J.; Kew, O. Genetic Inactivation of Poliovirus Infectivity by Increasing the Frequencies of CpG and UpA Dinucleotides within and across Synonymous Capsid Region Codons. J. Virol. 2009, 83, 9957–9969. [Google Scholar] [CrossRef]
- Gaunt, E.; Wise, H.M.; Zhang, H.; Ni Lee, L.; Atkinson, N.J.; Nicol, M.Q.; Highton, A.J.; Klenerman, P.; Beard, P.; Dutia, B.M.; et al. Elevation of CpG frequencies in influenza A genome attenuates pathogenicity but enhances host response to infection. eLife 2016, 5. [Google Scholar] [CrossRef]
- Takata, M.A.; Carneiro, D.G.; Zang, T.M.; Soll, S.J.; York, A.; Blanco-Melo, D.; Bieniasz, P.D. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 2017, 550, 124–127. [Google Scholar] [CrossRef]
- Meagher, J.L.; Takata, M.; Gonçalves-Carneiro, D.; Keane, S.C.; Rebendenne, A.; Ong, H.; Orr, V.K.; Macdonald, M.R.; Stuckey, J.A.; Bieniasz, P.D.; et al. Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences. Proc. Natl. Acad. Sci USA 2019, 116, 24303–24309. [Google Scholar] [CrossRef]
- Li, M.M.; Aguilar, E.G.; Michailidis, E.; Pabon, J.; Park, P.; Wu, X.; De Jong, Y.P.; Schneider, W.M.; Molina, H.; Rice, C.M.; et al. Characterization of Novel Splice Variants of Zinc Finger Antiviral Protein (ZAP). J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Schwerk, J.; Soveg, F.W.; Ryan, A.P.; Thomas, K.R.; Hatfield, L.D.; Ozarkar, S.; Forero, A.; Kell, A.M.; Roby, J.A.; So, L.; et al. RNA-binding protein isoforms ZAP-S and ZAP-L have distinct antiviral and immune resolution functions. Nat. Immunol. 2019, 20, 1610–1620. [Google Scholar] [CrossRef]
- Ficarelli, M.; Antzin-Anduetza, I.; Hugh-White, R.; Firth, A.E.; Sertkaya, H.; Wilson, H.; Neil, S.J.D.; Schulz, R.; Swanson, C.M. CpG Dinucleotides Inhibit HIV-1 Replication through Zinc Finger Antiviral Protein (ZAP)-Dependent and -Independent Mechanisms. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, X.; Tu, F.; Wang, Q.; Fan, Z.; Gao, G. TRIM25 Is Required for the Antiviral Activity of Zinc Finger Antiviral Protein. J. Virol. 2017, 91, e00088-17. [Google Scholar] [CrossRef]
- Li, M.M.; Lau, Z.; Cheung, P.; Aguilar, E.; Schneider, W.M.; Bozzacco, L.; Molina, H.; Buehler, E.; Takaoka, A.; Rice, C.M.; et al. TRIM25 Enhances the Antiviral Action of Zinc-Finger Antiviral Protein (ZAP). PLoS Pathog. 2017, 13, e1006145. [Google Scholar] [CrossRef]
- Ficarelli, M.; Wilson, H.; Galao, R.; Mazzon, M.; Antzin-Anduetza, I.; Marsh, M.; Neil, S.J.D.; Swanson, C.M. KHNYN is essential for the zinc finger antiviral protein (ZAP) to restrict HIV-1 containing clustered CpG dinucleotides. eLife 2019, 8. [Google Scholar] [CrossRef]
- Kmiec, D.; Nchioua, R.; Sherrill-Mix, S.; Stürzel, C.M.; Heusinger, E.; Braun, E.; Gondim, M.V.P.; Hotter, D.; Sparrer, K.M.J.; Hahn, B.H.; et al. CpG Frequency in the 5’ Third of the env Gene Determines Sensitivity of Primary HIV-1 Strains to the Zinc-Finger Antiviral Protein. mBio 2020, 11. [Google Scholar] [CrossRef]
- Cooper, D.N.; Gerber-Huber, S. DNA methylation and CpG suppression. Cell Differ. 1985, 17, 199–205. [Google Scholar] [CrossRef]
- Blazkova, J.; Trejbalova, K.; Gondois-Rey, F.; Halfon, P.; Philibert, P.; Guiguen, A.; Verdin, E.; Olive, D.; Van Lint, C.; Hejnar, J. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009, 5, e1000554. [Google Scholar] [CrossRef]
- Xuan, Y.; Liu, L.; Shen, S.; Deng, H.; Gao, G. Zinc Finger Antiviral Protein Inhibits Murine Gammaherpesvirus 68 M2 Expression and Regulates Viral Latency in Cultured Cells. J. Virol. 2012, 86, 12431–12434. [Google Scholar] [CrossRef]
- Miyazato, P.; Matsuo, M.; Tan, B.J.Y.; Tokunaga, M.; Katsuya, H.; Islam, S.; Ito, J.; Murakawa, Y.; Satou, Y. HTLV-1 contains a high CG dinucleotide content and is susceptible to the host antiviral protein ZAP. Retrovirology 2019, 16, 38. [Google Scholar] [CrossRef]
- Yamasoba, D.; Sato, K.; Ichinose, T.; Imamura, T.; Koepke, L.; Joas, S.; Reith, E.; Hotter, D.; Misawa, N.; Akaki, K.; et al. N4BP1 restricts HIV-1 and its inactivation by MALT1 promotes viral reactivation. Nat. Microbiol. 2019, 4, 1532–1544. [Google Scholar] [CrossRef]
- Marco, A.; Marín, I. CGIN1: A Retroviral Contribution to Mammalian Genomes. Mol. Biol. Evol. 2009, 26, 2167–2170. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Qiu, C.; Miao, R.; Zhou, J.; Lee, A.; Liu, B.; Lester, S.N.; Fu, W.; Zhu, L.; Zhang, L.; et al. MCPIP1 restricts HIV infection and is rapidly degraded in activated CD4+ T cells. Proc. Natl. Acad. Sci USA 2013, 110, 19083–19088. [Google Scholar] [CrossRef] [PubMed]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nchioua, R.; Bosso, M.; Kmiec, D.; Kirchhoff, F. Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts. Viruses 2020, 12, 495. https://doi.org/10.3390/v12050495
Nchioua R, Bosso M, Kmiec D, Kirchhoff F. Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts. Viruses. 2020; 12(5):495. https://doi.org/10.3390/v12050495
Chicago/Turabian StyleNchioua, Rayhane, Matteo Bosso, Dorota Kmiec, and Frank Kirchhoff. 2020. "Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts" Viruses 12, no. 5: 495. https://doi.org/10.3390/v12050495
APA StyleNchioua, R., Bosso, M., Kmiec, D., & Kirchhoff, F. (2020). Cellular Factors Targeting HIV-1 Transcription and Viral RNA Transcripts. Viruses, 12(5), 495. https://doi.org/10.3390/v12050495




