Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants
Abstract
1. Introduction
2. Materials and Methods
2.1. Compilation of Sequence Datasets
2.2. A/H1N1pdm09 Transmission within India
2.3. Predictors of Transmission within India
2.4. Positive Selection Analysis
3. Results
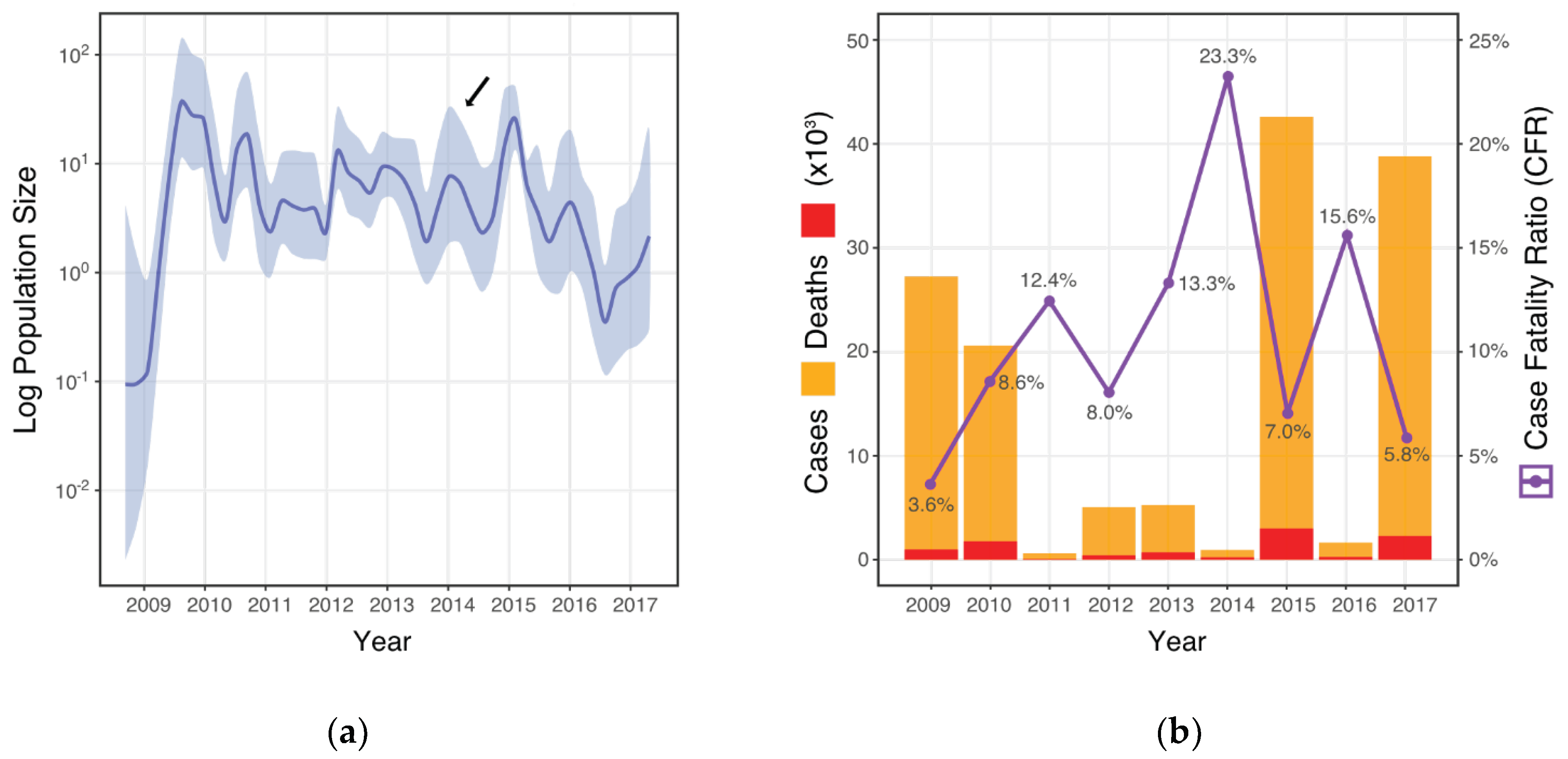
3.1. CFR and Viral Population Demographics
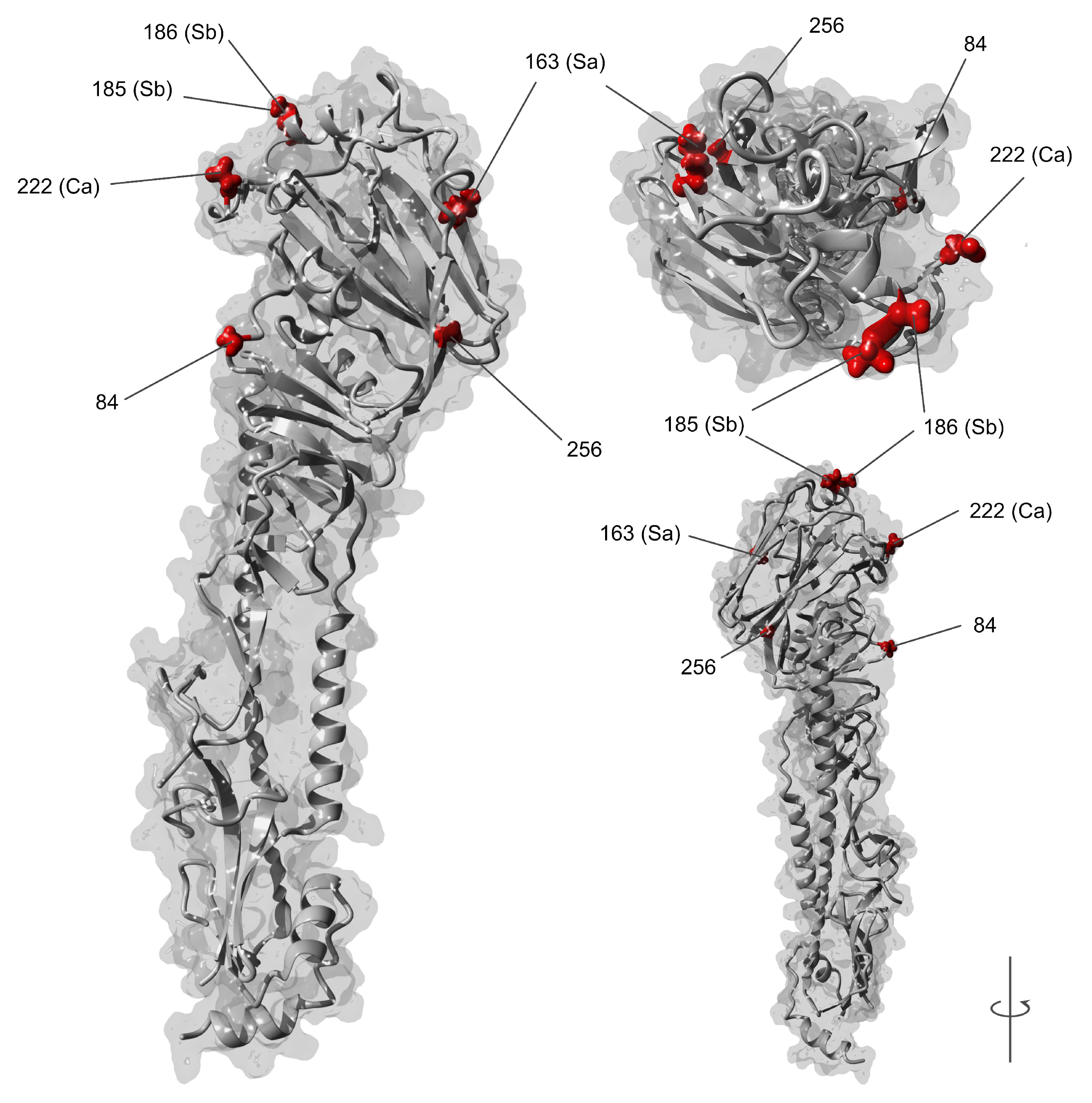
3.2. dN/dS Selection Analysis and Amino Acid Variations
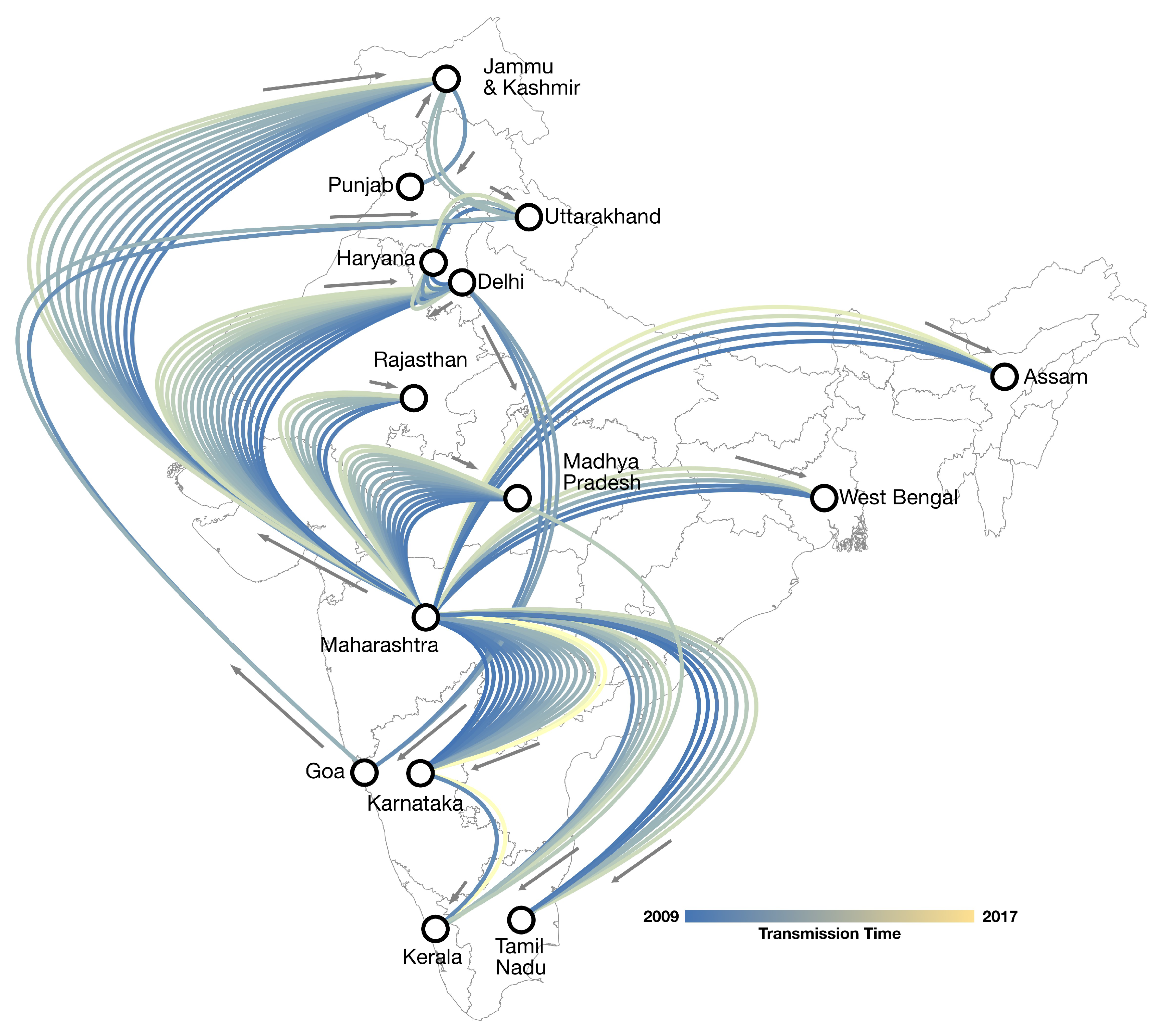
3.3. Phylogeography of A/H1N1pdm09
3.4. Generalized Linear Modelling
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Girard, M.P.; Tam, J.S.; Assossou, O.M.; Kieny, M.P. The 2009 A (H1N1) influenza virus pandemic: A review. Vaccine 2010, 28, 4895–4902. [Google Scholar] [CrossRef] [PubMed]
- Peiris, J.S.M.; Poon, L.L.M.; Guan, Y. Emergence of a novel swine-origin influenza a virus (S-OIV) H1N1 virus in humans. J. Clin. Virol. 2009, 45, 169–173. [Google Scholar] [CrossRef]
- Webb, S.A.R.; Pettilä, V.; Seppelt, I.; Bellomo, R.; Bailey, M.; Cooper, D.J.; Cretikos, M.; Davies, A.R.; Finfer, S.; Harrigan, P.W.J.; et al. Critical Care Services and 2009 H1N1 Influenza in Australia and New Zealand. N. Engl. J. Med. 2009, 361, 1925–1934. [Google Scholar] [PubMed]
- Mitchell, R.; Ogunremi, T.; Astrakianakis, G.; Bryce, E.; Gervais, R.; Gravel, D.; Johnston, L.; LeDuc, S.; Roth, V.; Taylor, G.; et al. Impact of the 2009 influenza A (H1N1) pandemic on Canadian health care workers: A survey on vaccination, illness, absenteeism, and personal protective equipment. Am. J. Infect. Control 2012, 40, 611–616. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.Y.; Kelly, H.; Ip, D.K.M.; Wu, J.T.; Leung, G.M.; Cowling, B.J. Case fatality risk of influenza a (H1N1pdm09): A systematic review. Epidemiology 2013, 24, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Dawood, F.S.; Iuliano, A.D.; Reed, C.; Meltzer, M.I.; Shay, D.K.; Cheng, P.-Y.; Bandaranayake, D.; Breiman, R.F.; Brooks, W.A.; Buchy, P.; et al. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Van Kerkhove, M.D.; Hirve, S.; Koukounari, A.; Mounts, A.W. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influ. Other Respir. Viruses 2013, 7, 872–886. [Google Scholar] [CrossRef]
- Simonsen, L.; Spreeuwenberg, P.; Lustig, R.; Taylor, R.J.; Fleming, D.M.; Kroneman, M.; Van Kerkhove, M.D.; Mounts, A.W.; Paget, W.J.; The GLaMOR Collaborating Teams. Global Mortality Estimates for the 2009 Influenza Pandemic from the GLaMOR Project: A Modeling Study. PLoS Med. 2013, 10, e1001558. [Google Scholar] [CrossRef]
- World Health Organization. H1N1 in Post-pandemic Period. Available online: https://www.who.int/mediacentre/news/statements/2010/h1n1_vpc_20100810/en/ (accessed on 31 December 2017).
- Broor, S.; Krishnan, A.; Roy, D.S.; Dhakad, S.; Kaushik, S.; Mir, M.A.; Singh, Y.; Moen, A.; Chadha, M.; Mishra, A.C.; et al. Dynamic Patterns of Circulating Seasonal and Pandemic A(H1N1)pdm09 Influenza Viruses From 2007–2010 in and around Delhi, India. PLOS ONE 2012, 7, e29129. [Google Scholar] [CrossRef][Green Version]
- Bedford, T.; Riley, S.; Barr, I.G.; Broor, S.; Chadha, M.; Cox, N.J.; Daniels, R.S.; Gunasekaran, C.P.; Hurt, A.C.; Kelso, A.; et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature 2015, 523, 217–220. [Google Scholar] [CrossRef]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. The Global Circulation of Seasonal Influenza A (H3N2) Viruses. Science 2008, 320, 340–346. [Google Scholar] [CrossRef]
- Choudhry, A.; Singh, S.; Khare, S.; Rai, A.; Rawat, D.; Aggarwal, R.; Chauhan, L. Emergence of pandemic 2009 influenza A H1N1, India. Indian J. Med. Res. 2012, 135, 534–537. [Google Scholar]
- Gurav, Y.K.; Pawar, S.D.; Chadha, M.S.; A Potdar, V.; Deshpande, A.S.; Koratkar, S.S.; Hosmani, A.H.; Mishra, A.C. Pandemic influenza A(H1N1) 2009 outbreak in a residential school at Panchgani, Maharashtra, India. Indian J. Med. Res. 2010, 132, 67–71. [Google Scholar]
- Cases of Influenza a H1N1 (swine flu)—State/ut- Wise, Year- Wise for 2009, 2010, 2011 and 2012. Available online: http://mohfw. nic.in/showfile.php?lid=2121 (accessed on 31 December 2017).
- Seasonal Influenza (h1n1)– State/ut- Wise, Year- Wise Number of Cases and Death from 2010 to 2017. Available online: http://idsp.nic.in/showfile.php?lid=3908 (accessed on 31 December 2017).
- Murray, C.J.; Lopez, A.D.; Chin, B.; Feehan, D.; Hill, K.H. Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: A quantitative analysis. Lancet 2006, 368, 2211–2218. [Google Scholar] [CrossRef]
- Kumar, T.; Bhatia, D.; Lakshmi, P.V.M.; Laserson, K.F.; Narain, J.P. Risk factors for death during a resurgence of influenza-A (H1N1) pdm09 in Punjab State in 2013. Indian J. Public Health 2017, 61, 9. [Google Scholar] [CrossRef]
- Kulkarni, S.; Narain, J.; Gupta, S.; Dhariwal, A.; Singh, S.; MacIntyre, C. Influenza A (H1N1) in India: Changing epidemiology and its implications. Natl. Med. J. India 2019. [Google Scholar] [CrossRef]
- Malhotra, B.; Singh, R.; Sharma, P.; Meena, D.; Gupta, J.; Atreya, A.; Meena, B.R. Epidemiological & clinical profile of influenza A (H1N1) 2009 virus infections during 2015 epidemic in Rajasthan. Indian J. Med. Res. 2016, 144, 918–923. [Google Scholar]
- Baillie, G.J.; Galiano, M.; Agapow, P.-M.; Myers, R.; Chiam, R.; Gall, A.; Palser, A.L.; Watson, S.J.; Hedge, J.; Underwood, A. Evolutionary dynamics of local pandemic H1N1/2009 influenza virus lineages revealed by whole-genome analysis. J. Virol. 2012, 86, 11–18. [Google Scholar] [CrossRef]
- Hedge, J.; Lycett, S.J.; Rambaut, A. Real-time characterization of the molecular epidemiology of an influenza pandemic. Boil. Lett. 2013, 9, 20130331. [Google Scholar] [CrossRef]
- Lemey, P.; Rambaut, A.; Bedford, T.; Faria, N.R.; Bielejec, F.; Baele, G.; Russell, C.A.; Smith, D.J.; Pybus, O.G.; Brockmann, D.; et al. Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2. PLOS Pathog. 2014, 10, e1003932. [Google Scholar] [CrossRef]
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Boil. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, 16. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the Accuracy of Demographic and Molecular Clock Model Comparison While Accommodating Phylogenetic Uncertainty. Mol. Boil. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef]
- Baele, G.; Li, W.L.S.; Drummond, A.J.; Suchard, M.A.; Lemey, P. Accurate Model Selection of Relaxed Molecular Clocks in Bayesian Phylogenetics. Mol. Boil. Evol. 2012, 30, 239–243. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.T.; Carvalho, L.M.; Pybus, O.G. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, ew007. [Google Scholar] [CrossRef]
- Rambaut, A.; Suchard, M.; Xie, D.; Drummond, A. Tracer v1.6 (Software). Available online: http://tree.bio.ed.ac.uk/software/tracer/ (accessed on 15 May 2019).
- Minin, V.N.; Bloomquist, E.W.; Suchard, M.A. Smooth Skyride through a Rough Skyline: Bayesian Coalescent-Based Inference of Population Dynamics. Mol. Boil. Evol. 2008, 25, 1459–1471. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, M.A. Improving Bayesian Population Dynamics Inference: A Coalescent-Based Model for Multiple Loci. Mol. Boil. Evol. 2012, 30, 713–724. [Google Scholar] [CrossRef]
- Bielejec, F.; Baele, G.; Vrancken, B.; Suchard, M.A.; Rambaut, A.; Lemey, P. SpreaD3: Interactive Visualization of Spatiotemporal History and Trait Evolutionary Processes. Mol. Boil. Evol. 2016, 33, 2167–2169. [Google Scholar] [CrossRef] [PubMed]
- Lemey, P.; Rambaut, A.; Drummond, A.J.; Suchard, M.A. Bayesian Phylogeography Finds Its Roots. PLoS Comput. Boil. 2009, 5, e1000520. [Google Scholar] [CrossRef] [PubMed]
- Jeffreys, H. The Theory of Probability, 3rd ed.; Oxford University Press: Oxford, UK, 1961. [Google Scholar]
- Dudas, G.; Carvalho, L.M.; Bedford, T.; Tatem, A.J.; Baele, G.; Faria, N.R.; Park, D.J.; Ladner, J.T.; Arias, A.; Asogun, D.; et al. Virus genomes reveal factors that spread and sustained the Ebola epidemic. Nature 2017, 544, 309. [Google Scholar] [CrossRef] [PubMed]
- Scotch, M.; Mei, C.; Makonnen, Y.J.; Pinto, J.; Ali, A.; Vegso, S.; Kane, M.; Sarkar, I.N.; Rabinowitz, P. Phylogeography of influenza A H5N1 clade 2.2.1.1 in Egypt. BMC Genom. 2013, 14, 871. [Google Scholar] [CrossRef] [PubMed]
- Domestic Traffic Reports. Available online: http://dgca.nic.in/reports/Traffic-ind.htm (accessed on 22 August 2019).
- Beard, R.; Magee, D.; Suchard, M.A.; Lemey, P.; Scotch, M. Generalized Linear Models for Identifying Predictors of the Evolutionary Diffusion of Viruses. AMIA Jt. Summits Transl. Sci. Proc. 2014, 2014, 23–28. [Google Scholar]
- Yang, Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J. Mol. Evol. 1994, 39, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Pond, S.L.K.; Frost, S.D.W. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol. Boil. Evol. 2005, 22, 1208–1222. [Google Scholar] [CrossRef]
- Pond, S.L.K.; Muse, S.V. HyPhy: Hypothesis Testing Using Phylogenies. In Practical Considerations for Adaptive Trial Design and Implementation; Springer Science and Business Media LLC: Berlin, Germany, 2005; pp. 125–181. [Google Scholar]
- Burke, D.F.; Smith, D.J. A Recommended Numbering Scheme for Influenza A HA Subtypes. PLOS ONE 2014, 9, e112302. [Google Scholar] [CrossRef]
- Yang, H.; Chang, J.C.; Guo, Z.; Carney, P.J.; Shore, D.A.; Donis, R.O.; Cox, N.J.; Villanueva, J.M.; Klimov, A.I.; Stevens, J. Structural Stability of Influenza A(H1N1)pdm09 Virus Hemagglutinins. J. Virol. 2014, 88, 4828–4838. [Google Scholar] [CrossRef]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef]
- Nishiura, H. Case fatality ratio of pandemic influenza. Lancet Infect. Dis. 2010, 10, 443–444. [Google Scholar] [CrossRef]
- Cousins, S. Death toll from swine flu in India exceeds 2500. BMJ 2015, 351, 4966. [Google Scholar] [CrossRef]
- India Struggles with Deadly Swine Flu Outbreak. Available online: https://www.bbc.com/news/world-asia-india-31547455 (accessed on 18 January 2019).
- Swine Flu Deaths at 1895; Number of Cases near 32k Mark. Available online: https://indianexpress.com/article/india/india-others/swine-flu-deaths-at-1895-cases-near-32k-mark/ (accessed on 18 January 2019).
- Summary of the 2015–2016 Influenza Season. Available online: https://www.cdc.gov/flu/about/season/flu-season-2015–2016.htm (accessed on 23 January 2019).
- Influenza in Europe—Season 2013–2014; European Centre for Disease Prevention and Control (ECDC): Stockholm. Available online: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/Influenza-2013-14-season-report.pdf (accessed on 23 January 2019).
- Petrova, V.N.; Russell, C.A. The evolution of seasonal influenza viruses. Nat. Rev. Microbiol. 2018, 16, 47. [Google Scholar] [CrossRef]
- Treanor, J.J. 167—influenza (including avian influenza and swine influenza). In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases, 8th ed.; Bennett, J.E., Dolin, R., Blaser, M.J., Eds.; Elsevier Saunders: Philadelphia, Pennsylvania, 2015; pp. 2000–2024. [Google Scholar]
- Hay, A.J.; Gregory, V.; Douglas, A.R.; Lin, Y.P. The evolution of human influenza viruses. Philos. Trans. R. Soc. B Boil. Sci. 2001, 356, 1861–1870. [Google Scholar] [CrossRef]
- Yasuhara, A.; Yamayoshi, S.; Soni, P.; Takenaga, T.; Kawakami, C.; Takashita, E.; Sakai-Tagawa, Y.; Uraki, R.; Ito, M.; Iwatsuki-Horimoto, K.; et al. Diversity of antigenic mutants of influenza A(H1N1)pdm09 virus escaped from human monoclonal antibodies. Sci. Rep. 2017, 7, 17735. [Google Scholar] [CrossRef]
- Horm, S.V.; Mardy, S.; Rith, S.; Ly, S.; Heng, S.; Vong, S.; Kitsutani, P.; Ieng, V.; Tarantola, A.; Ly, S. Epidemiological and virological characteristics of influenza viruses circulating in Cambodia from 2009 to 2011. PloS. ONE 2014, 9, e110713. [Google Scholar] [CrossRef]
- Nguyen, H.K.L.; Nguyen, P.T.K.; Nguyen, T.C.; Hoang, P.V.M.; Le, T.T.; Vuong, C.D.; Nguyen, A.P.; Tran, L.T.T.; Nguyen, B.G.; Le, M.Q. Virological characterization of influenza H1N1pdm09 in Vietnam, 2010-2013. Influ. Other Respir. Viruses 2015, 9, 216–224. [Google Scholar] [CrossRef]
- Pandey, S.; Sahu, M.; Potdar, V.; Barde, P. Molecular analysis of influenza A H1N1pdm09 virus circulating in Madhya Pradesh, India in the year 2017. VirusDisease 2018, 29, 380–384. [Google Scholar] [CrossRef]
- Ramos, A.P.; Herrera, B.A.; Ramírez, O.V.; García, A.A.; Jiménez, M.M.; Valdés, C.S.; Fernández, A.G.; González, G.; Fernández, S.I.O.; Báez, G.G.; et al. Molecular and phylogenetic analysis of influenza A H1N1 pandemic viruses in Cuba, May 2009 to August 2010. Int. J. Infect. Dis. 2013, 17, e565–e567. [Google Scholar] [CrossRef][Green Version]
- Houng, H.-S.H.; Garner, J.; Zhou, Y.; Lyons, A.; Kuschner, R.; Deye, G.; Clair, K.S.; Douce, R.W.; Chicaiza, W.; Blair, P.J.; et al. Emergent 2009 influenza A(H1N1) viruses containing HA D222N mutation associated with severe clinical outcomes in the Americas. J. Clin. Virol. 2012, 53, 12–15. [Google Scholar] [CrossRef]
- Ledesma, J.; Pozo, F.; Reina, G.; Blasco, M.; Rodríguez, G.; Montes, M.; López-Miragaya, I.; Salvador, C.; Reina, J.; De Lejarazu, R.O.; et al. Genetic diversity of influenza A(H1N1)2009 virus circulating during the season 2010–2011 in Spain. J. Clin. Virol. 2012, 53, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, T.; De Rosa, F.; Cerutti, F.; Pagani, N.; Allice, T.; Stella, M.L.; Milia, M.G.; Calcagno, A.; Burdino, E.; Gregori, G.; et al. A(H1N1)pdm09 hemagglutinin D222G and D222N variants are frequently harbored by patients requiring extracorporeal membrane oxygenation and advanced respiratory assistance for severe A(H1N1)pdm09 infection. Influ. Other Respir. Viruses 2013, 7, 1416–1426. [Google Scholar] [CrossRef] [PubMed]
- Koel, B.F.; Burke, D.F.; Bestebroer, T.M.; Van Der Vliet, S.; Zondag, G.C.M.; Vervaet, G.; Skepner, E.; Lewis, N.S.; Spronken, M.I.J.; Russell, C.A.; et al. Substitutions Near the Receptor Binding Site Determine Major Antigenic Change During Influenza Virus Evolution. Science 2013, 342, 976–979. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Llamas, R.; Alfaro-Ruiz, L.; Canon, C.A.; Rosshandler, I.I.; Cruz-Lagunas, A.; Zúñiga, J.; Vega, R.R.; Wong, C.W.; Maurer-Stroh, S.; Córdoba, S.R. Molecular features of influenza A (H1N1) pdm09 prevalent in Mexico during winter seasons 2012–2014. PloS. ONE 2017, 12, e0180419. [Google Scholar] [CrossRef] [PubMed]
- Parida, M.; Dash, P.K.; Kumar, J.S.; Joshi, G.; Tandel, K.; Sharma, S.; Srivastava, A.; Agarwal, A.; Saha, A.; Saraswat, S. Emergence of influenza A (H1N1) pdm09 Genogroup 6B and drug resistant virus, India, January to May 2015. Eurosurveillance 2016, 21, 30124. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Shirakura, M.; Fujisaki, S.; Kishida, N.; Burke, D.F.; Smith, D.J.; Kuwahara, T.; Takashita, E.; Takayama, I.; Nakauchi, M.; et al. Characterization of influenza A(H1N1)pdm09 viruses isolated from Nepalese and Indian outbreak patients in early 2015. Influ. Other Respir. Viruses 2017, 11, 399–403. [Google Scholar] [CrossRef] [PubMed]
- Linderman, S.L.; Chambers, B.S.; Zost, S.J.; Parkhouse, K.; Li, Y.; Herrmann, C.; Ellebedy, A.H.; Carter, D.M.; Andrews, S.F.; Zheng, N.Y.; et al. Potential antigenic explanation for atypical H1N1 infections among middle-aged adults during the 2013–2014 influenza season. Proc. Natl. Acad. Sci. USA 2014, 111, 15798–15803. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2017 southern hemisphere influenza. In Weekly Epidemiological Record; World Health Organization: Geneva, Switzerland, 2016; Volume 91. [Google Scholar]
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2019 southern hemisphere influenza. In Weekly Epidemiological Record; World Health Organization: Geneva, Switzerland, 2018; Volume 93, pp. 553–576. [Google Scholar]
- World Health Organization. Recommended composition of influenza virus vaccines for use in the 2019–2020 northern hemisphere influenza. In Weekly Epidemiological Record; World Health Organization: Geneva, Switzerland, 2019; Volume 94, pp. 141–160. [Google Scholar]
- Abed, Y.; Pizzorno, A.; Hamelin, M.-E.; Leung, A.; Joubert, P.; Couture, C.; Kobasa, D.; Boivin, G.; Pizzorno, M.A. The 2009 Pandemic H1N1 D222G Hemagglutinin Mutation Alters Receptor Specificity and Increases Virulence in Mice but Not in Ferrets. J. Infect. Dis. 2011, 204, 1008–1016. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Belser, J.A.; Jayaraman, A.; Raman, R.; Pappas, C.; Zeng, H.; Cox, N.J.; Katz, J.M.; Sasisekharan, R.; Tumpey, T.M. Effect of D222G Mutation in the Hemagglutinin Protein on Receptor Binding, Pathogenesis and Transmissibility of the 2009 Pandemic H1N1 Influenza Virus. PLOS ONE 2011, 6, e25091. [Google Scholar] [CrossRef] [PubMed]
- A Vazquez-Perez, J.; Isa, P.; Kobasa, D.; E Ormsby, C.; E Ramírez-Gonzalez, J.; Romero-Rodríguez, D.P.; Ranadheera, C.; Li, Y.; Bastien, N.; Embury-Hyatt, C.; et al. A (H1N1) pdm09 HA D222 variants associated with severity and mortality in patients during a second wave in Mexico. Virol. J. 2013, 10, 41. [Google Scholar] [CrossRef] [PubMed]
- Goka, E.A.; Vallely, P.J.; Mutton, K.J.; Klapper, P.E.; Goka, E. Mutations associated with severity of the pandemic influenza A(H1N1)pdm09 in humans: a systematic review and meta-analysis of epidemiological evidence. Arch. Virol. 2014, 159, 3167–3183. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Nayak, M.K.; Dutta, S.; Panda, S.; Satpathi, B.R.; Chawla-Sarkar, M. Genetic Characterization of Circulating 2015 A(H1N1)pdm09 Influenza Viruses from Eastern India. PLoS ONE 2016, 11, 0168464. [Google Scholar] [CrossRef] [PubMed]
- Tharakaraman, K.; Sasisekharan, R. Influenza Surveillance: 2014–2015 H1N1 “Swine”-Derived Influenza Viruses from India. Cell Host Microbe 2015, 17, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Press note 116939. Available online: http://pib.nic.in/newsite/printrelease.aspx?Relid=116939 (accessed on 2 May 2019).
- Nelson, M.I.; Simonsen, L.; Viboud, C.; Miller, M.A.; Taylor, J.; George, K.S.; Griesemer, S.B.; Ghedin, E.; Sengamalay, N.A.; Spiro, D.J.; et al. Stochastic Processes Are Key Determinants of Short-Term Evolution in Influenza A Virus. PLOS Pathog. 2006, 2, e125. [Google Scholar]
- Lowen, A.C.; Mubareka, S.; Steel, J.; Palese, P. Influenza Virus Transmission Is Dependent on Relative Humidity and Temperature. PLOS Pathog. 2007, 3, e151. [Google Scholar] [CrossRef] [PubMed]
- Lowen, A.C.; Steel, J. Roles of Humidity and Temperature in Shaping Influenza Seasonality. J. Virol. 2014, 88, 7692–7695. [Google Scholar] [CrossRef]
- Koul, P.A.; Broor, S.; Saha, S.; Barnes, J.; Smith, C.; Shaw, M.; Chadha, M.; Lal, R.B. Differences in Influenza Seasonality by Latitude, Northern India. Emerg. Infect. Dis. 2014, 20, 1746–1749. [Google Scholar] [CrossRef]
- Chadha, M.S.; Potdar, V.A.; Saha, S.; Koul, P.A.; Broor, S.; Dar, L.; Chawla-Sarkar, M.; Biswas, D.; Gunasekaran, P.; Abraham, A.M.; et al. Dynamics of Influenza Seasonality at Sub-Regional Levels in India and Implications for Vaccination Timing. PLOS ONE 2015, 10, 0124122. [Google Scholar] [CrossRef]
- Magee, D.; Suchard, M.A.; Scotch, M. Bayesian phylogeography of influenza A/H3N2 for the 2014-15 season in the United States using three frameworks of ancestral state reconstruction. PLoS Comput. Biol 2017, 13, 1005389. [Google Scholar] [CrossRef]
- De Maio, N.; Wu, C.-H.; O’Reilly, K.M.; Wilson, D. New Routes to Phylogeography: A Bayesian Structured Coalescent Approximation. PLoS Genet. 2015, 11, 1005421. [Google Scholar] [CrossRef]
- Indian Railways Statistical Publications 2016–17: Statistical summary—Indian Railways. Available online: http://www.indianrailways.gov.in/railwayboard/uploads/directorate/stat_econ/IRSP_2016–17/Annual_Report_Accounts_Eng/Statistical_Summary.pdf (accessed on 15 August 2019).
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of Amino Acids in HA and PB2 Critical for the Transmission of H5N1 Avian Influenza Viruses in a Mammalian Host. PLOS Pathog. 2009, 5, e1000709. [Google Scholar] [CrossRef] [PubMed]
| Year | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| S/UT (Population 106) | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 | Total | % |
| Assam (31.2) | 4 | 4 | 1 | 11 | 20 | 3.3 | |||||
| Delhi (16.8) | 10 | 4 | 7 | 14 | 6 | 2 | 20 | 63 | 10.3 | ||
| Goa (1.5) | 1 | 3 | 3 | 3 | 10 | 1.6 | |||||
| Haryana (25.4) | 1 | 2 | 1 | 3 | 3 | 2 | 12 | 2.0 | |||
| Jammu and Kashmir (12.5) | 1 | 2 | 8 | 29 | 7 | 11 | 58 | 9.5 | |||
| Karnataka (61.1) | 19 | 16 | 5 | 15 | 6 | 61 | 10.0 | ||||
| Kerala (33.4) | 1 | 2 | 9 | 2 | 3 | 3 | 2 | 22 | 3.6 | ||
| Madhya Pradesh (72.6) | 4 | 8 | 4 | 5 | 5 | 18 | 44 | 7.2 | |||
| Maharashtra (112.4) | 42 | 8 | 5 | 54 | 66 | 6 | 53 | 8 | 242 | 39.5 | |
| Punjab (27.7) | 1 | 2 | 4 | 7 | 1.1 | ||||||
| Rajasthan (68.6) | 3 | 1 | 3 | 5 | 6 | 18 | 2.9 | ||||
| Tamil Nadu (72.15) | 5 | 2 | 2 | 6 | 1 | 16 | 2.6 | ||||
| Uttarakhand (10.1) | 1 | 1 | 1 | 2 | 1 | 6 | 1.0 | ||||
| West Bengal (91.3) | 3 | 5 | 26 | 34 | 5.5 | ||||||
| Year Total | 96 | 53 | 37 | 143 | 96 | 13 | 147 | 19 | 9 | 613 | 100.0 |
| Site (H3 a) | Ag b | India Taxa (n = 613) | International (S1) | International (S2) | |||
|---|---|---|---|---|---|---|---|
| dN/dS | 95% BCI | dN/dS | 95% BCI | dN/dS | 95% BCI | ||
| 84 (92) | n/a | 8.14 | (5.68–10.92) | 2.53 | (1.69–3.45) | 3.51 | (2.33–4.79) |
| 163 (166) | Sa | 3.83 | (2.68–5.35) | 4.54 | (2.98–6.38) | 2.62 | (1.79–3.68) |
| 185 (188) | Sb | 4.50 | (3.18–6.21) | 2.14 | (1.41–2.94) | 1.13 | (0.76–1.54) |
| 186 (189) | Sb | 3.35 | (2.26–4.51) | 1.44 | (0.97–1.94) | 1.47 | (0.93–1.97) |
| 222 (225) | Ca | 13.42 | (9.43–18.42) | 3.42 | (2.30–4.81) | 4.32 | (2.87–5.85) |
| 256 (259) | n/a | 4.39 | (3.12–6.09) | 1.16 | (0.74–1.58) | 1.08 | (0.75–1.48) |
| Predictor | E(δ) Probability | (β| δ) Coefficient | 95% BCI | BF a |
|---|---|---|---|---|
| Distance | 0.88 | −0.53 | −0.79 to −0.28 | 269.3 |
| SS origin | 0.87 | 1.46 | 0.96 to 2.12 | 237.2 |
| P Flux destination | 0.31 | 0.35 | 0.15 to 0.56 | 15.67 |
| SS destination | 0.22 | 0.26 | 0.11 to 0.42 | 10.03 |
| Pop Dense origin | 0.2 | −1.43 | −2.82 to −0.28 | 8.93 |
| P Flux origin | 0.14 | 1.86 | −0.46 to 2.95 | 5.84 |
| Average temp origin | 0.12 | −0.49 | −0.8 to −0.18 | 4.69 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adam, D.C.; Scotch, M.; MacIntyre, C.R. Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants. Viruses 2019, 11, 791. https://doi.org/10.3390/v11090791
Adam DC, Scotch M, MacIntyre CR. Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants. Viruses. 2019; 11(9):791. https://doi.org/10.3390/v11090791
Chicago/Turabian StyleAdam, Dillon C., Matthew Scotch, and C Raina. MacIntyre. 2019. "Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants" Viruses 11, no. 9: 791. https://doi.org/10.3390/v11090791
APA StyleAdam, D. C., Scotch, M., & MacIntyre, C. R. (2019). Phylodynamics of Influenza A/H1N1pdm09 in India Reveals Circulation Patterns and Increased Selection for Clade 6b Residues and Other High Mortality Mutants. Viruses, 11(9), 791. https://doi.org/10.3390/v11090791






