Comparative Transcriptome Analysis Provides Molecular Insights into the Interaction of Beet necrotic yellow vein virus and Beet soil-borne mosaic virus with Their Host Sugar Beet
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Virus Inoculation
2.2. RNA Extraction, Library Preparation and Sequencing
2.3. Transcriptome Analysis
2.4. RT-qPCR
3. Results
3.1. Sequencing Statistic and RT-qPCR Validation
3.2. Functional Classification of DEGs
3.3. Interaction with Auxin Biosynthesis and Signalling Pathways
3.4. Activation of Auxin-Regulated LR Development
3.5. Reprogramming of the Plant Transcriptional Network
3.6. Plant Defence Response Against BNYVV and BSBMV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gilmer, D.; Ratti, C.; Ictv, R.C. ICTV Virus Taxonomy Profile: Benyviridae. J. Gen. Virol. 2017, 98, 1571–1572. [Google Scholar] [CrossRef]
- McGrann, G.R.D.; Grimmer, M.K.; Mutasa-Göttgens, E.S.; Stevens, M. Progress towards the understanding and control of sugar beet rhizomania disease. Mol. Plant Pathol. 2009, 10, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Heidel, G.B.; Rush, C.M.; Kendall, T.L.; Lommel, S.A.; French, R.C. Characteristics of beet soilborne mosaic virus, a furo-like virus infecting sugar beet. Plant Dis. 1997, 81, 1070–1076. [Google Scholar] [CrossRef] [PubMed]
- Lee, L.; Telford, E.B.; Batten, J.S.; Scholthof, K.B.; Rush, C.M. Complete nucleotide sequence and genome organization of Beet soilborne mosaic virus, a proposed member of the genus Benyvirus. Arch. Virol. 2001, 146, 2443–2453. [Google Scholar] [CrossRef] [PubMed]
- Keskin, B. Polymyxa betae n.sp., ein Parasit in den Wurzeln von Beta vulgaris Tournefort, besonders während der Jugendentwicklung der Zuckerrübe. Archiv. Mikrobiol. 1964, 49, 348–374. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T.; Kondo, H. Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors. J. Gen. Plant Pathol. 2013, 79, 307–320. [Google Scholar] [CrossRef]
- Workneh, F.; Villanueva, E.; Steddom, K.; Rush, C.M. Spatial association and distribution of Beet necrotic yellow vein virus and Beet soilborne mosaic virus in sugar beet fields. Plant Dis. 2003, 87, 707–711. [Google Scholar] [CrossRef][Green Version]
- Piccinni, G.; Rush, C.M. Determination of optimum irrigation regime and water use efficiency of sugar beet grown in pathogen-infested soil. Plant Dis. 2000, 84, 1067–1072. [Google Scholar] [CrossRef]
- Fernando Gil, J.; Liebe, S.; Thiel, H.; Lennefors, B.-L.; Kraft, T.; Gilmer, D.; Maiss, E.; Varrelmann, M.; Savenkov, E.I. Massive up-regulation of LBD transcription factors and EXPANSINs highlights the regulatory programs of rhizomania disease. Mol. Plant Pathol. 2018, 19, 2333–2348. [Google Scholar] [CrossRef]
- Ratti, C.; Hleibieh, K.; Bianchi, L.; Schirmer, A.; Autonell, C.R.; Gilmer, D. Beet soil-borne mosaic virus RNA-3 is replicated and encapsidated in the presence of BNYVV RNA-1 and -2 and allows long distance movement in Beta macrocarpa. Virology 2009, 385, 392–399. [Google Scholar] [CrossRef]
- Tamada, T.; Kusume, T. Evidence that the 75K readthrough protein of beet necrotic yellow vein virus RNA-2 is essential for transmission by the fungus Polymyxa betae. J. Gen. Virol. 1991, 72, 1497–1504. [Google Scholar] [CrossRef] [PubMed]
- Gilmer, D.; Bouzoubaa, S.; Hehn, A.; Guilley, H.; Richards, K.; Jonard, G. Efficient cell-to-cell movement of beet necrotic yellow vein virus requires 3′ proximal genes located on RNA 2. Virology 1992, 189, 40–47. [Google Scholar] [CrossRef]
- Chiba, S.; Hleibieh, K.; Delbianco, A.; Klein, E.; Ratti, C.; Ziegler-Graff, V.; Bouzoubaa, S.; Gilmer, D. The benyvirus RNA silencing suppressor is essential for long-distance movement, requires both zinc-finger and NoLS basic residues but not a nucleolar localization for its silencing-suppression activity. Mol. Plant Microbe Interact. 2013, 26, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Dunoyer, P.; Pfeffer, S.; Fritsch, C.; Hemmer, O.; Voinnet, O.; Richards, K.E. Identification, subcellular localization and some properties of a cysteine-rich suppressor of gene silencing encoded by peanut clump virus. Plant J. 2002, 29, 555–567. [Google Scholar] [CrossRef] [PubMed]
- Lauber, E.; Guilley, H.; Tamada, T.; Richards, K.E.; Jonard, G. Vascular movement of beet necrotic yellow vein virus in Beta macrocarpa is probably dependent on an RNA 3 sequence domain rather than a gene product. J. Gen. Virol. 1998, 79, 385–393. [Google Scholar] [CrossRef]
- Tamada, T.; Uchino, H.; Kusume, T.; Saito, M. RNA 3 Deletion mutants of Beet necrotic yellow vein virus do not cause rhizomania disease in sugar beets. Phytopathology 1999, 89, 1000–1006. [Google Scholar] [CrossRef]
- D’Alonzo, M.; Delbianco, A.; Lanzoni, C.; Autonell, C.R.; Gilmer, D.; Ratti, C. Beet soil-borne mosaic virus RNA-4 encodes a 32 kDa protein involved in symptom expression and in virus transmission through Polymyxa betae. Virology 2012, 423, 187–194. [Google Scholar] [CrossRef]
- Tamada, T.; Abe, H. Evidence that Beet necrotic yellow vein virus RNA-4 is essential for efficient transmission by the fungus Polymyxa betae. J. Gen. Virol. 1989, 70, 3391–3398. [Google Scholar] [CrossRef]
- Lavenus, J.; Goh, T.; Roberts, I.; Guyomarc’h, S.; Lucas, M.; de Smet, I.; Fukaki, H.; Beeckman, T.; Bennett, M.; Laplaze, L. Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 2013, 18, 450–458. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Estelle, M. Auxin signaling and regulated protein degradation. Trends Plant Sci. 2004, 9, 302–308. [Google Scholar] [CrossRef]
- Dharmasiri, N.; Dharmasiri, S.; Weijers, D.; Lechner, E.; Yamada, M.; Hobbie, L.; Ehrismann, J.S.; Jürgens, G.; Estelle, M. Plant development is regulated by a family of auxin receptor F box proteins. Dev. Cell 2005, 9, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Trinh, C.D.; Laplaze, L.; Guyomarc’h, S. Lateral root formation: Building a meristem de novo. Annu. Plant Rev. 2018, 1, 1–44. [Google Scholar]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Peltier, C.; Schmidlin, L.; Klein, E.; Taconnat, L.; Prinsen, E.; Erhardt, M.; Heintz, D.; Weyens, G.; Lefebvre, M.; Renou, J.-P.; et al. Expression of the Beet necrotic yellow vein virus p25 protein induces hormonal changes and a root branching phenotype in Arabidopsis thaliana. Transgenic Res. 2011, 20, 443–466. [Google Scholar] [CrossRef]
- Schmidlin, L.; de Bruyne, E.; Weyens, G.; Lefebvre, M.; Gilmer, D. Identification of differentially expressed root genes upon rhizomania disease. Mol. Plant Pathol. 2008, 9, 741–751. [Google Scholar] [CrossRef]
- Meunier, A.; Schmit, J.-F.; Stas, A.; Kutluk, N.; Bragard, C. Multiplex reverse transcription-PCR for simultaneous detection of Beet necrotic yellow vein virus, Beet soilborne virus, and Beet virus Q and their vector Polymyxa betae KESKIN on sugar beet. Appl. Environ. Microbiol. 2003, 69, 2356–2360. [Google Scholar] [CrossRef]
- Laufer, M.; Mohammad, H.; Maiss, E.; Richert-Pöggeler, K.; Dall’Ara, M.; Ratti, C.; Gilmer, D.; Liebe, S.; Varrelmann, M. Biological properties of Beet soil-borne mosaic virus and Beet necrotic yellow vein virus cDNA clones produced by isothermal in vitro recombination: Insights for reassortant appearance. Virology 2018, 518, 25–33. [Google Scholar] [CrossRef]
- Bornemann, K.; Varrelmann, M. Analysis of the resistance-breaking ability of different beet necrotic yellow vein virus isolates loaded into a single Polymyxa betae population in soil. Phytopathology 2011, 101, 718–724. [Google Scholar] [CrossRef]
- Voinnet, O.; Vain, P.; Angell, S.; Baulcombe, D.C. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 1998, 95, 177–187. [Google Scholar] [CrossRef]
- Pferdmenges, F.; Korf, H.; Varrelmann, M. Identification of rhizomania-infected soil in Europe able to overcome Rz1 resistance in sugar beet and comparison with other resistance-breaking soils from different geographic origins. Eur. J. Plant Pathol. 2009, 124, 31–43. [Google Scholar] [CrossRef]
- Verwaaijen, B.; Wibberg, D.; Kröber, M.; Winkler, A.; Zrenner, R.; Bednarz, H.; Niehaus, K.; Grosch, R.; Pühler, A.; Schlüter, A. The Rhizoctonia solani AG1-IB (isolate 7/3/14) transcriptome during interaction with the host plant lettuce (Lactuca sativa L.). PLoS ONE 2017, 12, e0177278. [Google Scholar] [CrossRef] [PubMed]
- Verwaaijen, B.; Wibberg, D.; Winkler, A.; Zrenner, R.; Bednarz, H.; Niehaus, K.; Grosch, R.; Pühler, A.; Schlüter, A. A comprehensive analysis of the Lactuca sativa, L. transcriptome during different stages of the compatible interaction with Rhizoctonia solani. Sci. Rep. 2019, 9, 7221. [Google Scholar] [CrossRef]
- Wibberg, D.; Andersson, L.; Tzelepis, G.; Rupp, O.; Blom, J.; Jelonek, L.; Pühler, A.; Fogelqvist, J.; Varrelmann, M.; Schlüter, A.; et al. Genome analysis of the sugar beet pathogen Rhizoctonia solani AG2-2IIIB revealed high numbers in secreted proteins and cell wall degrading enzymes. BMC Genom. 2016, 17, 245. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Hilker, R.; Stadermann, K.B.; Doppmeier, D.; Kalinowski, J.; Stoye, J.; Straube, J.; Winnebald, J.; Goesmann, A. ReadXplorer--visualization and analysis of mapped sequences. Bioinformatics 2014, 30, 2247–2254. [Google Scholar] [CrossRef]
- Hilker, R.; Stadermann, K.B.; Schwengers, O.; Anisiforov, E.; Jaenicke, S.; Weisshaar, B.; Zimmermann, T.; Goesmann, A. ReadXplorer 2-detailed read mapping analysis and visualization from one single source. Bioinformatics 2016, 32, 3702–3708. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ai, C.; Kong, L. CGPS: A machine learning-based approach integrating multiple gene set analysis tools for better prioritization of biologically relevant pathways. J. Genet. Genom. 2018, 45, 489–504. [Google Scholar] [CrossRef]
- Wu, J.; Mao, X.; Cai, T.; Luo, J.; Wei, L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006, 34, W720–W724. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.-Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef]
- Jin, J.; He, K.; Tang, X.; Li, Z.; Lv, L.; Zhao, Y.; Luo, J.; Gao, G. An Arabidopsis transcriptional regulatory map reveals distinct functional and evolutionary features of novel transcription factors. Mol. Biol. Evol. 2015, 32, 1767–1773. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef]
- Jin, J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC BioInform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Boutilier, K.; Offringa, R.; Sharma, V.K.; Kieft, H.; Ouellet, T.; Zhang, L.; Hattori, J.; Liu, C.-M.; van Lammeren, A.A.M.; Miki, B.L.A.; et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 2002, 14, 1737–1749. [Google Scholar] [CrossRef] [PubMed]
- Ohto, M.-A.; Hayashi, S.; Sawa, S.; Hashimoto-Ohta, A.; Nakamura, K. Involvement of HLS1 in sugar and auxin signaling in Arabidopsis leaves. Plant Cell Physiol. 2006, 47, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Tiryaki, I.; Rowe, M.L. Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 2002, 14, 1405–1415. [Google Scholar] [CrossRef]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef]
- Berckmans, B.; Vassileva, V.; Schmid, S.P.C.; Maes, S.; Parizot, B.; Naramoto, S.; Magyar, Z.; Alvim Kamei, C.L.; Koncz, C.; Bögre, L.; et al. Auxin-dependent cell cycle reactivation through transcriptional regulation of Arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 2011, 23, 3671–3683. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, J. EXPANSINA17 up-regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol. 2013, 54, 1600–1611. [Google Scholar] [CrossRef]
- Lee, H.W.; Kim, M.-J.; Kim, N.Y.; Lee, S.H.; Kim, J. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013, 73, 212–224. [Google Scholar] [CrossRef]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; de Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription factor functional protein-protein interactions in plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, K.; Somssich, I.E. Transcriptional networks in plant immunity. New Phytol. 2015, 206, 932–947. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Glazebrook, J.; Clarke, J.D.; Volko, S.; Dong, X. The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 1997, 88, 57–63. [Google Scholar] [CrossRef]
- Delaney, T.P.; Friedrich, L.; Ryals, J.A. Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. USA 1995, 92, 6602–6606. [Google Scholar] [CrossRef]
- Rochon, A.; Boyle, P.; Wignes, T.; Fobert, P.R.; Després, C. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. Plant Cell 2006, 18, 3670–3685. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, D.; Chu, J.Y.; Boyle, P.; Wang, Y.; Bridle, I.D.; de Luca, V.; Després, C. The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 2012, 1, 639–647. [Google Scholar] [CrossRef]
- Zhang, Y.; Tessaro, M.J.; Lassner, M.; Li, X. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. Plant Cell 2003, 15, 2647–2653. [Google Scholar] [CrossRef]
- Foley, R.C.; Singh, K.B. TGA5 acts as a positive and TGA4 acts as a negative regulator of ocs element activity in Arabidopsis roots in response to defence signals. FEBS Lett. 2004, 563, 141–145. [Google Scholar] [CrossRef]
- Forouhar, F.; Yang, Y.; Kumar, D.; Chen, Y.; Fridman, E.; Park, S.W.; Chiang, Y.; Acton, T.B.; Montelione, G.T.; Pichersky, E.; et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. USA 2005, 102, 1773–1778. [Google Scholar] [CrossRef]
- Ding, C.-K.; Wang, C.Y.; Gross, K.C.; Smith, D.L. Jasmonate and salicylate induce the expression of pathogenesis-related-protein genes and increase resistance to chilling injury in tomato fruit. Planta 2002, 214, 895–901. [Google Scholar] [CrossRef]
- Niki, T.; Mitsuhara, I.; Seo, S.; Ohtsubo, N.; Ohashi, Y. Antagonistic effect of salicylic acid and jasmonic acid on the expression of pathogenesis-related (PR) protein genes in wounded mature tobacco leaves. Plant Cell Physiol. 1998, 39, 500–507. [Google Scholar] [CrossRef]
- Jin Kim, Y.; Kook Hwang, B. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant 2000, 108, 51–60. [Google Scholar] [CrossRef]
- Xu, Y.; Chang, P.; Liu, D.; Narasimhan, M.L.; Raghothama, K.G.; Hasegawa, P.M.; Bressan, R.A. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell 1994, 6, 1077–1085. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.E.; Findell, J.L.; Schaller, G.E.; Sisler, E.C.; Bleecker, A.B. Ethylene perception by the ERS1 protein in Arabidopsis. Plant Physiol. 2000, 123, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Potuschak, T.; Lechner, E.; Parmentier, Y.; Yanagisawa, S.; Grava, S.; Koncz, C.; Genschik, P. EIN3-dependent regulation of plant ethylene hormone signaling by two arabidopsis F box proteins: EBF1 and EBF2. Cell 2003, 115, 679–689. [Google Scholar] [CrossRef]
- Solano, R.; Stepanova, A.; Chao, Q.; Ecker, J.R. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998, 12, 3703–3714. [Google Scholar] [CrossRef]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef]
- Fan, H.; Sun, H.; Wang, Y.; Zhang, Y.; Wang, X.; Li, D.; Yu, J.; Han, C. Deep sequencing-based transcriptome profiling reveals comprehensive insights into the responses of Nicotiana benthamiana to beet necrotic yellow vein virus infections containing or lacking RNA4. PLoS ONE 2014, 9, e85284. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Y.; Sun, H.; Liu, J.; Wang, Y.; Wang, X.; Li, D.; Yu, J.; Han, C. Transcriptome analysis of Beta macrocarpa and identification of differentially expressed transcripts in response to Beet necrotic yellow vein virus infection. PLoS ONE 2015, 10, e0132277. [Google Scholar] [CrossRef]
- Zheng, Y.; Ding, B.; Fei, Z.; Wang, Y. Comprehensive transcriptome analyses reveal tomato plant responses to tobacco rattle virus-based gene silencing vectors. Sci. Rep. 2017, 7, 9771. [Google Scholar] [CrossRef]
- Zuluaga, A.P.; Vega-Arreguín, J.C.; Fei, Z.; Matas, A.J.; Patev, S.; Fry, W.E.; Rose, J.K.C. Analysis of the tomato leaf transcriptome during successive hemibiotrophic stages of a compatible interaction with the oomycete pathogen Phytophthora infestans. Mol. Plant Pathol. 2016, 17, 42–54. [Google Scholar] [CrossRef]
- Fang, J.; Lin, A.; Qiu, W.; Cai, H.; Umar, M.; Chen, R.; Ming, R. Transcriptome profiling revealed stress-induced and disease resistance genes up-regulated in PRSV resistant transgenic papaya. Front. Plant Sci. 2016, 7, 855. [Google Scholar] [CrossRef]
- Pollini, C.P.; Giunchedi, L. Comparative histopathology of sugar beets that are susceptible and partially resistant to Rhizomania. Phytopathol. Mediterr. 1989, 28, 16–21. [Google Scholar]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benková, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Zhu, J.; Du, X.; Cui, X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta 2012, 236, 1227–1237. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, N.Y.; Lee, D.J.; Kim, J. LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009, 151, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishna, P.; Ruiz Duarte, P.; Rance, G.A.; Schubert, M.; Vordermaier, V.; Vu, L.D.; Murphy, E.; Vilches Barro, A.; Swarup, K.; Moirangthem, K.; et al. EXPANSIN A1-mediated radial swelling of pericycle cells positions anticlinal cell divisions during lateral root initiation. Proc. Natl. Acad. Sci. USA 2019, 116, 8597–8602. [Google Scholar] [CrossRef]
- Sampedro, J.; Cosgrove, D.J. The expansin superfamily. Genome Biol. 2005, 6, 242. [Google Scholar] [CrossRef][Green Version]
- Che, J.; Yamaji, N.; Shen, R.F.; Ma, J.F. An Al-inducible expansin gene, OsEXPA10 is involved in root cell elongation of rice. Plant J. 2016, 88, 132–142. [Google Scholar] [CrossRef]
- Park, S.-H.; Li, F.; Renaud, J.; Shen, W.; Li, Y.; Guo, L.; Cui, H.; Sumarah, M.; Wang, A. NbEXPA1, an α-expansin, is plasmodesmata-specific and a novel host factor for potyviral infection. Plant J. 2017, 92, 846–861. [Google Scholar] [CrossRef]
- Tan, J.; Wang, M.; Shi, Z.; Miao, X. OsEXPA10 mediates the balance between growth and resistance to biotic stress in rice. Plant Cell Rep. 2018, 37, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Miedes, E.; Vanholme, R.; Boerjan, W.; Molina, A. The role of the secondary cell wall in plant resistance to pathogens. Front. Plant Sci. 2014, 5, 358. [Google Scholar] [CrossRef] [PubMed]
- Nyalugwe, E.P.; Barbetti, M.J.; Clode, P.L.; Jones, R.A.C. Systemic hypersensitive resistance to turnip mosaic virus in Brassica juncea is associated with multiple defense responses, especially phloem necrosis and xylem occlusion. Plant Dis. 2016, 100, 1261–1270. [Google Scholar] [CrossRef] [PubMed]
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.-Q.; Fan, H.-Y.; Jiang, N.; Wang, Y.; Zhang, Z.-Y.; Zhang, Y.-L.; Wang, X.-B.; Li, D.-W.; Yu, J.-L.; Han, C.-G. Infection of Beet necrotic yellow vein virus with RNA4-encoded P31 specifically up-regulates pathogenesis-related protein 10 in Nicotiana benthamiana. Virol. J. 2014, 11, 118. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Klessig, D.F. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc. Natl. Acad. Sci. USA 2003, 100, 16101–16106. [Google Scholar] [CrossRef]
- Zhu, X.; Qi, L.; Liu, X.; Cai, S.; Xu, H.; Huang, R.; Li, J.; Wei, X.; Zhang, Z. The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 2014, 164, 1499–1514. [Google Scholar] [CrossRef]
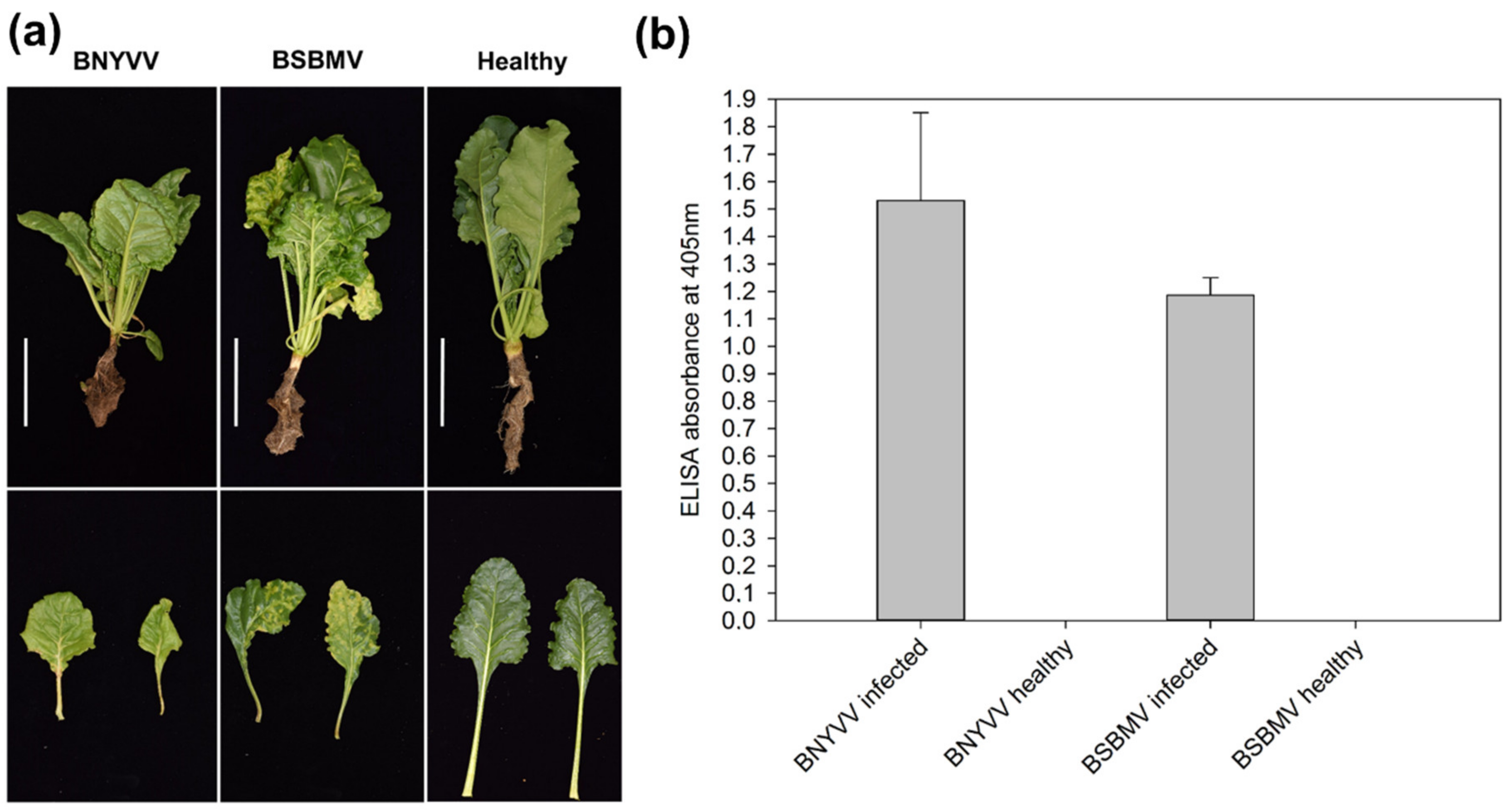
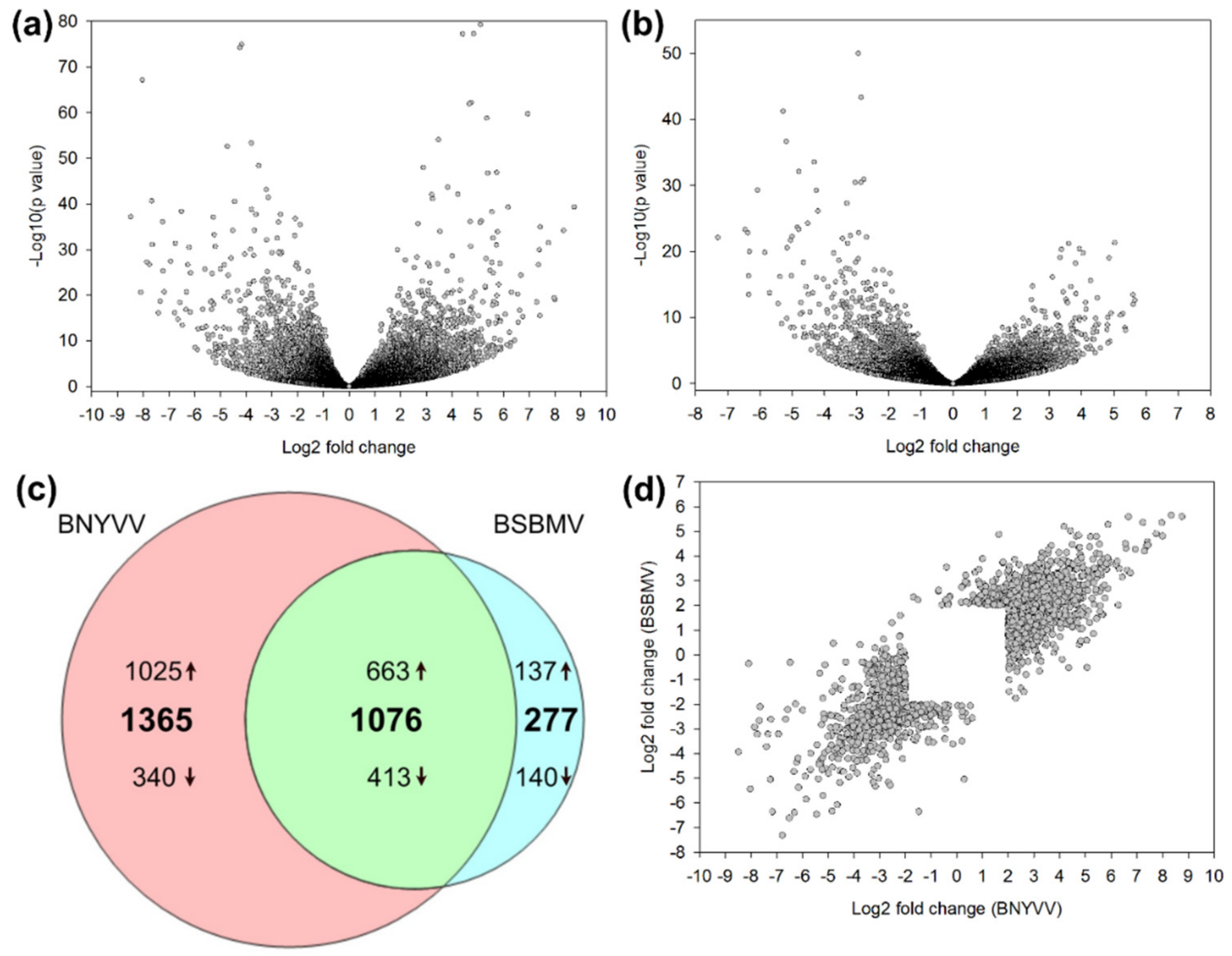
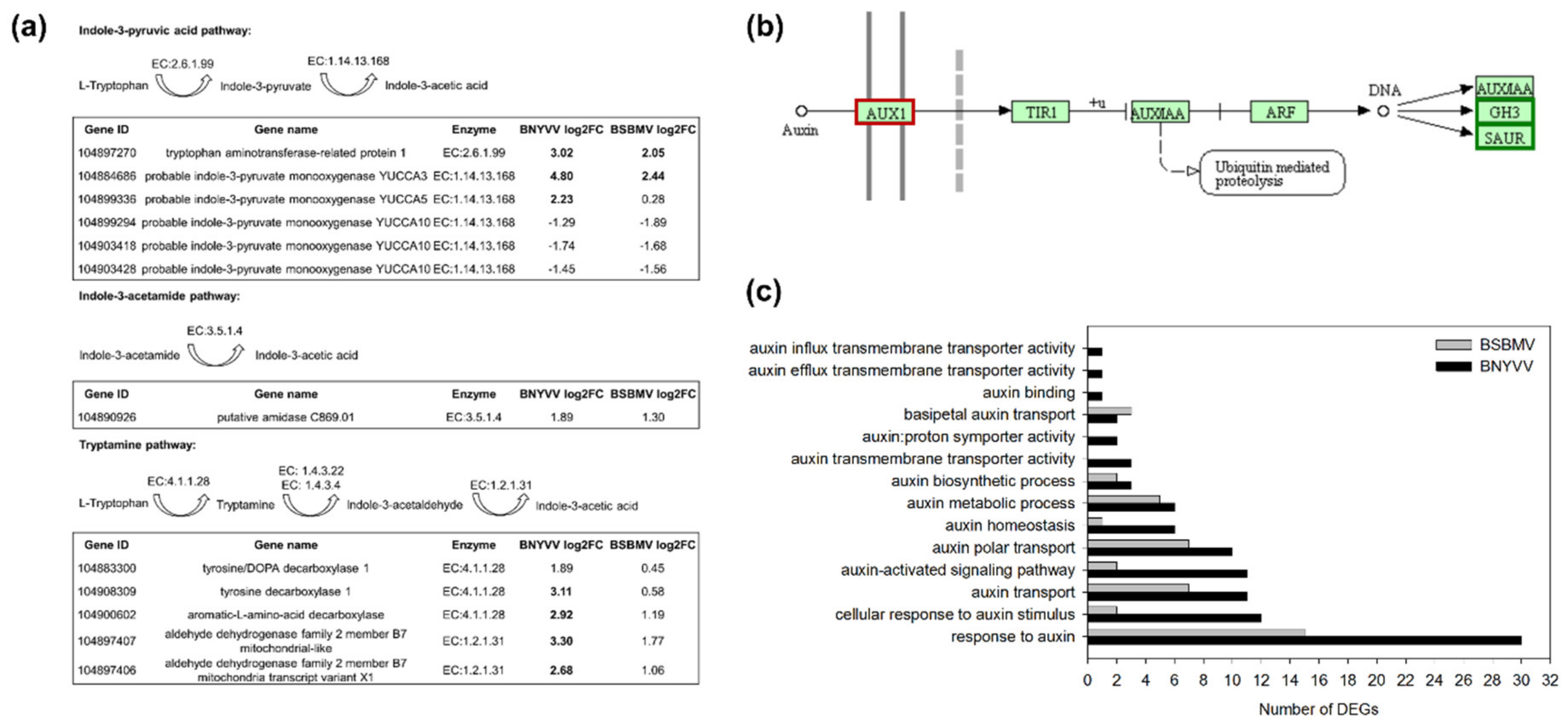

| Virus | Repetition | Sequenced Reads | Filtered Reads (%) | Mapping Virus (%) | Mapping Sugar Beet (%) |
|---|---|---|---|---|---|
| BNYVV | 1 | 21,958,505 | 99.99 | 4.12 | 79.97 |
| 2 | 21,892,827 | 99.98 | 7.32 | 77.43 | |
| 3 | 23,448,634 | 99.99 | 3.98 | 80.30 | |
| BSBMV | 1 | 27,134,031 | 99.99 | 24.92 | 62.65 |
| 2 | 24,148,407 | 99.99 | 23.22 | 63.35 | |
| 3 | 19,896,214 | 99.99 | 25.2 | 62.51 | |
| Healthy | 1 | 23,734,086 | 100 | 0 | 84.32 |
| 2 | 21,551,604 | 99.99 | 0 | 84.44 | |
| 3 | 18,678,083 | 99.99 | 0 | 84.13 |
| GOID | Term | BNYVV DEGs | BSBMV DEGs | ||||
|---|---|---|---|---|---|---|---|
| Total | Up | Down | Total | Up | Down | ||
| GO:0048046 | apoplast | 99 | 75 | 24 | 54 | 36 | 18 |
| GO:0003824 | catalytic activity | 883 | 657 | 226 | 480 | 311 | 169 |
| GO:0071944 | cell periphery | 531 | 393 | 138 | 282 | 181 | 101 |
| GO:0005618 | cell wall | 173 | 123 | 50 | 102 | 64 | 38 |
| GO:0071554 | cell wall organization or biogenesis | 128 | 112 | 16 | 57 | 50 | 7 |
| GO:0030312 | external encapsulating structure | 173 | 123 | 50 | 102 | 64 | 38 |
| GO:0005576 | extracellular region | 387 | 301 | 86 | 216 | 155 | 61 |
| GO:0009813 | flavonoid biosynthetic process | 45 | 32 | 13 | 29 | 14 | 15 |
| GO:0009812 | flavonoid metabolic process | 52 | 36 | 16 | 33 | 16 | 17 |
| GO:0016020 | membrane | 774 | 566 | 208 | 430 | 266 | 164 |
| GO:0008152 | metabolic process | 991 | 711 | 280 | 542 | 330 | 212 |
| GO:0055114 | oxidation-reduction process | 221 | 148 | 73 | 127 | 72 | 55 |
| GO:0016491 | oxidoreductase activity | 217 | 151 | 66 | 128 | 78 | 50 |
| GO:0009505 | plant-type cell wall | 99 | 72 | 27 | 59 | 36 | 23 |
| GO:0009628 | response to abiotic stimulus | 246 | 133 | 113 | 159 | 66 | 93 |
| GO:0042221 | response to chemical | 305 | 189 | 116 | 179 | 77 | 102 |
| GO:1901700 | response to oxygen-containing compound | 199 | 125 | 74 | 113 | 49 | 64 |
| GO:0050896 | response to stimulus | 624 | 398 | 226 | 371 | 174 | 197 |
| GO:0006950 | response to stress | 405 | 263 | 142 | 249 | 118 | 131 |
| GO:0044763 | single-organism cellular process | 663 | 469 | 194 | 373 | 226 | 147 |
| GO:0044710 | single-organism metabolic process | 489 | 344 | 145 | 272 | 164 | 108 |
| GO:0044699 | single-organism process | 961 | 690 | 271 | 529 | 325 | 204 |
| KEGG Pathway | BNYVV | BSBMV | ||
|---|---|---|---|---|
| Genes | p-Value | Genes | p-Value | |
| Phenylpropanoid biosynthesis | 48 | 2.55 × 10−11 | 24 | 3.90 × 10−05 |
| Biosynthesis of secondary metabolites | 157 | 9.85 × 10−11 | 82 | 4.00 × 10−05 |
| Metabolic pathways | 236 | 9.39 × 10−10 | 127 | 4.00 × 10−05 |
| Flavonoid biosynthesis | 10 | 0.0011 | 5 | 0.0572 |
| Starch and sucrose metabolism | 32 | 0.0080 | 16 | 0.1663 |
| Glycolysis/gluconeogenesis | 21 | 0.0165 | 10 | 0.2386 |
| Steroid biosynthesis | 10 | 0.0178 | 3 | 0.4754 |
| Pentose and glucuronate interconversions | 16 | 0.0205 | 6 | 0.3923 |
| Stilbenoid, diarylheptanoid and gingerol biosynthesis | 11 | 0.0271 | 3 | 0.5831 |
| Tyrosine metabolism | 10 | 0.0290 | 4 | 0.3607 |
| Alanine, aspartate and glutamate metabolism | 11 | 0.0295 | 3 | 0.5831 |
| Circadian rhythm-plant | 9 | 0.0388 | 7 | 0.0314 |
| Sesquiterpenoid and triterpenoid biosynthesis | 7 | 0.0391 | 5 | 0.0633 |
| Arginine and proline metabolism | 11 | 0.0443 | 6 | 0.2386 |
| NCBI GENE ID | Gene Name | BNYVV log2FC | p-Value | BSBMV log2FC | p-Value |
|---|---|---|---|---|---|
| LBDs | |||||
| 104904514 | LBD 33 | 4.82 | 0.0000 | 0.57 | 0.6849 |
| 104890156 | LBD 6 | 4.05 | 0.0000 | 1.09 | 0.4096 |
| 104905420 | LBD 19 | 2.44 | 0.0000 | 1.14 | 0.1231 |
| 104908367 | LBD 15 | 2.43 | 0.0000 | 0.88 | 0.1850 |
| 104905421 | LBD 18 | 2.30 | 0.0000 | 0.83 | 0.1930 |
| 104903105 | LBD 21 | −0.70 | 0.5803 | −2.32 | 0.0316 |
| 104906092 | LBD 41 | −2.77 | 0.0000 | −2.42 | 0.0000 |
| 104893389 | LBD 20 | −3.42 | 0.0000 | −2.72 | 0.0015 |
| EXPs | |||||
| 104905343 | EXPB15L | 6.31 | 0.0000 | 3.48 | 0.0001 |
| 104892376 | EXPA7 | 4.64 | 0.0000 | 2.03 | 0.0646 |
| 104893284 | EXPA7L | 4.39 | 0.0001 | 2.11 | 0.0771 |
| 104904256 | EXPA4c | 3.52 | 0.0000 | 2.85 | 0.0000 |
| 104887636 | EXPA10 | 3.07 | 0.0005 | 3.65 | 0.0000 |
| 104903052 | EXPB3 | 3.05 | 0.0000 | 3.06 | 0.0000 |
| 104903845 | EXLA1b | 2.9 | 0.0000 | 1.71 | 0.0456 |
| 104894031 | EXPA10L-2 | 2.43 | 0.0002 | 2.34 | 0.0005 |
| 104887634 | EXPA4a | 2.14 | 0.0021 | 2.02 | 0.0063 |
| 104903843 | EXLA1a | 2.09 | 0.0005 | 1.76 | 0.0108 |
| 104887635 | EXPA2L | 1.43 | 0.2301 | 2.95 | 0.0008 |
| 104892824 | EXLB1a | −2.35 | 0.0000 | −2.09 | 0.0000 |
| 104908292 | EXLB1c | −3.30 | 0.0001 | −1.92 | 0.0523 |
| 104904178 | EXLA3c | −5.5 | 0.0000 | −3.63 | 1.0000 |
| 104904176 | EXLA3b | −6.57 | 0.0000 | −2.22 | 0.0606 |
| Transcription Factor Family | Downregulated | Upregulated | ||
|---|---|---|---|---|
| BNYVV | BSBMV | BNYVV | BSBMV | |
| AP2 | 1 | 1 | 3 | 3 |
| ARF | 0 | 0 | 1 | 0 |
| ARF-B | 0 | 0 | 1 | 0 |
| B3 | 1 | 0 | 3 | 3 |
| bHLH | 9 | 6 | 16 | 5 |
| bZIP | 3 | 3 | 1 | 0 |
| C2H2 | 2 | 0 | 5 | 1 |
| C3H | 2 | 1 | 1 | 0 |
| CO-like | 2 | 2 | 0 | 0 |
| DBB | 1 | 1 | 0 | 0 |
| DoF | 1 | 1 | 4 | 0 |
| E2F/DP | 0 | 0 | 1 | 0 |
| ERF | 5 | 6 | 11 | 4 |
| FAR1 | 1 | 1 | 0 | 0 |
| G2-like | 2 | 0 | 0 | 0 |
| GATA | 0 | 0 | 1 | 1 |
| GRAS | 0 | 0 | 2 | 1 |
| HD-ZIP | 3 | 0 | 2 | 1 |
| HRT-like | 0 | 0 | 1 | 1 |
| HSF | 1 | 1 | 0 | 0 |
| LBD | 2 | 3 | 5 | 0 |
| MIKC_MADS | 3 | 1 | 3 | 0 |
| MYB | 1 | 2 | 8 | 3 |
| MYB_related | 1 | 0 | 3 | 2 |
| NAC | 0 | 1 | 8 | 0 |
| NF-YA | 1 | 0 | 0 | 0 |
| NF-YC | 0 | 0 | 0 | 1 |
| RAV | 1 | 2 | 0 | 0 |
| TALE | 1 | 0 | 1 | 0 |
| TCP | 0 | 0 | 2 | 0 |
| Trihelix | 1 | 1 | 1 | 0 |
| WOX | 0 | 0 | 2 | 0 |
| WRKY | 2 | 4 | 5 | 1 |
| ZF-HD | 0 | 0 | 1 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernando Gil, J.; Wibberg, D.; Eini, O.; Savenkov, E.I.; Varrelmann, M.; Liebe, S. Comparative Transcriptome Analysis Provides Molecular Insights into the Interaction of Beet necrotic yellow vein virus and Beet soil-borne mosaic virus with Their Host Sugar Beet. Viruses 2020, 12, 76. https://doi.org/10.3390/v12010076
Fernando Gil J, Wibberg D, Eini O, Savenkov EI, Varrelmann M, Liebe S. Comparative Transcriptome Analysis Provides Molecular Insights into the Interaction of Beet necrotic yellow vein virus and Beet soil-borne mosaic virus with Their Host Sugar Beet. Viruses. 2020; 12(1):76. https://doi.org/10.3390/v12010076
Chicago/Turabian StyleFernando Gil, Jose, Daniel Wibberg, Omid Eini, Eugene I. Savenkov, Mark Varrelmann, and Sebastian Liebe. 2020. "Comparative Transcriptome Analysis Provides Molecular Insights into the Interaction of Beet necrotic yellow vein virus and Beet soil-borne mosaic virus with Their Host Sugar Beet" Viruses 12, no. 1: 76. https://doi.org/10.3390/v12010076
APA StyleFernando Gil, J., Wibberg, D., Eini, O., Savenkov, E. I., Varrelmann, M., & Liebe, S. (2020). Comparative Transcriptome Analysis Provides Molecular Insights into the Interaction of Beet necrotic yellow vein virus and Beet soil-borne mosaic virus with Their Host Sugar Beet. Viruses, 12(1), 76. https://doi.org/10.3390/v12010076





