Generation of Virus- and dsRNA-Derived siRNAs with Species-Dependent Length in Insects
Abstract
1. Introduction
2. Material and Methods
2.1. Insect Rearing
2.2. Cell Culture
2.3. Viral Production and Quantification
2.4. Viral Infections and Sample Collection
2.5. dsRNA Treatment and Sample Collection
2.6. RNA Extraction
2.7. Small RNA Sequencing and Data Analysis
3. Results
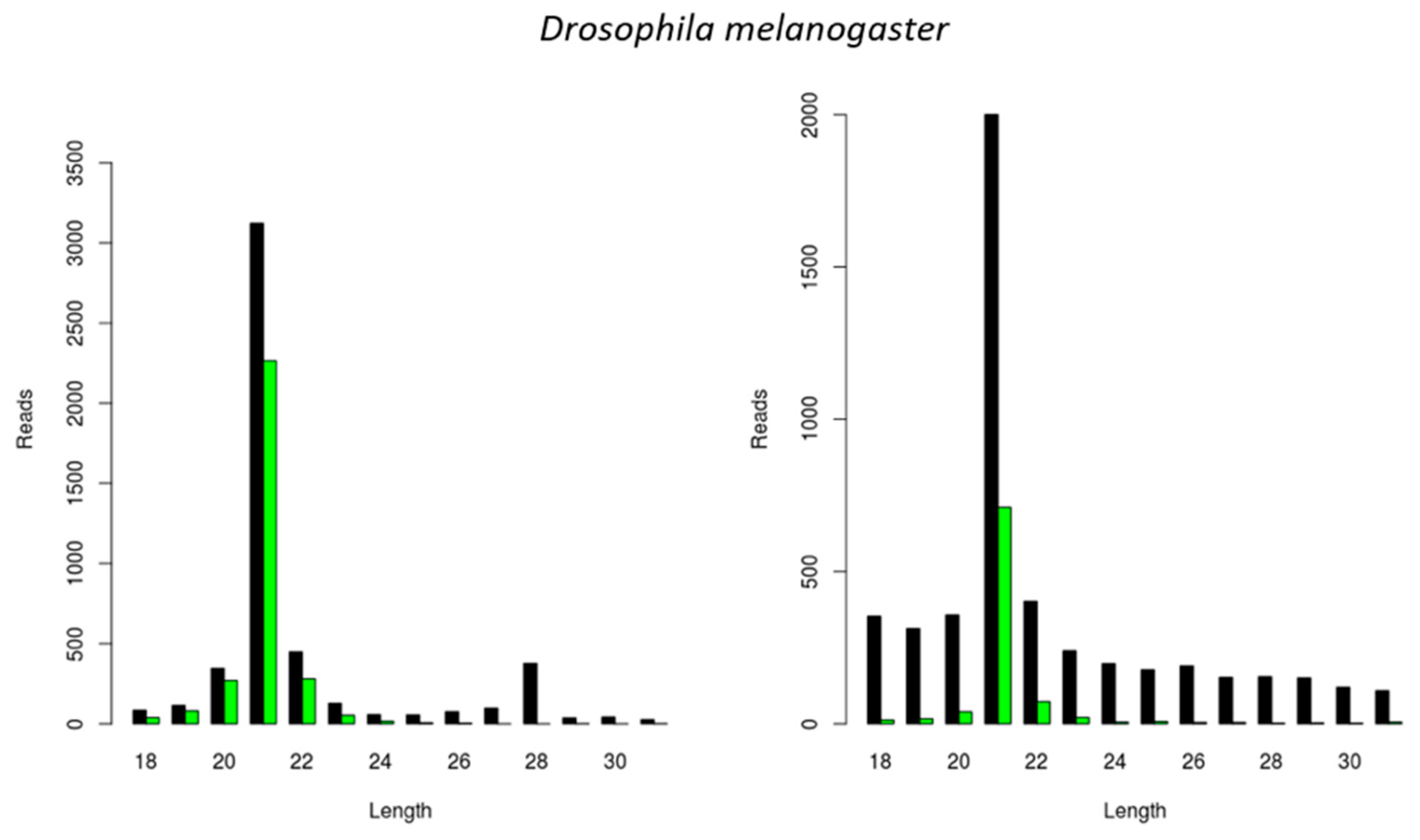
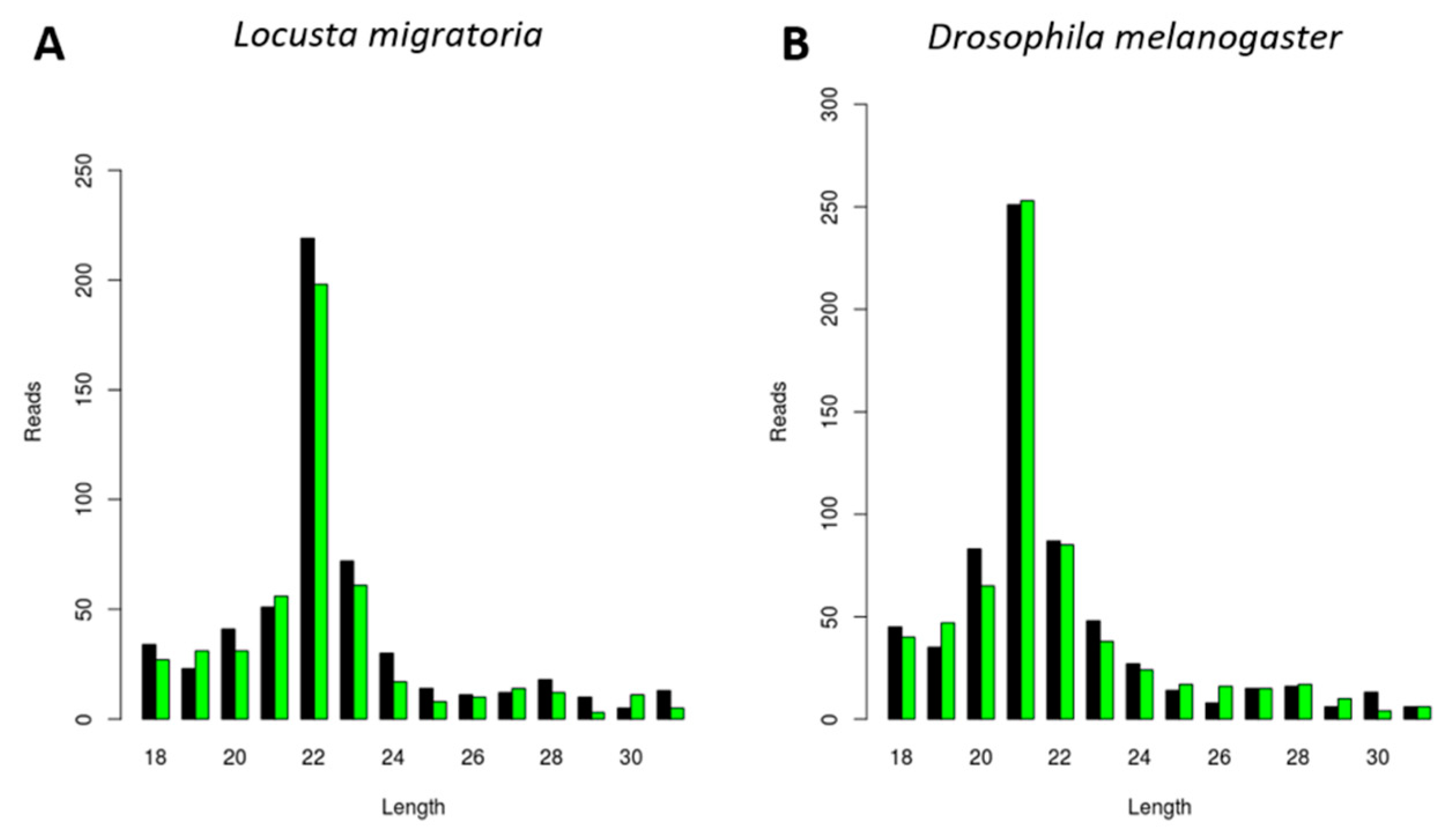
3.1. Generation of CrPV-Derived siRNAs with Species-Dependent Length Distribution
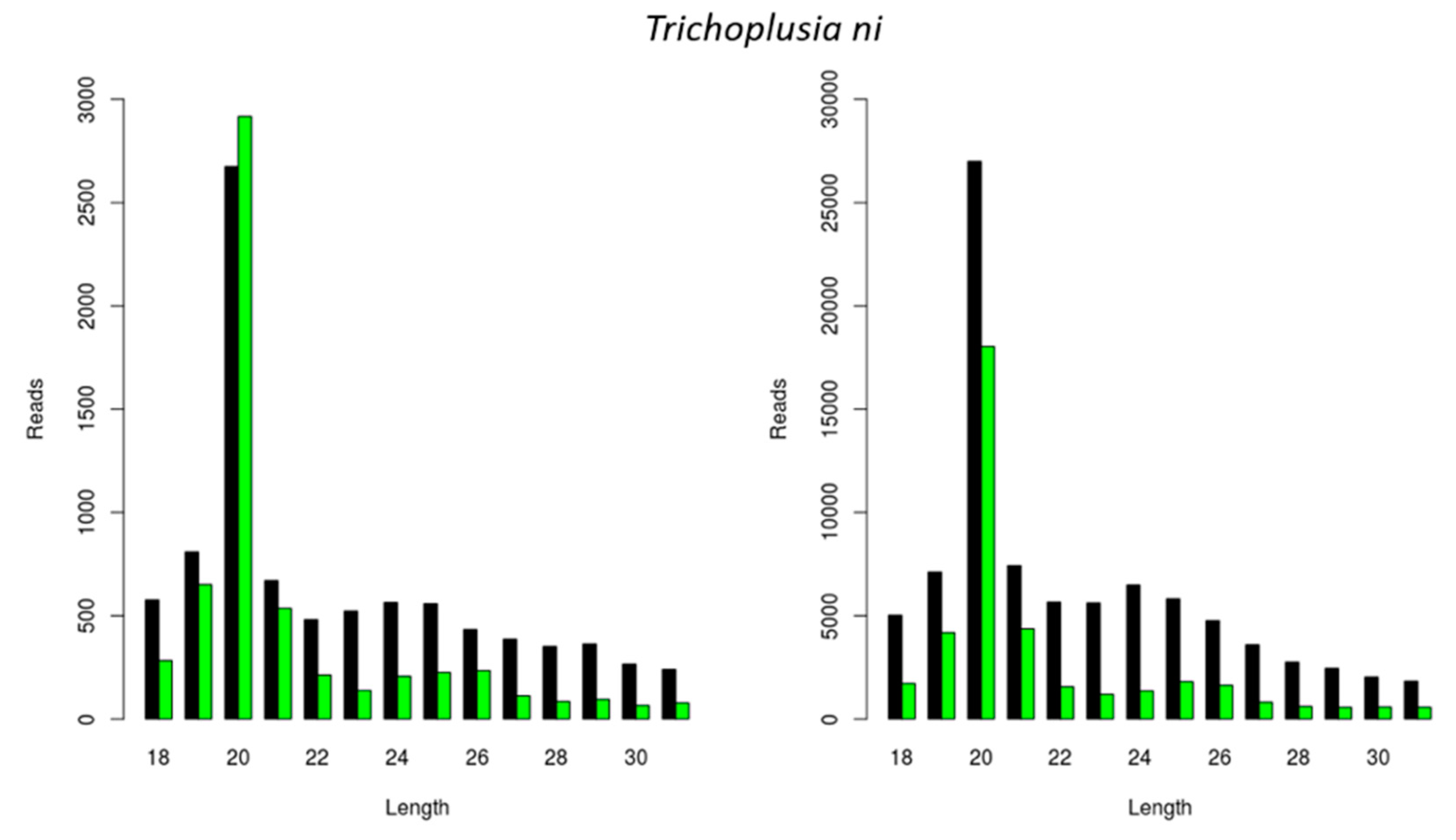
3.2. Generation of FHV-Derived siRNAs with Species-Dependent Length Distribution
3.3. Generation of Other Virally Derived siRNAs with Species-Dependent Length Distribution
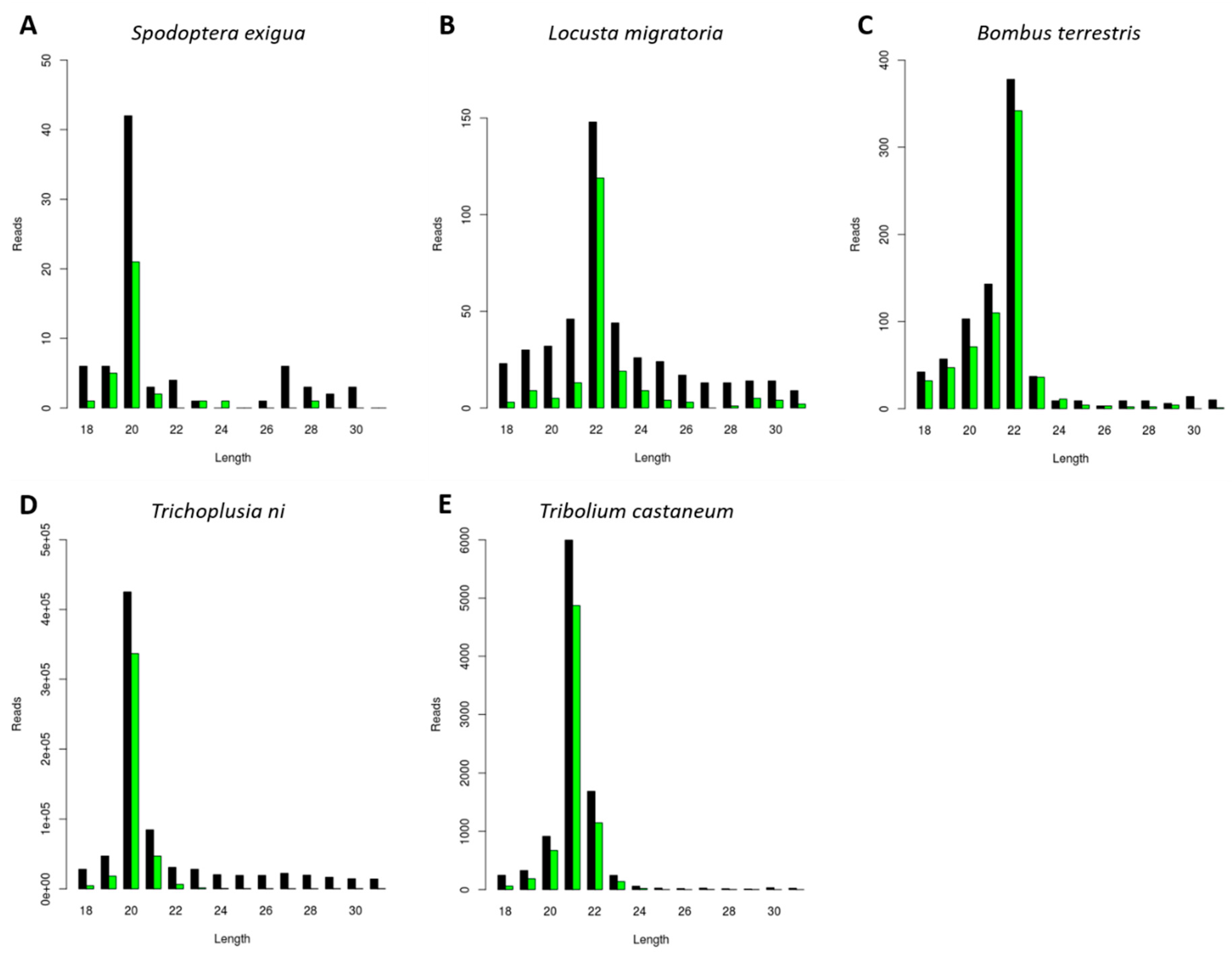
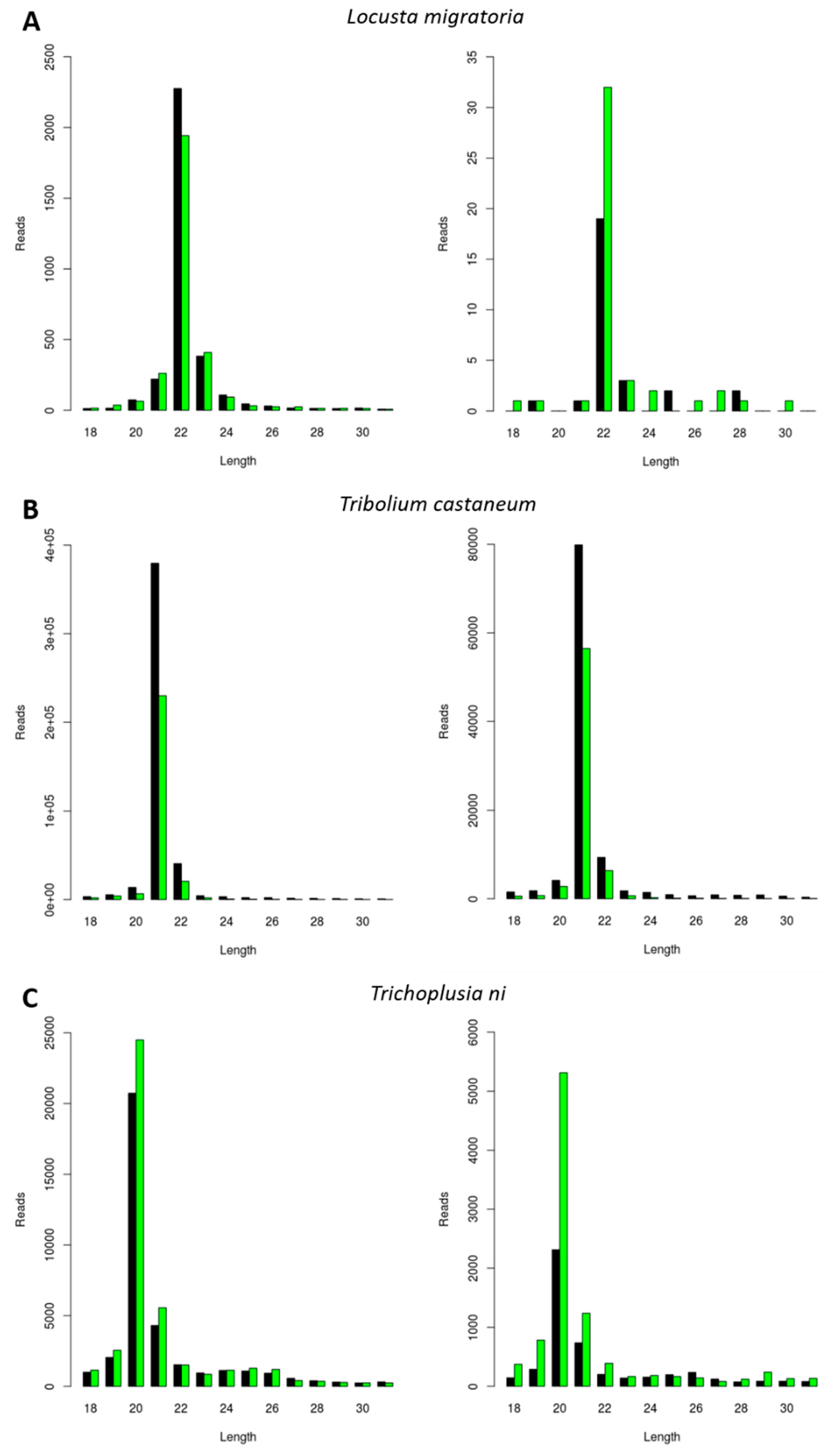
3.4. Generation of dsRNA-Derived siRNAs with Species-Dependent Length Distribution
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Martinez, J.; Patkaniowska, A.; Lendeckel, W.; Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001, 20, 6877–6888. [Google Scholar] [CrossRef] [PubMed]
- Nykänen, A.; Haley, B.; Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 2001, 3, 309–321. [Google Scholar] [CrossRef]
- Tomari, Y.; Matranga, C.; Haley, B.; Martinez, N.; Zamore, P.D. A protein sensor for siRNA asymmetry. Science 2004, 306, 1377–1380. [Google Scholar] [CrossRef] [PubMed]
- Bronkhorst, A.W.; van Cleef, K.W.R.; Vodovar, N.; Ince, I.A.; Blanc, H.; Vlak, J.M.; Saleh, M.-C.; van Rij, R.P. The DNA virus Invertebrate iridescent virus 6 is a target of the Drosophila RNAi machinery. Proc. Natl. Acad. Sci. USA 2012, 109, E3604–E3613. [Google Scholar] [CrossRef]
- Mongelli, V.; Saleh, M.-C. Bugs Are Not to Be Silenced: Small RNA Pathways and Antiviral Responses in Insects. Annu. Rev. Virol. 2016, 3, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Vogel, E.; Santos, D.; Mingels, L.; Verdonckt, T.W.; Vanden Broeck, J. RNA interference in insects: Protecting beneficials and controlling pests. Front. Physiol. 2019, 9, 1912. [Google Scholar] [CrossRef]
- Head, G.P.; Carroll, M.W.; Evans, S.P.; Rule, D.M.; Willse, A.R.; Clark, T.L.; Storer, N.P.; Flannagan, R.D.; Samuel, L.W.; Meinke, L.J. Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: Efficacy and resistance management. Pest Manag. Sci. 2017, 73, 1883–1899. [Google Scholar] [CrossRef]
- Kolliopoulou, A.; Taning, C.N.T.; Smagghe, G.; Swevers, L. Viral Delivery of dsRNA for Control of Insect Agricultural Pests and Vectors of Human Disease: Prospects and Challenges. Front. Physiol. 2017, 8, 399. [Google Scholar] [CrossRef]
- Taning, C.N.T.; Christiaens, O.; Li, X.X.; Swevers, L.; Casteels, H.; Maes, M.; Smagghe, G. Engineered flock house virus for targeted gene suppression through RNAi in fruit flies (Drosophila melanogaster) in vitro and in Vivo. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef]
- Niu, J.; Taning, C.N.T.; Christiaens, O.; Smagghe, G.; Wang, J.J. Rethink RNAi in Insect Pest Control: Challenges and Perspectives. In Advances in Insect Physiology; Smagghe, G., Ed.; Academic Press Inc.: Cambridge, MA, USA, 2018; pp. 1–17. ISBN 9780128150795. [Google Scholar]
- Smagghe, G.; Degheele, D. Action of a novel nonsteroidal ecdysteroid mimic, tebufenozide (RH-5992), on insects of different orders. Pestic. Sci. 1994, 42, 85–92. [Google Scholar] [CrossRef]
- Wang, H.; Smagghe, G.; Meeus, I. The role of a single gene encoding the Single von Willebrand factor C-domain protein (SVC) in bumblebee immunity extends beyond antiviral defense. Insect Biochem. Mol. Biol. 2017, 91, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Garrey, J.L.; Lee, Y.-Y.; Au, H.H.T.; Bushell, M.; Jan, E. Host and viral translational mechanisms during cricket paralysis virus infection. J. Virol. 2010, 84, 1124–1138. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. 2014. Available online: sourceforge.net/projects/bbmap/ (accessed on 11 August 2019).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 11 August 2019).
- Wu, Q.; Luo, Y.; Lu, R.; Lau, N.; Lai, E.C.; Li, W.-X.; Ding, S.-W. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 1606–1611. [Google Scholar] [CrossRef] [PubMed]
- Santos, D.; Wynant, N.; Van den Brande, S.; Verdonckt, T.-W.; Mingels, L.; Peeters, P.; Kolliopoulou, A.; Swevers, L.; Vanden Broeck, J. Insights into RNAi-based antiviral immunity in Lepidoptera: Acute and persistent infections in Bombyx mori and Trichoplusia ni cell lines. Sci. Rep. 2018, 8, 2423. [Google Scholar] [CrossRef] [PubMed]
- Zerbino, D.R. Using the Velvet de novo assembler for short-read sequencing technologies. Curr. Protoc. Bioinform. 2010, 31, 11. [Google Scholar]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- R Core Team R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Watson, M.; Schnettler, E.; Kohl, A. viRome: An R package for the visualization and analysis of viral small RNA sequence datasets. Bioinformatics 2013, 29, 1902–1903. [Google Scholar] [CrossRef]
- Huvenne, H.; Smagghe, G. Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 2010, 56, 227–235. [Google Scholar] [CrossRef]
- Song, M.-S.; Rossi, J.J. Molecular mechanisms of Dicer: Endonuclease and enzymatic activity. Biochem. J. 2017, 474, 1603–1618. [Google Scholar] [CrossRef]
- Kolliopoulou, A.; Van Nieuwerburgh, F.; Stravopodis, D.J.; Deforce, D.; Swevers, L.; Smagghe, G. Transcriptome analysis of bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. PLoS ONE 2015, 10, e0121447. [Google Scholar] [CrossRef]
- Zografidis, A.; Van Nieuwerburgh, F.; Kolliopoulou, A.; Apostolou-Karampelis, K.; Head, S.R.; Deforce, D.; Smagghe, G.; Swevers, L. Viral small RNA analysis of Bombyx mori larval midgut during persistent and pathogenic cytoplasmic polyhedrosis virus infection. J. Virol. 2015, 89, 11473–11486. [Google Scholar] [CrossRef]
- Chejanovsky, N.; Ophir, R.; Schwager, M.S.; Slabezki, Y.; Grossman, S.; Cox-Foster, D. Characterization of viral siRNA populations in honey bee colony collapse disorder. Virology 2014, 454–455, 176–183. [Google Scholar] [CrossRef]
- Wei, Y.; Chen, S.; Yang, P.; Ma, Z.; Kang, L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009, 10, R6. [Google Scholar] [CrossRef]
- Mueller, S.; Gausson, V.; Vodovar, N.; Deddouche, S.; Troxler, L.; Perot, J.; Pfeffer, S.; Hoffmann, J.A.; Saleh, M.-C.; Imler, J.-L. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila. Proc. Natl. Acad. Sci. USA 2010, 107, 19390–19395. [Google Scholar] [CrossRef]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA Interference-Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J. Immunol. 2013, 190, 650–658. [Google Scholar] [CrossRef]
- Petit, M.; Mongelli, V.; Frangeul, L.; Blanc, H.; Jiggins, F.; Saleh, M.-C. piRNA pathway is not required for antiviral defense in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2016, 113, E4218–E4227. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Sabin, L.R.; Zheng, Q.; Thekkat, P.; Yang, J.; Hannon, G.J.; Gregory, B.D.; Tudor, M.; Cherry, S. Dicer-2 Processes Diverse Viral RNA Species. PLoS ONE 2013, 8, e55458. [Google Scholar] [CrossRef]
- Miesen, P.; Ivens, A.; Buck, A.H.; van Rij, R.P. Small RNA Profiling in Dengue Virus 2-Infected Aedes Mosquito Cells Reveals Viral piRNAs and Novel Host miRNAs. PLoS Negl. Trop. Dis. 2016, 10, e0004452. [Google Scholar] [CrossRef]
- Rückert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single processing center models for human Dicer and bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef]
- MacRae, I.J.; Zhou, K.; Li, F.; Repic, A.; Brooks, A.N.; Cande, W.Z.; Adams, P.D.; Doudna, J.A. Structural basis for double-stranded RNA processing by Dicer. Science 2006, 311, 195–198. [Google Scholar] [CrossRef]
- Lau, P.W.; Guiley, K.Z.; De, N.; Potter, C.S.; Carragher, B.; MacRae, I.J. The molecular architecture of human Dicer. Nat. Struct. Mol. Biol. 2012, 19, 436–440. [Google Scholar] [CrossRef]
- Ma, E.; Zhou, K.; Kidwell, M.A.; Doudna, J.A. Coordinated activities of human dicer domains in regulatory RNA processing. J. Mol. Biol. 2012, 422, 466–476. [Google Scholar] [CrossRef]
- Obbard, D.J.; Jiggins, F.M.; Halligan, D.L.; Little, T.J. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr. Biol. 2006, 16, 580–585. [Google Scholar] [CrossRef]
- Mukherjee, K.; Campos, H.; Kolaczkowski, B. Evolution of animal and plant dicers: Early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 2013, 30, 627–641. [Google Scholar] [CrossRef]
- Wynant, N.; Santos, D.; Vanden Broeck, J. The evolution of animal Argonautes: Evidence for the absence of antiviral AGO Argonautes in vertebrates. Sci. Rep. 2017, 7, 9230. [Google Scholar] [CrossRef]
- Whyard, S.; Singh, A.D.; Wong, S. Ingested double-stranded RNAs can act as species-specific insecticides. Insect Biochem. Mol. Biol. 2009, 39, 824–832. [Google Scholar] [CrossRef]
- Gong, L.; Yang, X.; Zhang, B.; Zhong, G.; Hu, M. Silencing of Rieske iron-sulfur protein using chemically synthesised siRNA as a potential biopesticide against Plutella xylostella. Pest Manag. Sci. 2011, 67, 514–520. [Google Scholar] [CrossRef]
- Gong, L.; Chen, Y.; Hu, Z.; Hu, M. Testing Insecticidal Activity of Novel Chemically Synthesized siRNA against Plutella xylostella under Laboratory and Field Conditions. PLoS ONE 2013, 8, e62990. [Google Scholar] [CrossRef]
- Jakubowska, A.K.; Murillo, R.; Carballo, A.; Williams, T.; van Lent, J.W.M.; Caballero, P.; Herrero, S. Iflavirus increases its infectivity and physical stability in association with baculovirus. PeerJ 2016, 4, e1687. [Google Scholar] [CrossRef]
- Carballo, A.; Murillo, R.; Jakubowska, A.; Herrero, S.; Williams, T.; Caballero, P. Co-infection with iflaviruses influences the insecticidal properties of Spodoptera exigua multiple nucleopolyhedrovirus occlusion bodies: Implications for the production and biosecurity of baculovirus insecticides. PLoS ONE 2017, 12, e0177301. [Google Scholar] [CrossRef]
- Cappelle, K.; Smagghe, G.; Dhaenens, M.; Meeus, I. Israeli Acute Paralysis Virus Infection Leads to an Enhanced RNA Interference Response and Not Its Suppression in the Bumblebee Bombus terrestris. Viruses 2016, 8, 334. [Google Scholar] [CrossRef]
- Katsuma, S.; Tanaka, S.; Omuro, N.; Takabuchi, L.; Daimon, T.; Imanishi, S.; Yamashita, S.; Iwanaga, M.; Mita, K.; Maeda, S.; et al. Novel macula-like virus identified in Bombyx mori cultured cells. J. Virol. 2005, 79, 5577–5584. [Google Scholar] [CrossRef]
- Habayeb, M.S.; Ekengren, S.K.; Hultmark, D. Nora virus, a persistent vitus in Drosophila, defines a new picorna-like virus family. J. Gen. Virol. 2006, 87, 3045–3051. [Google Scholar] [CrossRef]
- Li, T.-C.; Scotti, P.D.; Miyamura, T.; Takeda, N. Latent infection of a new alphanodavirus in an insect cell line. J. Virol. 2007, 81, 10890–10896. [Google Scholar] [CrossRef]
- Jovel, J.; Schneemann, A. Molecular characterization of Drosophila cells persistently infected with Flock House virus. Virology 2011, 419, 43–53. [Google Scholar] [CrossRef][Green Version]
- Iwanaga, M.; Hitotsuyama, T.; Katsuma, S.; Ishihara, G.; Daimon, T.; Shimada, T.; Imanishi, S.; Kawasaki, H. Infection study of Bombyx mori macula-like virus (BmMLV) using a BmMLV-negative cell line and an infectious cDNA clone. J. Virol. Methods 2012, 179, 316–324. [Google Scholar] [CrossRef]
- Suzuki, T.; Takeshima, Y.; Mikamoto, T.; Saeki, J.-D.; Kato, T.; Park, E.Y.; Kawagishi, H.; Dohra, H. Genome Sequence of a Novel Iflavirus from mRNA Sequencing of the Pupa of Bombyx mori Inoculated with Cordyceps militaris. Genome Announc. 2015, 3, e01039-15. [Google Scholar] [CrossRef]
- Swevers, L.; Ioannidis, K.; Kolovou, M.; Zografidis, A.; Labropoulou, V.; Santos, D.; Wynant, N.; Vanden Broeck, J.; Wang, L.; Cappelle, K.; et al. Persistent RNA virus infection of lepidopteran cell lines: Interactions with the RNAi machinery. J. Insect Physiol. 2016, 93–94, 81–93. [Google Scholar] [CrossRef]
- Millán-Leiva, A.; Jakubowska, A.K.; Ferré, J.; Herrero, S. Genome sequence of SeIV-1, a novel virus from the Iflaviridae family infective to Spodoptera exigua. J. Invertebr. Pathol. 2012, 109, 127–133. [Google Scholar] [CrossRef]
- Silva, L.A.; Ardisson-Araujo, D.M.P.; Tinoco, R.S.; Fernandes, O.A.; Melo, F.L.; Ribeiro, B.M. Complete genome sequence and structural characterization of a novel iflavirus isolated from Opsiphanes invirae (Lepidoptera: Nymphalidae). J. Invertebr. Pathol. 2015, 130, 136–140. [Google Scholar] [CrossRef]
- Goodman, C.L.; Stanley, D.; Ringbauer, J.A.; Beeman, R.W.; Silver, K.; Park, Y. A cell line derived from the red flour beetle Tribolium castaneum (Coleoptera: Tenebrionidae). In Vitro Cell. Dev. Biol. Anim. 2012, 48, 426–433. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santos, D.; Mingels, L.; Vogel, E.; Wang, L.; Christiaens, O.; Cappelle, K.; Wynant, N.; Gansemans, Y.; Van Nieuwerburgh, F.; Smagghe, G.; et al. Generation of Virus- and dsRNA-Derived siRNAs with Species-Dependent Length in Insects. Viruses 2019, 11, 738. https://doi.org/10.3390/v11080738
Santos D, Mingels L, Vogel E, Wang L, Christiaens O, Cappelle K, Wynant N, Gansemans Y, Van Nieuwerburgh F, Smagghe G, et al. Generation of Virus- and dsRNA-Derived siRNAs with Species-Dependent Length in Insects. Viruses. 2019; 11(8):738. https://doi.org/10.3390/v11080738
Chicago/Turabian StyleSantos, Dulce, Lina Mingels, Elise Vogel, Luoluo Wang, Olivier Christiaens, Kaat Cappelle, Niels Wynant, Yannick Gansemans, Filip Van Nieuwerburgh, Guy Smagghe, and et al. 2019. "Generation of Virus- and dsRNA-Derived siRNAs with Species-Dependent Length in Insects" Viruses 11, no. 8: 738. https://doi.org/10.3390/v11080738
APA StyleSantos, D., Mingels, L., Vogel, E., Wang, L., Christiaens, O., Cappelle, K., Wynant, N., Gansemans, Y., Van Nieuwerburgh, F., Smagghe, G., Swevers, L., & Vanden Broeck, J. (2019). Generation of Virus- and dsRNA-Derived siRNAs with Species-Dependent Length in Insects. Viruses, 11(8), 738. https://doi.org/10.3390/v11080738






