Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades
Abstract
1. Introduction
2. Materials and Methods
2.1. Iridovirus Isolates Used
2.2. Diagnostic Screening of Vertebrate and Invertebrate Samples for IIVs
2.3. Cell Culture-Based Methods (Isolation, Propagation and Purification)
2.4. Animal Infection Studies
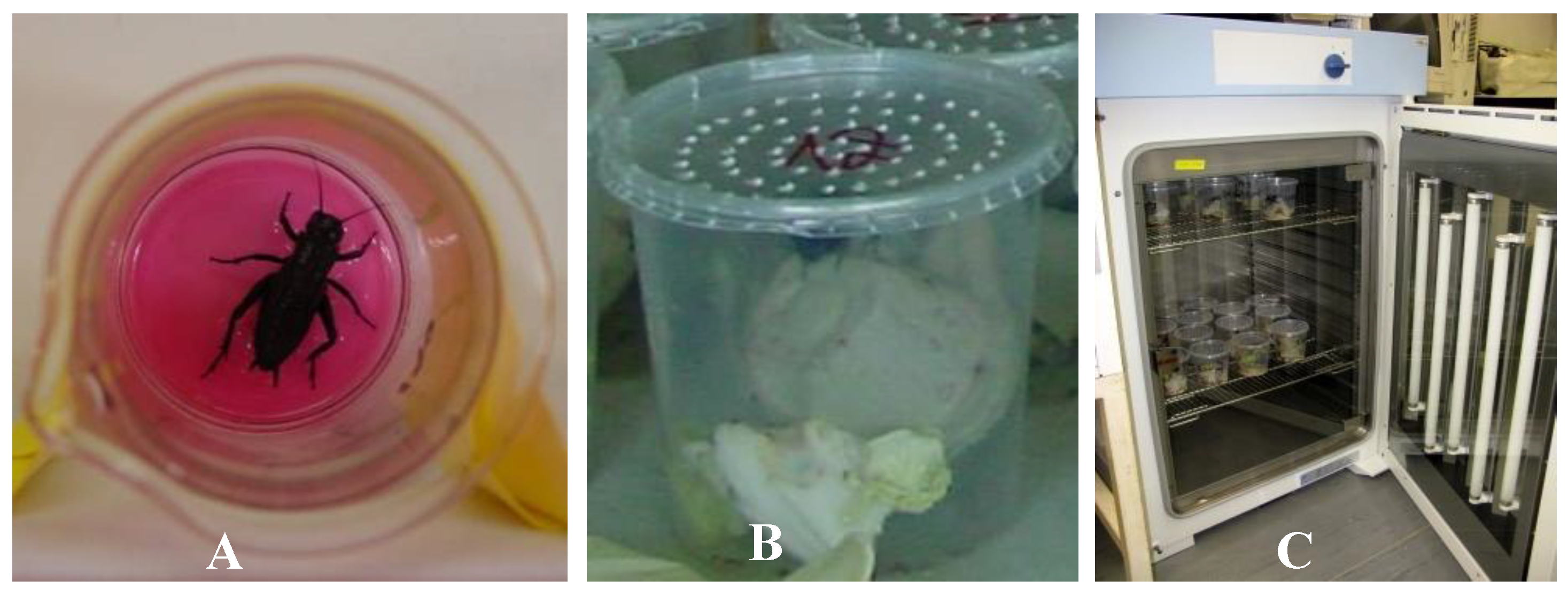
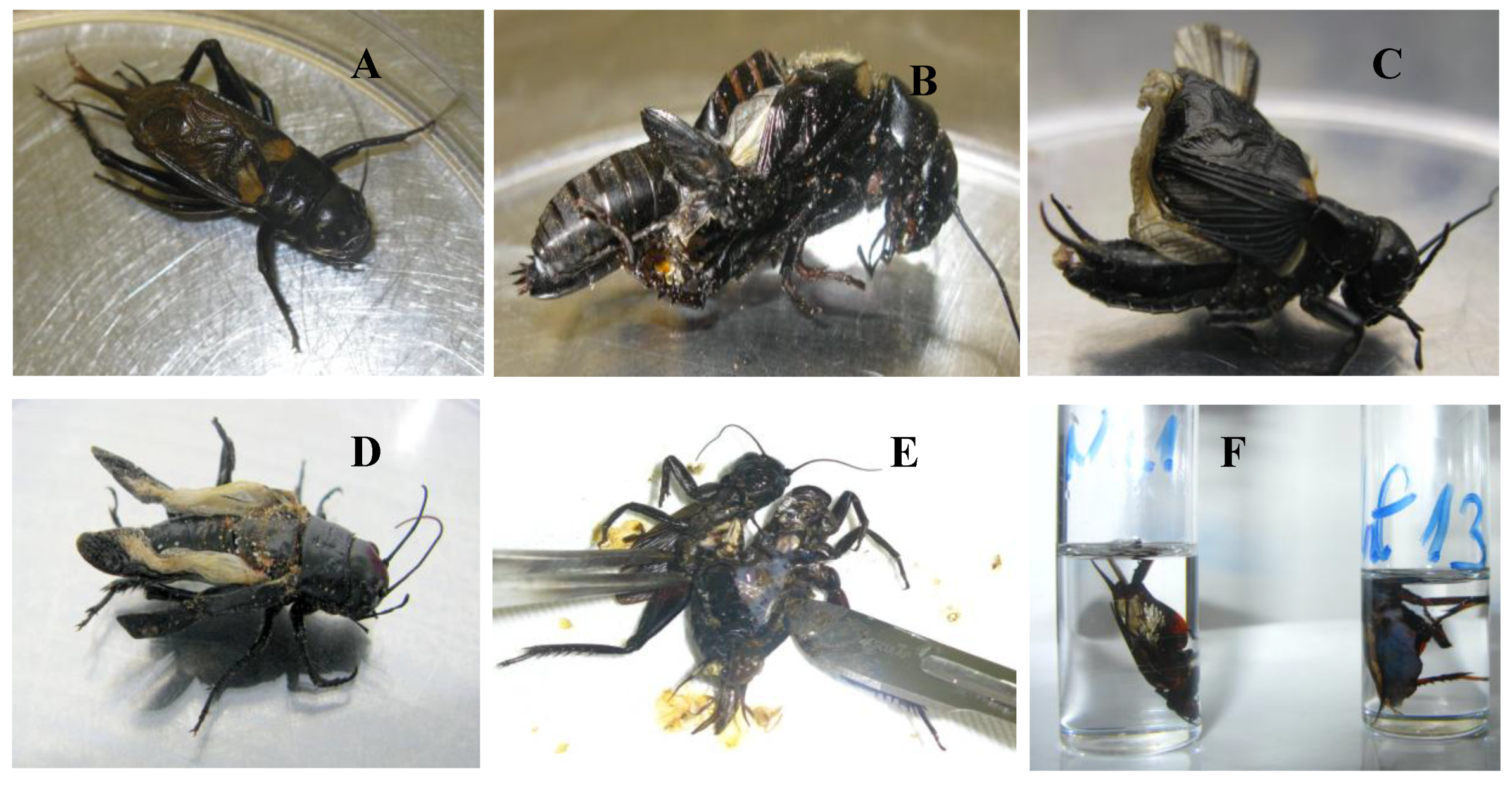
2.4.1. Cricket Bioassay
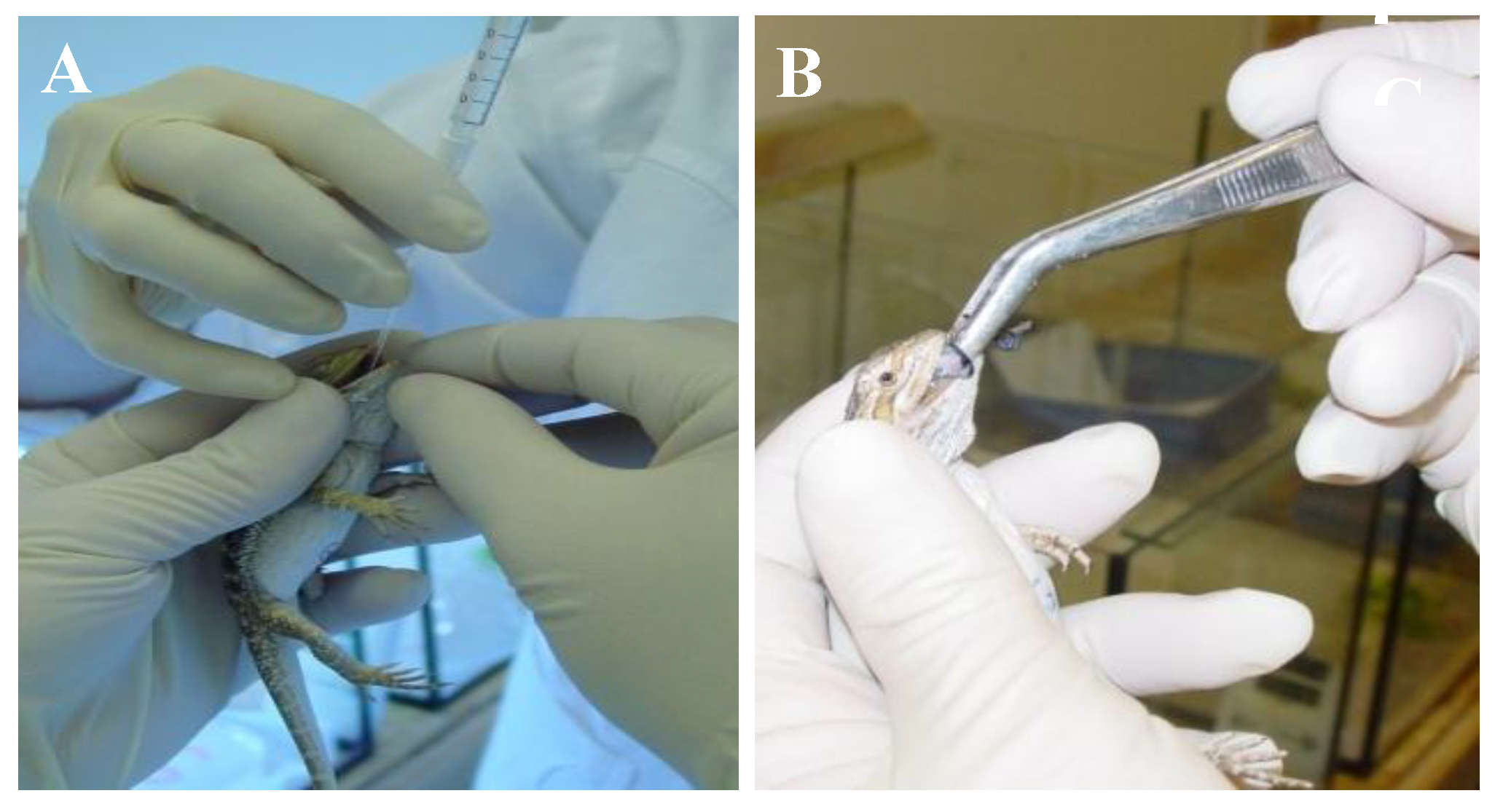
2.4.2. Bearded Dragon Transmission Study
2.4.3. Microscopic Techniques
2.5. Molecular Biological Techniques
2.5.1. DNA Extraction
2.5.2. Conventional PCRs (nPCR)
2.5.3. DNA sequencing (Sanger-, MPS)
2.5.4. Analysis of Sequences
2.5.5. Real-Time PCR (qPCR)
3. Results
3.1. Diagnostic Testing
3.2. Sanger Sequencing Comparison of Isolates
3.3. Genome Sequencing of the Chameleon Isolate
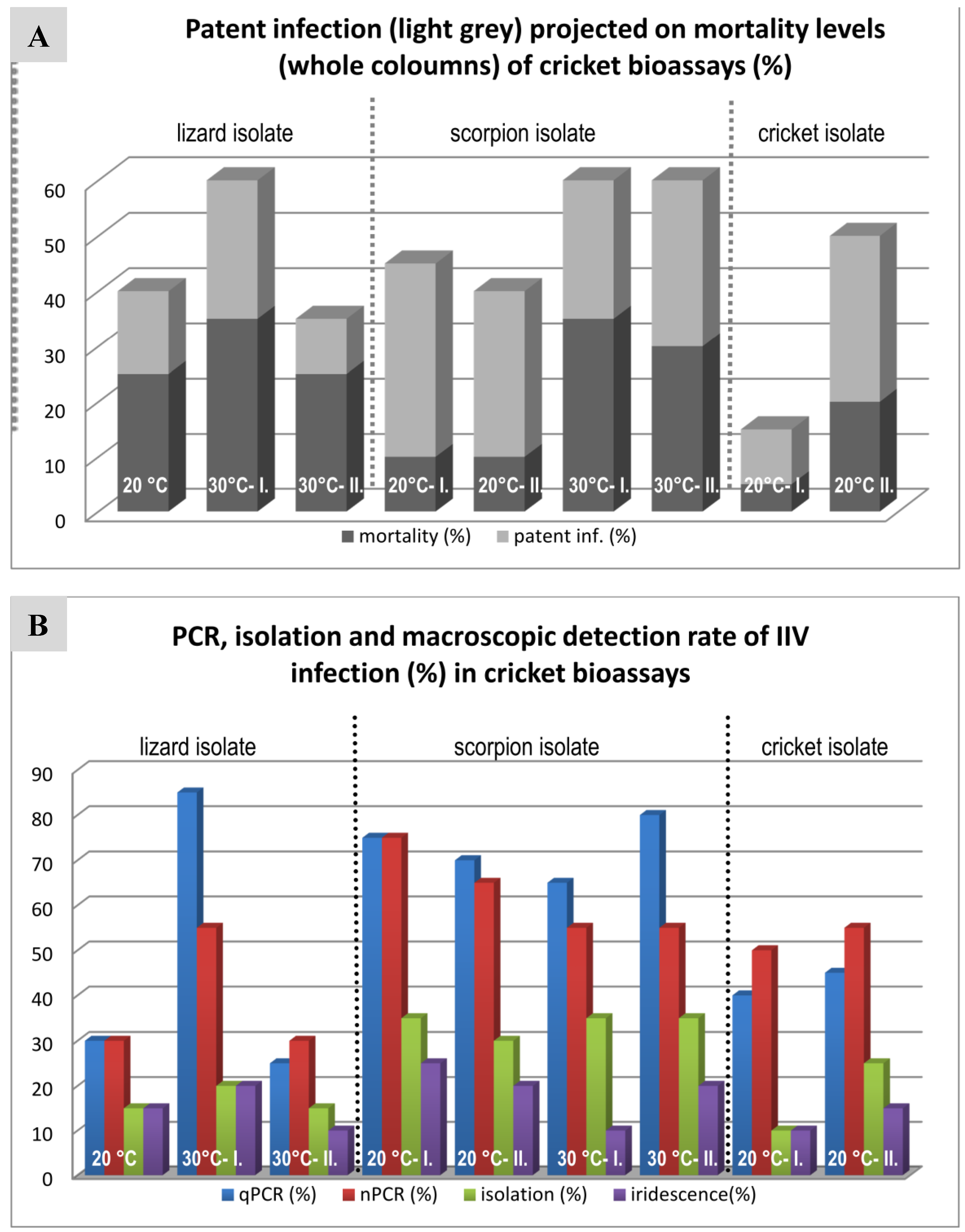
3.4. Cricket Infection Study
3.5. Transmission Study with Bearded Dragons
4. Discussion
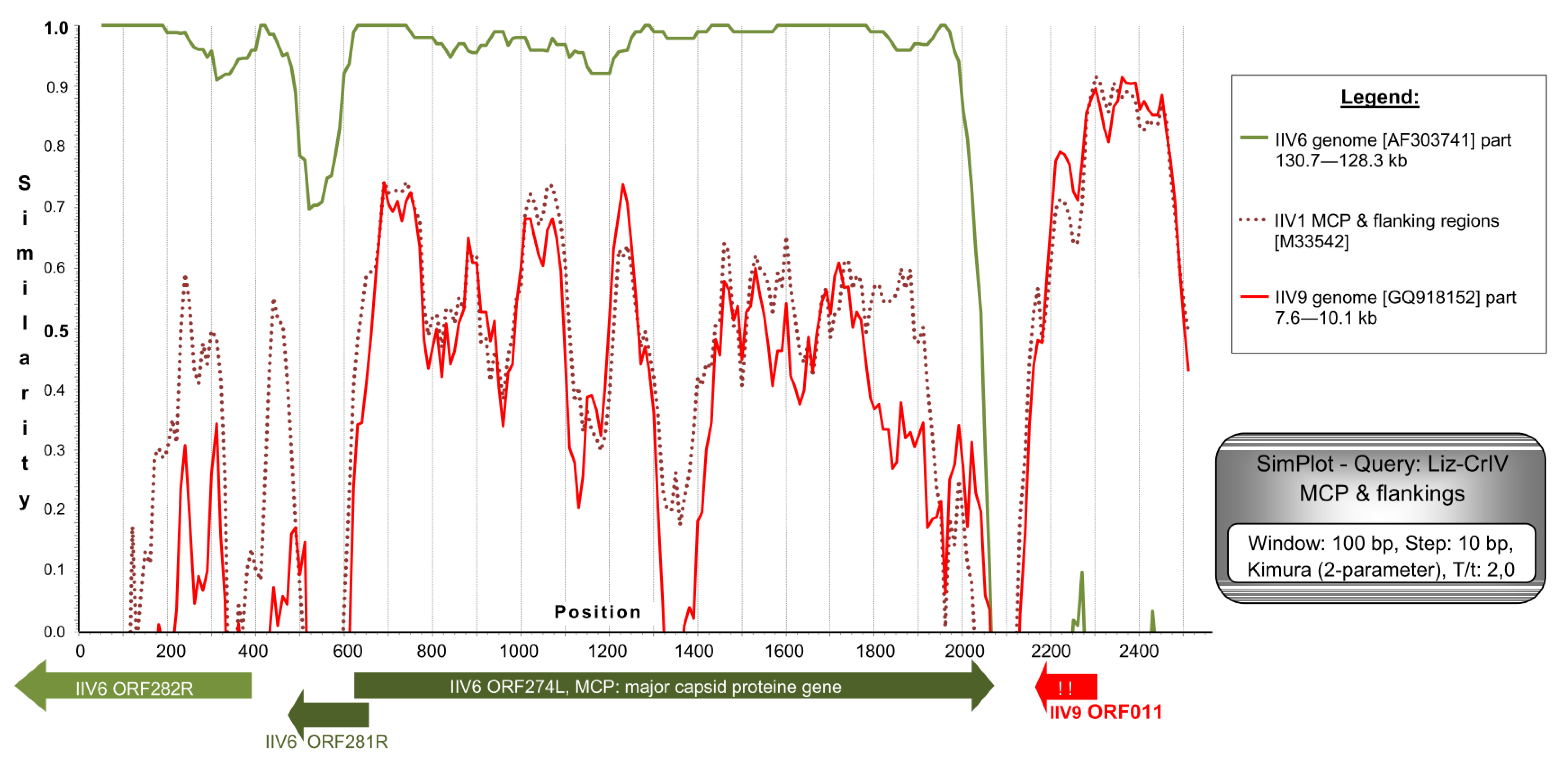
4.1. Comparison of Genome Fragments of Different IIV Isolates
4.2. Genome Sequencing of the Chameleon Isolate, Differences from the IIV6 Genome
4.3. Comparison of Three IIV Isolates in a Cricket Bioassay
4.4. Bearded Dragon Transmission Study
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chinchar, V.G.; Hick, P.; Ince, I.A.; Jancovich, J.K.; Marschang, R.; Qin, Q.; Subramaniam, K.; Waltzek, T.B.; Whittington, R.; Williams, T.; et al. ICTV virus taxonomy profile: Iridoviridae. J. Gen. Virol. 2017, 98, 890–891. [Google Scholar] [CrossRef] [PubMed]
- Chinchar, V.G.; Yang, F.; Huang, J.; Williams, T.; Whittington, R.; Jancovich, J.; Subramaniam, K.; Waltzek, T.; Hick, P.; Ince, I.A.; et al. ICTV proposal 2018.004d: Short title: One new genus with one new species in the subfamily Betairidovirinae. 2018. Available online: https://talk.ictvonline.org/ (accessed on 10 April 2019).
- Vetten, H.J.; Haenni, A.L. Taxon-specific suffixes for vernacular names. Arch. Virol. 2006, 151, 1249–1250. [Google Scholar] [CrossRef] [PubMed]
- Williams, T. The iridoviruses. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 1996. [Google Scholar]
- Williams, T.; Barbosa-Solomieu, V.; Chinchar, V.G. A decade of advances in iridovirus research. Adv. Virus Res. 2005, 65, 173–248. [Google Scholar] [PubMed]
- İnce, İ.; Özcan, O.; Ilter-Akulke, A.Z.; Scully, E.D.; Özgen, A. Invertebrate iridoviruses: A glance over the last decade. Viruses 2018, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Xeros, N. An unusual virus disease of a dipterous larva. Nature 1954, 173, 866–867. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Williams, R.C. A crystallizable insect virus. Nature 1957, 179, 119–120. [Google Scholar] [PubMed]
- Williams, T. Iridoviruses of invertebrates. In Encyclopedia of Virology; Mahy, B.W.J., Van Regenmortel, M.H.V., Eds.; Elsevier: Oxford, UK, 2008; pp. 161–167. [Google Scholar]
- Williams, T. Natural invertebrate hosts of iridoviruses (Iridoviridae). Neotrop. Entomol. 2008, 37, 615–632. [Google Scholar] [CrossRef]
- Tinsley, T.W.; Kelly, D.C. An interim nomenclature system for the iridescent group of insect viruses. J. Invertebr. Pathol. 1970, 16, 470–472. [Google Scholar] [CrossRef]
- Williams, T. Comparative studies of iridoviruses: Further support for a new classification. Virus Res. 1994, 33, 99–121. [Google Scholar] [CrossRef]
- Williams, T.; Cory, J.S. Proposals for a new classification of iridescent viruses. J. Gen. Virol. 1994, 75 Pt 6, 1291–1301. [Google Scholar] [CrossRef]
- Webby, R.; Kalmakoff, J. Sequence comparison of the major capsid protein gene from 18 diverse iridoviruses. Arch. Virol. 1998, 143, 1949–1966. [Google Scholar] [CrossRef] [PubMed]
- Tang, K.F.; Redman, R.M.; Pantoja, C.R.; Groumellec, M.L.; Duraisamy, P.; Lightner, D.V. Identification of an iridovirus in Acetes erythraeus (Sergestidae) and the development of in situ hybridization and PCR method for its detection. J. Invertebr. Pathol. 2007, 96, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Delhon, G.; Tulman, E.R.; Afonso, C.L.; Lu, Z.; Becnel, J.J.; Moser, B.A.; Kutish, G.F.; Rock, D.L. Genome of invertebrate iridescent virus type 3 (mosquito iridescent virus). J. Virol. 2006, 80, 8439–8449. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.K.; Young, V.L.; Kleffmann, T.; Ward, V.K. Genomic and proteomic analysis of invertebrate iridovirus type 9. J. Virol. 2011, 85, 7900–7911. [Google Scholar] [CrossRef] [PubMed]
- Eaton, H.E.; Ring, B.A.; Brunetti, C.R. The genomic diversity and phylogenetic relationship in the family Iridoviridae. Viruses 2010, 2, 1458–1475. [Google Scholar] [CrossRef]
- Muttis, E.; Miele, S.A.; Belaich, M.N.; Micieli, M.V.; Becnel, J.J.; Ghiringhelli, P.D.; García, J.J. First record of a mosquito iridescent virus in Culex pipiens l. (Diptera: Culicidae). Arch. Virol. 2012, 157, 1569–1571. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Li, S.; Zhao, Q.; Pei, G.; An, X.; Guo, X.; Zhou, H.; Zhang, Z.; Zhang, J.; Tong, Y. Isolation and characterization of a novel invertebrate iridovirus from adult Anopheles minimus (AMIV) in China. J. Invertebr. Pathol. 2015, 127, 1–5. [Google Scholar] [CrossRef]
- Qiu, L.; Chen, M.M.; Wang, R.Y.; Wan, X.Y.; Li, C.; Zhang, Q.L.; Dong, X.; Yang, B.; Xiang, J.H.; Huang, J. Complete genome sequence of shrimp hemocyte iridescent virus (SHIV) isolated from white leg shrimp, Litopenaeus vannamei. Arch. Virol. 2018, 163, 781–785. [Google Scholar] [CrossRef]
- Available online: Https://talk.Ictvonline.Org//taxonomy/p/taxonomy-history?Taxnode_id=201856294 (accessed on 10 April 2019).
- Li, F.; Xu, L.; Yang, F. Genomic characterization of a novel iridovirus from redclaw crayfish Cherax quadricarinatus: Evidence for a new genus within the family Iridoviridae. J. Gen. Virol. 2017, 98, 2589–2595. [Google Scholar] [CrossRef]
- Toenshoff, E.R.; Fields, P.D.; Bourgeois, Y.X.; Ebert, D. The end of a 60-year riddle: Identification and genomic characterization of an iridovirus, the causative agent of white fat cell disease in zooplankton. G3 Genes Genomes Genet. 2018, 8, 1259–1272. [Google Scholar] [CrossRef]
- Kleespies, R.G.; Tidona, C.A.; Darai, G. Characterization of a new iridovirus isolated from crickets and investigations on the host range. J. Invertebr. Pathol. 1999, 73, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Hernández, O.; Maldonado, G.; Williams, T. An epizootic of patent iridescent virus disease in multiple species of blackflies in chiapas, mexico. Med. Vet. Entomol. 2000, 14, 458–462. [Google Scholar] [CrossRef] [PubMed]
- Jakob, N.J.; Müller, K.; Bahr, U.; Darai, G. Analysis of the first complete DNA sequence of an invertebrate iridovirus: Coding strategy of the genome of chilo iridescent virus. Virology 2001, 286, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Henderson, C.W.; Johnson, C.L.; Lodhi, S.A.; Bilimoria, S.L. Replication of chilo iridescent virus in the cotton boll weevil, Anthonomus grandis, and development of an infectivity assay. Arch. Virol. 2001, 146, 767–775. [Google Scholar] [CrossRef] [PubMed]
- Nalcacioglu, R.; Muratoglu, H.; Yesilyurt, A.; van Oers, M.M.; Vlak, J.M.; Demirbag, Z. Enhanced insecticidal activity of chilo iridescent virus expressing an insect specific neurotoxin. J. Invertebr. Pathol. 2016, 138, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Jakob, N.J.; Darai, G. Molecular anatomy of chilo iridescent virus genome and the evolution of viral genes. Virus Genes 2002, 25, 299–316. [Google Scholar] [CrossRef] [PubMed]
- Eaton, H.E.; Metcalf, J.; Penny, E.; Tcherepanov, V.; Upton, C.; Brunetti, C.R. Comparative genomic analysis of the family Iridoviridae: Re-annotating and defining the core set of iridovirus genes. Virol. J. 2007, 4, 11. [Google Scholar] [CrossRef]
- Just, F.T.; Essbauer, S.S. Characterization of an iridescent virus isolated from Gryllus bimaculatus (Orthoptera: Gryllidae). J. Invertebr. Pathol. 2001, 77, 51–61. [Google Scholar] [CrossRef]
- Just, F.; Essbauer, S.; Ahne, W.; Blahak, S. Occurrence of an invertebrate iridescent-like virus (Iridoviridae) in reptiles. J. Vet. Med. 2001, 48, 685–694. [Google Scholar] [CrossRef]
- Marschang, R.E.; Becher, P. Isolation and characterization of iridoviruses from lizards. In Proceedings of the 5th International Symposium on Viruses of Lower Vertebrates, Seattle, WA, USA, 27–30 August 2002. [Google Scholar]
- Weinmann, N.; Papp, T.; Pedro Alves de Matos, A.; Teifke, J.P.; Marschang, R.E. Experimental infection of crickets (Gryllus bimaculatus) with an invertebrate iridovirus isolated from a high-casqued chameleon (Chamaeleo hoehnelii). J. Vet. Diagn. Investig. 2007, 19, 674–679. [Google Scholar] [CrossRef]
- Stöhr, A.C.; Papp, T.; Marschang, R.E. Repeated detection of an invertebrate iridovirus in amphibians. J. Herpetol. Med. Surg. 2013, 26, 54–58. [Google Scholar] [CrossRef]
- Papp, T.; Spann, D.; Marschang, R.E. Development and use of a real-time polymerase chain reaction for the detection of group ii invertebrate iridoviruses in pet lizards and prey insects. J. Zoo Wildl. Med. 2014, 45, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Hierholzer, J.C.; Killington, R.A. Virus isolation and quantitation. In Virology Methods Manual; Mahy, B.W., Kangro, H.O., Eds.; Academic Press: London, UK, 1996; pp. 25–46. [Google Scholar]
- Sambrook, J.; Russell, D.W. Purification of Nucleic Acids by Extraction with Phenol:Chloroform; Cold Spring Harbor Laboratory Press: Suffolk, NY, USA, 2006. [Google Scholar]
- Jakob, N.J.; Kleespies, R.G.; Tidona, C.A.; Müller, K.; Gelderblom, H.R.; Darai, G. Comparative analysis of the genome and host range characteristics of two insect iridoviruses: Chilo iridescent virus and a cricket iridovirus isolate. J. Gen. Virol. 2002, 83, 463–470. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory Press: Suffolk, NY, USA, 1989. [Google Scholar]
- Bonfield, J.K.; Smith, K.; Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 1995, 23, 4992–4999. [Google Scholar] [CrossRef] [PubMed]
- Golubchik, T.; Wise, M.J.; Easteal, S.; Jermiin, L.S. Mind the gaps: Evidence of bias in estimates of multiple sequence alignments. Mol. Biol. Evol. 2007, 24, 2433–2442. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. Mega x: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Milne, I.; Lindner, D.; Bayer, M.; Husmeier, D.; McGuire, G.; Marshall, D.F.; Wright, F. Topali v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics 2009, 25, 126–127. [Google Scholar] [CrossRef]
- Aberer, A.J.; Kobert, K.; Stamatakis, A. Exabayes: Massively parallel Bayesian tree inference for the whole-genome era. Mol. Biol. Evol. 2014, 31, 2553–2556. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. Mrbayes: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. Mrbayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Shields, E.J.; Sheng, L.; Weiner, A.K.; Garcia, B.A.; Bonasio, R. High-quality genome assemblies reveal long non-coding rnas expressed in ant brains. Cell Rep. 2018, 23, 3078–3090. [Google Scholar] [CrossRef] [PubMed]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Tidona, C.A.; Schnitzler, P.; Kehm, R.; Darai, G. Is the major capsid protein of iridoviruses a suitable target for the study of viral evolution? Virus Genes 1998, 16, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Piégu, B.; Guizard, S.; Yeping, T.; Cruaud, C.; Couloux, A.; Bideshi, D.K.; Federici, B.A.; Bigot, Y. Complete genome sequence of invertebrate iridovirus IIV22A, a variant of IIV22, isolated originally from a blackfly larva. Stand. Genomic Sci. 2014, 9, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.J.; Waksman, G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005, 71, 401–440. [Google Scholar] [PubMed]
- Chinchar, V.; Hick, J.; Jancovich, J.; Subramaniam, K.; Waltzek, T.; Whittington, R.; Williams, T. ICTV proposal 2018.007d: Short title: 8 new species in the family Iridoviridae; removal of 3 existing species. 2018. Available online: https://talk.ictvonline.org/ (accessed on 10 April 2019).
- Shackelton, L.A.; Holmes, E.C. The evolution of large DNA viruses: Combining genomic information of viruses and their hosts. Trends Microbiol. 2004, 12, 458–465. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P.; Witzany, G. Viruses are essential agents within the roots and stem of the tree of life. J. Theor. Biol. 2010, 262, 698–710. [Google Scholar] [CrossRef] [PubMed]
- Yesilyurt, A.; Muratoglu, H.; Demirbag, Z.; Nalcacioglu, R. Chilo iridescent virus encodes two functional metalloproteases. Arch. Virol. 2019, 164, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Filée, J.; Pouget, N.; Chandler, M. Phylogenetic evidence for extensive lateral acquisition of cellular genes by nucleocytoplasmic large DNA viruses. BMC Evol. Biol. 2008, 8, 320. [Google Scholar] [CrossRef]
- Filée, J.; Chandler, M. Gene exchange and the origin of giant viruses. Intervirology 2010, 53, 354–361. [Google Scholar] [CrossRef]
- Behncke, H.; Stöhr, A.C.; Heckers, K.O.; Ball, I.; Marschang, R.E. Mass-mortality in green striped tree dragons (Japalura splendida) associated with multiple viral infections. Vet. Rec. 2013, 173, 248. [Google Scholar] [CrossRef] [PubMed]
- Stöhr, A.C.; Blahak, S.; Heckers, K.O.; Wiechert, J.; Behncke, H.; Mathes, K.; Günther, P.; Zwart, P.; Ball, I.; Rüschoff, B.; et al. Ranavirus infections associated with skin lesions in lizards. Vet. Res. 2013, 44, 84. [Google Scholar] [CrossRef] [PubMed]
- McIntosh, A.H.; Kimura, M. Replication of the insect chilo iridescent virus (CIV) in a poikilothermic vertebrate cell line. Intervirology 1974, 4, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Cerutti, M.; Devauchelle, G. Inhibition of macromolecular synthesis in cells infected with an invertebrate virus (iridovirus type 6 or CIV). Arch. Virol. 1980, 63, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Ahlers, L.R.; Bastos, R.G.; Hiroyasu, A.; Goodman, A.G. Invertebrate iridescent virus 6, a DNA virus, stimulates a mammalian innate immune response through RIG-I-like receptors. PLoS ONE 2016, 11, e0166088. [Google Scholar] [CrossRef] [PubMed]
- Lorbacher de Ruiz, H.; Gelderblom, H.; Hofmann, W.; Darai, G. Insect iridescent virus type 6 induced toxic degenerative hepatitis in mice. Med. Microbiol. Immunol. 1986, 175, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Weinmann, N. Experimental infection of crickets (Ggryllus bimaculatus) with an invertebrate iridovirus isolated from a high-casqued chameleon (Chamaeleo hoehnelii), and trial on infecting bearded dragons (Pogona vitticeps) by feeding with infected crickets (Vet. Med. dissertation in German); Justus-Liebig-Universität: Giessen, Germany, 2007. [Google Scholar]
 ORF highly similiar (85%–100%) to IIV6 homolog.
ORF highly similiar (85%–100%) to IIV6 homolog.  ORF broken or much shorter (>20%) than IIV6 homolog.
ORF broken or much shorter (>20%) than IIV6 homolog.  ORF is joined into one ORF from two IIV6 homologs.
ORF is joined into one ORF from two IIV6 homologs.  ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.
ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.  ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.
ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.  ORF most similar to a gene from a non-IV large DNA virus.
ORF most similar to a gene from a non-IV large DNA virus.  ORF most similar to a gene from a eukaryotic organism.
ORF most similar to a gene from a eukaryotic organism.  ORF most similar to a gene from a prokaryote.
ORF most similar to a gene from a prokaryote.  ORF with no homology to current GenBank entries.
ORF with no homology to current GenBank entries.  GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).
GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).
 ORF highly similiar (85%–100%) to IIV6 homolog.
ORF highly similiar (85%–100%) to IIV6 homolog.  ORF broken or much shorter (>20%) than IIV6 homolog.
ORF broken or much shorter (>20%) than IIV6 homolog.  ORF is joined into one ORF from two IIV6 homologs.
ORF is joined into one ORF from two IIV6 homologs.  ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.
ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.  ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.
ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.  ORF most similar to a gene from a non-IV large DNA virus.
ORF most similar to a gene from a non-IV large DNA virus.  ORF most similar to a gene from a eukaryotic organism.
ORF most similar to a gene from a eukaryotic organism.  ORF most similar to a gene from a prokaryote.
ORF most similar to a gene from a prokaryote.  ORF with no homology to current GenBank entries.
ORF with no homology to current GenBank entries.  GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).
GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).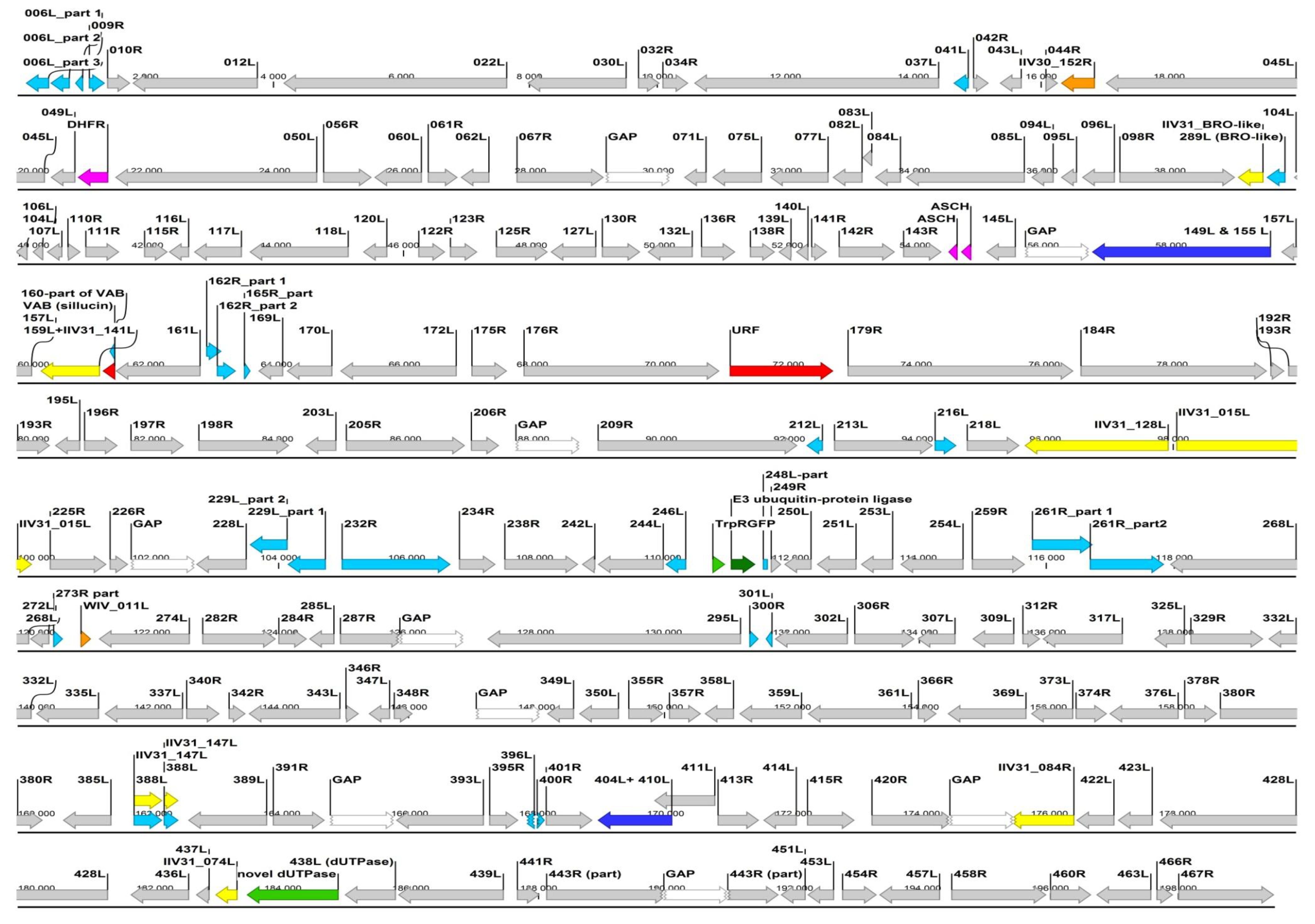
| Host Species | ID, No. | Owner | IIV Positive Organs | Case History | |
|---|---|---|---|---|---|
| Squamata, | High-casqued chameleon (Trioceros /Chamaeleo/ hoehnelii) | Ir.iso.1 100/2001 | A | Kidney, liver, spleen, lung, intestine | Emaciation, kerato-conjunctivitis |
| Bearded dragon (Pogona vitticeps) | Ir.iso.2 66/2003 | B | Lung, heart, tongue | Unknown | |
| Bearded dragon (Pogona vitticeps) | Ir.iso.364/2003 | B | Lung, brain, tongue, stomach, intestine | Unknown | |
| Spiny tailed lizard (Uromastyx sp.) | Ir.iso.4 08/2004 | B | Skin | Hyperkeratosis | |
| Four-horned chameleon (Trioceros quadricornis) | Ir.iso.5 626/2000 | C | Liver | Emaciation, several animals died suddenly | |
| Green iguana (Iguana iguana) | Ir.iso.6 1125/2000 | D | Skin | Hyperkeratosis | |
| Arthropoda | House cricket (Acheta domesticus) | Ir.iso.752/2003 | E | Whole body | Prey insects, died suddenly |
| Emperor scorpion (Pandinus imperator) | Ir.iso.825/2006 | F | Abdominal organs | Loss of UV colouration |
| Name | Length in Liz–CrIV (aa) | Length of Homolog ORF (aa) | aa Identity/ Similarity (%) | Comment /Function |
|---|---|---|---|---|
| IIV30_152R | 176 | 173 | 55/68 | Hypothetical protein of IIV30 |
| DHRF | 158 | 159 | 50/73 | Dihidrofolate reductase of Labilithrix luteola (Protobacterium) |
| IIV31_BRO-like | 133 | 416 | 89/85 | Ortholog of IIV31_198R and IIV6_189L C-terminal regions |
| ASCH containing /1 | 48 | 137 | 50/66 | Thermococcus sp. ASCH and Gonium pectorale (alga) hypothetical protein |
| ASCH containing /2 | 53 | 137 | 53/69 | ASCH containing protein (RNA binding) of Dependentiae bacterium |
| VAB (sillucin) | 65 | NA | NA | NO BLAST homology (only N-terminal part is similar to IIV6_160L (VAB = viral antibiotic peptide)) |
| URF | 539 | NA | NA | NO BLAST homology to any GenBank entry (ca. 2kbp insert of foreign DNA, compared to IIV6 genome) |
| IIV31_128L | 748 | 771 | 44/62 | 35%/55% identity/similarity to a hypothetical protein of Klosneuvirus |
| IIV31_015L | 699 | 676 | 62/78 | Polynucleotide kinase/ligase (PK) and a pseT superfamily domain |
| TrpRGFP | 66 | 260 | 54/69 | Tryptophan repeat gene family protein of an entomopoxvirus and a Kaumoebavirus |
| E3 ubiquitin protein ligase (RNFT1) | 127 | 252 | 33/48 | C-terminal half (63 aa) shows highest similarity to homolog gene of Python bivittatus and other snake homologs, it contains a ring-finger domain |
| IIV31_084R | 318 | 269 | 57/74 | Not completely covered, contains ABC type AA transport/ signal transduction system domain |
| IIV31_074L | 115 | 116 | 37/52 | Hypothetical protein of IIV31 |
| Novel dUTPase | 478 | 349 | 46/63 | Most similar homolog found in Pithovirus; contains an intein splicing domain (N-terminal part), and a trimeric dUTPase domain (C-terminal part) |
| A. | |||||||||
| Lizard Isolate | Scorpion Isolate | Cricket Isolate | |||||||
| 20 °C | 30 °C I | 30 °C II | 20 °C I. | 20 °C II. | 30 °C I. | 30 °C II. | 20 °C I. | 20 °C II. | |
| Mortality (%) | 40 | 60 | 35 | 45 | 40 | 60 | 60 | 15 | 50 |
| Patent inf. (%) | 15 | 25 | 10 | 35 | 30 | 25 | 30 | 10 | 30 |
| MST* (days) | 42 | 38.6 | 36.5 | 34 | 24.2 | 24.4 | 34.2 | 35 | 42 |
| qPCR (%) | 30 (15) | 85 (15) | 25 (5) | 75 (10) | 70 (5) | 65 (15) | 80 (20) | 40 (40) | 45 (20) |
| nPCR (%) | 30 (15) | 55 (15) | 30 | 75 (5) | 65 (5) | 55 (25) | 55 (25) | 50 (10) | 55 (10) |
| Isolation (%) | 15 | 20 (5) | 15 | 35 | 30 | 35 | 35 | 10 (5) | 25 |
| Iridescence (%) | 15 | 20 | 10 | 25 | 20 | 10 | 20 | 10 | 15 |
| B. | |||||||||
| Weinmann et al. [35] | |||||||||
| I. | II. | III. | |||||||
| Mortality (%) | 35 | 20 | 20 | ||||||
| Patent inf. (%) | NA | NA | NA | ||||||
| MST (days) | NA | NA | NA | ||||||
| qPCR (%) | NA | NA | NA | ||||||
| nPCR (%) | 75 | 15 | 30 | ||||||
| Isolation (%) | 45 | 5 | 25 | ||||||
| Iridescence (%) | 25 | 5 | 10 | ||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papp, T.; Marschang, R.E. Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades. Viruses 2019, 11, 600. https://doi.org/10.3390/v11070600
Papp T, Marschang RE. Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades. Viruses. 2019; 11(7):600. https://doi.org/10.3390/v11070600
Chicago/Turabian StylePapp, Tibor, and Rachel E. Marschang. 2019. "Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades" Viruses 11, no. 7: 600. https://doi.org/10.3390/v11070600
APA StylePapp, T., & Marschang, R. E. (2019). Detection and Characterization of Invertebrate Iridoviruses Found in Reptiles and Prey Insects in Europe over the Past Two Decades. Viruses, 11(7), 600. https://doi.org/10.3390/v11070600





