Quantifying Anti-HIV Envelope-Specific Antibodies in Plasma from HIV Infected Individuals
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Study Subjects
2.3. Gp120 Capture Plate-Based ELISA
2.4. Flow Cytometry-Based Env-Specific Ab Quantification Assay Using rgp120 Coated CEM (cCEM) Cells
2.5. Preparation of HIV-Infected CEM (iCEM) Cells
2.6. Flow Cytometry-Based Env-Specific Ab Quantification Assay Using iCEM
2.7. Data Analysis
2.8. Statistical Analysis
3. Results
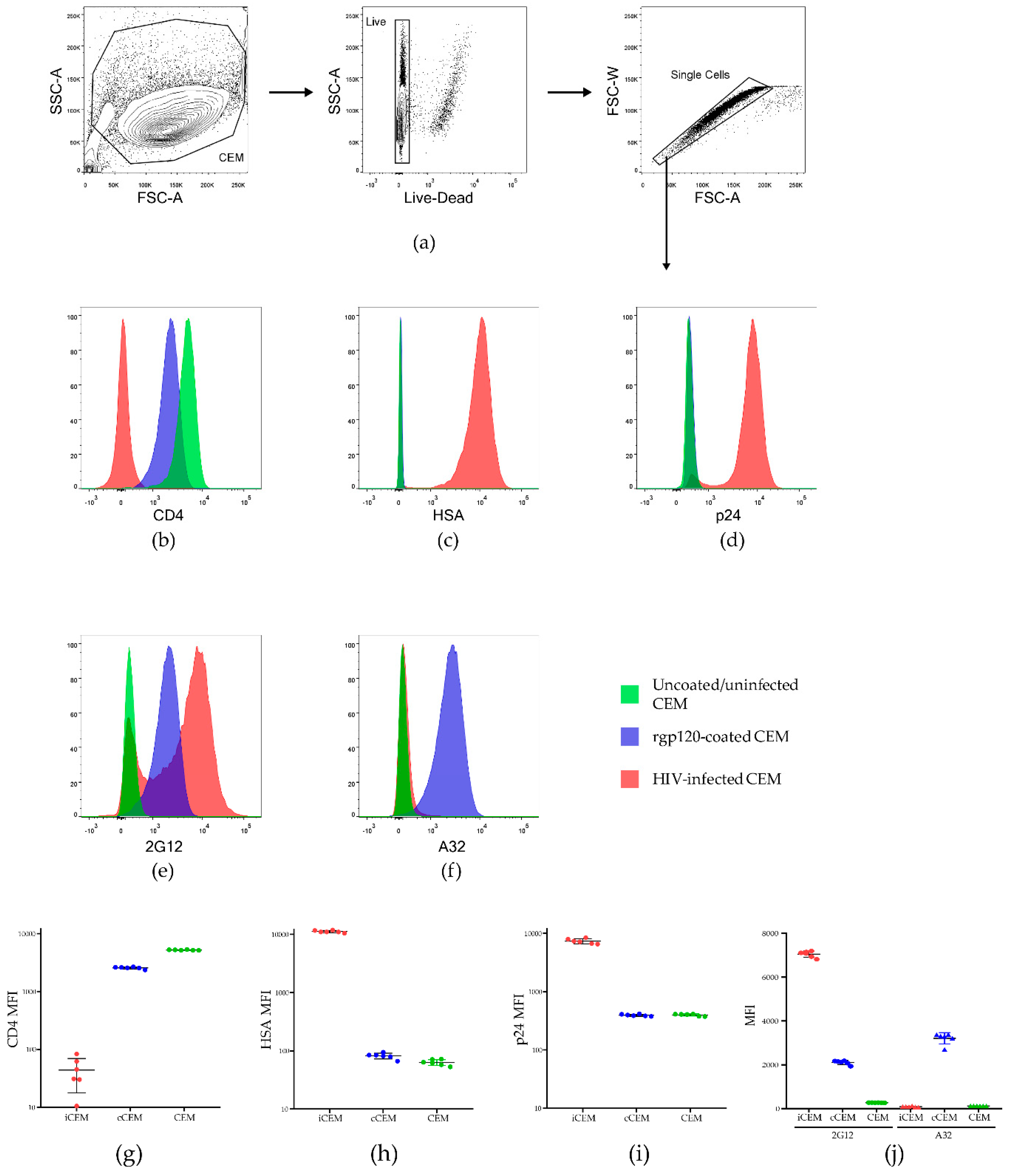
3.1. Characterization of iCEM Cells
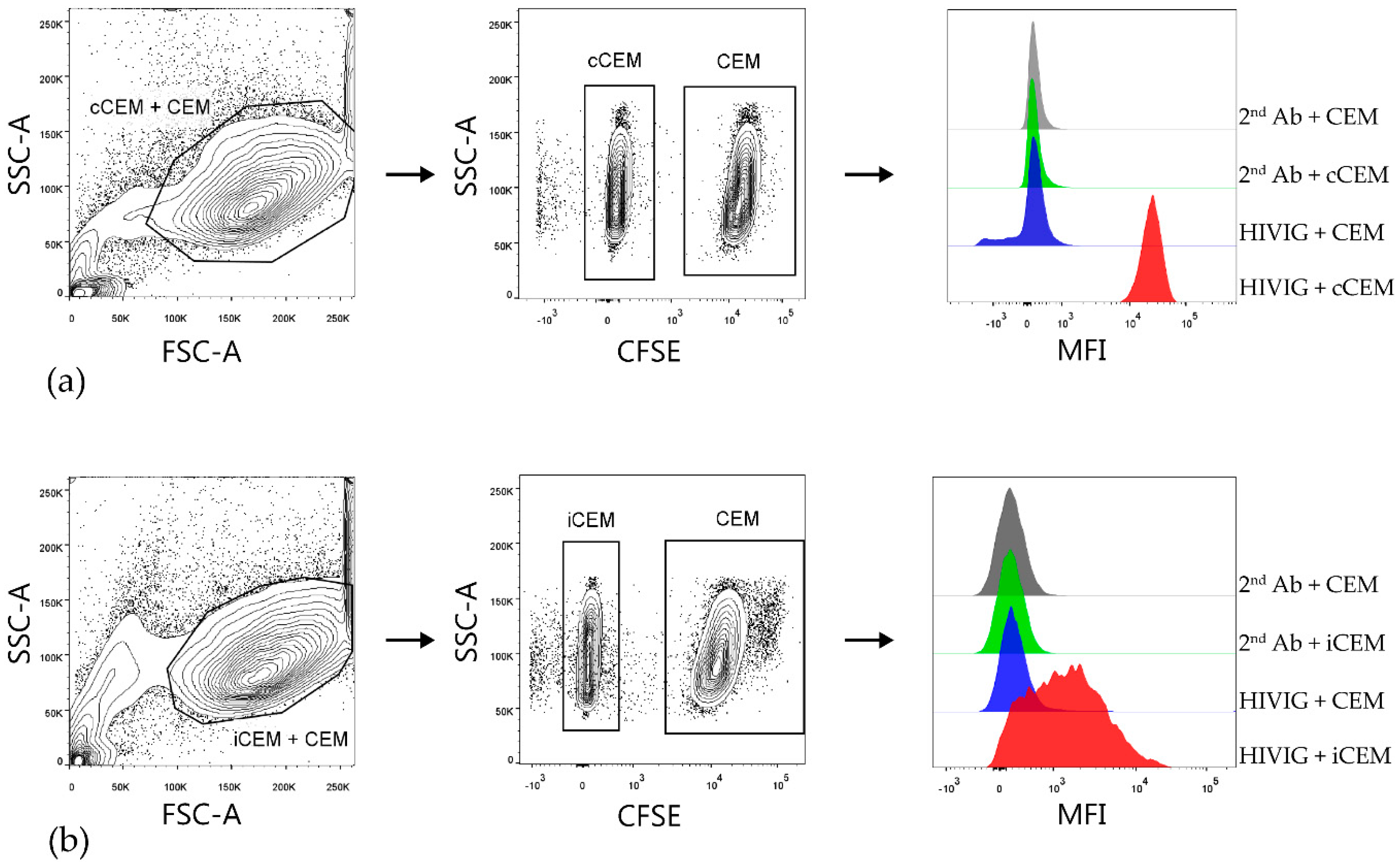
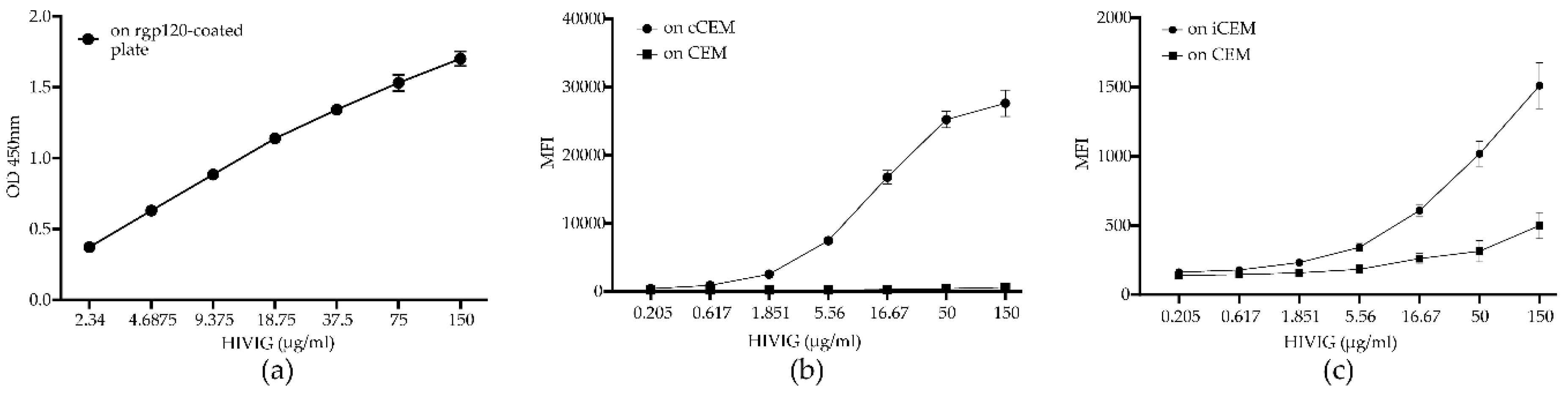
3.2. Flow Cytometry-Based Env-Specific Ab Quantification Assay
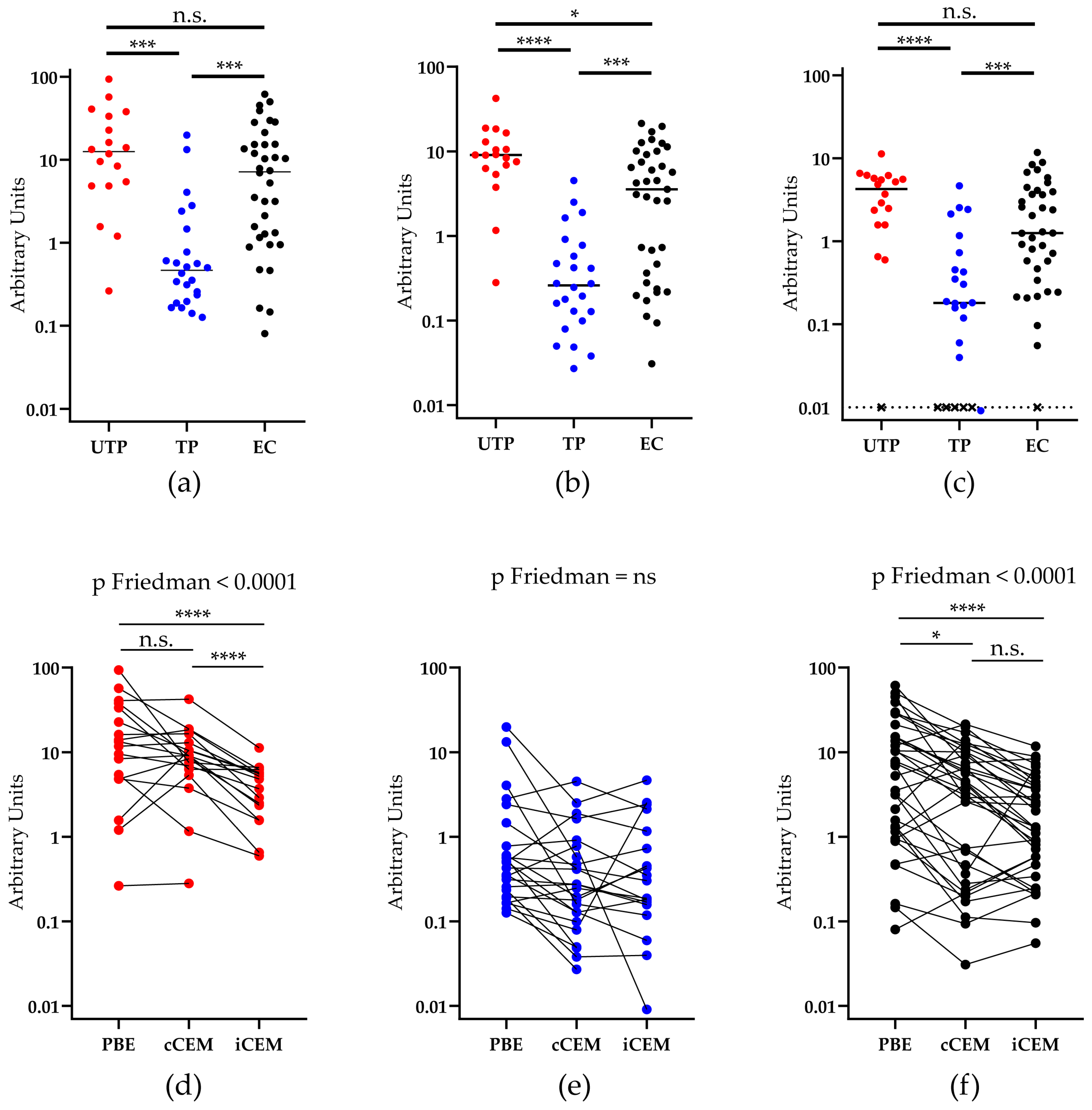
3.3. HIV Env-Specific Ab Quantification in Plasma Samples from HIV+ Subjects
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de Souza, M.; Adams, E.; et al. Vaccination with alvac and aidsvax to prevent HIV-1 infection in thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.X.; Bonsignori, M.; Alam, S.M.; McLellan, J.S.; Tomaras, G.D.; Moody, M.A.; Kozink, D.M.; Hwang, K.K.; Chen, X.; Tsao, C.Y.; et al. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 2013, 38, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Tomaras, G.D.; Ferrari, G.; Shen, X.; Alam, S.M.; Liao, H.X.; Pollara, J.; Bonsignori, M.; Moody, M.A.; Fong, Y.; Chen, X.; et al. Vaccine-induced plasma iga specific for the c1 region of the hiv-1 envelope blocks binding and effector function of igg. Proc. Natl. Acad. Sci. USA 2013, 110, 9019–9024. [Google Scholar] [CrossRef]
- Forthal, D.N.; Gilbert, P.B.; Landucci, G.; Phan, T. Recombinant gp120 vaccine-induced antibodies inhibit clinical strains of hiv-1 in the presence of fc receptor-bearing effector cells and correlate inversely with HIV infection rate. J. Immunol. 2007, 178, 6596–6603. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.A.-O.; Alter, G. Systems serology: Profiling vaccine induced humoral immunity against HIV. Retrovirology 2017, 14, 57. [Google Scholar] [CrossRef]
- Mayr, L.M.; Su, B.; Moog, C. Non-neutralizing antibodies directed against HIV and their functions. Front. Immunol. 2017. [Google Scholar] [CrossRef]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. Hiv-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011, 410, 582–608. [Google Scholar] [CrossRef]
- Merk, A.; Subramaniam, S. Hiv-1 envelope glycoprotein structure. Curr. Opin. Struct. Biol. 2013, 23, 268–276. [Google Scholar] [CrossRef]
- Wyatt, R.; Kwong, P.D.; Desjardins, E.; Sweet, R.W.; Robinson, J.; Hendrickson, W.A.; Sodroski, J.G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 1998, 393, 705–711. [Google Scholar] [CrossRef]
- Allan, J.S.; Coligan, J.E.; Barin, F.; McLane, M.F.; Sodroski, J.G.; Rosen, C.A.; Haseltine, W.A.; Lee, T.H.; Essex, M. Major glycoprotein antigens that induce antibodies in aids patients are encoded by HTLV-III. Science 1985, 228, 1091–1094. [Google Scholar] [CrossRef] [PubMed]
- Robey, W.G.; Safai, B.; Oroszlan, S.; Arthur, L.O.; Gonda, M.A.; Gallo, R.C.; Fischinger, P.J. Characterization of envelope and core structural gene products of HTLV-III with sera from aids patients. Science 1985, 228, 593–595. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Bartesaghi, A.; Borgnia, M.J.; Sapiro, G.; Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 2008, 455, 109–113. [Google Scholar] [CrossRef]
- Zhu, P.; Chertova, E.; Bess, J., Jr.; Lifson, J.D.; Arthur, L.O.; Liu, J.; Taylor, K.A.; Roux, K.H. Electron tomography analysis of envelope glycoprotein trimers on hiv and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 2003, 100, 15812–15817. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.B.; Gorman, J.; Ma, X.; Zhou, Z.; Arthos, J.; Burton, D.R.; Koff, W.C.; Courter, J.R.; Smith, A.B., 3rd; Kwong, P.D.; et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science 2014, 346, 759–763. [Google Scholar] [CrossRef]
- Herschhorn, A.; Ma, X.; Gu, C.; Ventura, J.D.; Castillo-Menendez, L.; Melillo, B.; Terry, D.S.; Smith, A.B., 3rd; Blanchard, S.C.; Munro, J.B.; et al. Release of gp120 restraints leads to an entry-competent intermediate state of the HIV-1 envelope glycoproteins. MBio 2016, 7. [Google Scholar] [CrossRef]
- Veillette, M.; Coutu, M.; Richard, J.; Batraville, L.A.; Dagher, O.; Bernard, N.; Tremblay, C.; Kaufmann, D.E.; Roger, M.; Finzi, A. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by antibody-dependent cellular cytotoxicity-mediating antibodies in sera from HIV-1-infected individuals. J. Virol. 2015, 89, 545–551. [Google Scholar] [CrossRef]
- Veillette, M.; Desormeaux, A.; Medjahed, H.; Gharsallah, N.E.; Coutu, M.; Baalwa, J.; Guan, Y.; Lewis, G.; Ferrari, G.; Hahn, B.H.; et al. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J. Virol. 2014, 88, 2633–2644. [Google Scholar] [CrossRef]
- Moore, J.P.; McKeating, J.A.; Weiss, R.A.; Sattentau, Q.J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science 1990, 250, 1139–1142. [Google Scholar] [CrossRef]
- Richard, J.; Prevost, J.; Baxter, A.E.; von Bredow, B.; Ding, S.; Medjahed, H.; Delgado, G.G.; Brassard, N.; Sturzel, C.M.; Kirchhoff, F.; et al. Uninfected bystander cells impact the measurement of HIV-specific antibody-dependent cellular cytotoxicity responses. MBio 2018, 9. [Google Scholar] [CrossRef]
- Guan, Y.; Pazgier, M.; Sajadi, M.M.; Kamin-Lewis, R.; Al-Darmarki, S.; Flinko, R.; Lovo, E.; Wu, X.; Robinson, J.E.; Seaman, M.S.; et al. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc. Natl. Acad. Sci. USA 2013, 110, E69–E78. [Google Scholar] [CrossRef] [PubMed]
- Von Bredow, B.; Arias, J.F.; Heyer, L.N.; Moldt, B.; Le, K.; Robinson, J.E.; Zolla-Pazner, S.; Burton, D.R.; Evans, D.T. Comparison of antibody-dependent cell-mediated cytotoxicity and virus neutralization by HIV-1 env-specific monoclonal antibodies. J. Virol. 2016, 90, 6127–6139. [Google Scholar] [CrossRef]
- Bruel, T.; Guivel-Benhassine, F.; Amraoui, S.; Malbec, M.; Richard, L.; Bourdic, K.; Donahue, D.A.; Lorin, V.; Casartelli, N.; Noel, N.; et al. Elimination of HIV-1-infected cells by broadly neutralizing antibodies. Nat. Commun. 2016, 7, 10844. [Google Scholar] [CrossRef]
- Bruel, T.; Guivel-Benhassine, F.; Lorin, V.; Lortat-Jacob, H.; Baleux, F.; Bourdic, K.; Noel, N.; Lambotte, O.; Mouquet, H.; Schwartz, O. Lack of adcc breadth of human nonneutralizing anti-HIV-1 antibodies. J. Virol. 2017, 91, e02440-16. [Google Scholar] [CrossRef]
- Pollara, J.; Hart, L.; Brewer, F.; Pickeral, J.; Packard, B.Z.; Hoxie, J.A.; Komoriya, A.; Ochsenbauer, C.; Kappes, J.C.; Roederer, M.; et al. High-throughput quantitative analysis of HIV-1 and SIV-specific adcc-mediating antibody responses. Cytom. A 2011, 79, 603–612. [Google Scholar] [CrossRef]
- Richard, J.; Veillette, M.; Batraville, L.A.; Coutu, M.; Chapleau, J.P.; Bonsignori, M.; Bernard, N.; Tremblay, C.; Roger, M.; Kaufmann, D.E.; et al. Flow cytometry-based assay to study HIV-1 gp120 specific antibody-dependent cellular cytotoxicity responses. J. Virol. Methods 2014, 208, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Smalls-Mantey, A.; Doria-Rose, N.; Klein, R.; Patamawenu, A.; Migueles, S.A.; Ko, S.Y.; Hallahan, C.W.; Wong, H.; Liu, B.; You, L.; et al. Antibody-dependent cellular cytotoxicity against primary HIV-infected CD4+ t cells is directly associated with the magnitude of surface igg binding. J. Virol. 2012, 86, 8672–8680. [Google Scholar] [CrossRef]
- Gomez-Roman, V.R.; Florese, R.H.; Patterson, L.J.; Peng, B.; Venzon, D.; Aldrich, K.; Robert-Guroff, M. A simplified method for the rapid fluorometric assessment of antibody-dependent cell-mediated cytotoxicity. J. Immunol. Methods 2006, 308, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Kramski, M.; Schorcht, A.; Johnston, A.P.; Lichtfuss, G.F.; Jegaskanda, S.; De, R.R.; Stratov, I.; Kelleher, A.D.; French, M.A.; Center, R.J.; et al. Role of monocytes in mediating HIV-specific antibody-dependent cellular cytotoxicity. J. Immunol. Methods 2012, 384, 51–61. [Google Scholar] [CrossRef]
- Ferrari, G.; Pollara, J.; Kozink, D.; Harms, T.; Drinker, M.; Freel, S.; Moody, A.M.; Alam, M.S.; Tomaras, G.D.; Ochsenbauer, C.; et al. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (adcc) activity and defines a common adcc epitope in human HIV-1 serum. J. Virol. 2011, 85, 7029–7036. [Google Scholar] [CrossRef]
- Konstantinus, I.N.; Gamieldien, H.; Mkhize, N.N.; Kriek, J.-M.; Passmore, J.-A.S. Comparing high-throughput methods to measure nk cell-mediated antibody dependent cellular cytotoxicity during HIV-infection. J. Immunol. Methods 2016, 434, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, M.E.; Mikhailova, A.; Brown, E.P.; Dowell, K.G.; Walker, B.D.; Bailey-Kellogg, C.; Suscovich, T.J.; Alter, G. Polyfunctional HIV-specific antibody responses are associated with spontaneous HIV control. PLOS Pathog. 2016, 12, e1005315. [Google Scholar] [CrossRef] [PubMed]
- Gooneratne, S.L.; Richard, J.; Lee, W.S.; Finzi, A.; Kent, S.J.; Parsons, M.S. Slaying the trojan horse: Natural killer cells exhibit robust anti-HIV-1 antibody-dependent activation and cytolysis against allogeneic T cells. J. Virol. 2015, 89, 97–109. [Google Scholar] [CrossRef]
- Parsons, M.S.; Wren, L.; Isitman, G.; Navis, M.; Stratov, I.; Bernard, N.F.; Kent, S.J. Hiv infection abrogates the functional advantage of natural killer cells educated through kir3dl1/hla-bw4 interactions to mediate anti-HIV antibody-dependent cellular cytotoxicity. J. Virol. 2012, 86, 4488–4495. [Google Scholar] [CrossRef]
- Banerjee, K.; Klasse, P.J.; Sanders, R.W.; Pereyra, F.; Michael, E.; Lu, M.; Walker, B.D.; Moore, J.P. Igg subclass profiles in infected HIV type 1 controllers and chronic progressors and in uninfected recipients of env vaccines. AIDS Res. Hum. Retroviruses 2010, 26, 445–458. [Google Scholar] [CrossRef]
- El-Far, M.; Kouassi, P.; Sylla, M.; Zhang, Y.; Fouda, A.; Fabre, T.; Goulet, J.-P.; van Grevenynghe, J.; Lee, T.; Singer, J.; et al. Proinflammatory isoforms of il-32 as novel and robust biomarkers for control failure in HIV-infected slow progressors. Sci. Rep. 2016, 6, 22902. [Google Scholar] [CrossRef] [PubMed]
- Imbeault, M.; Lodge, R.; Ouellet, M.; Tremblay, M.J. Efficient magnetic bead-based separation of HIV-1-infected cells using an improved reporter virus system reveals that p53 up-regulation occurs exclusively in the virus-expressing cell population. Virology 2009, 393, 160–167. [Google Scholar] [CrossRef]
- Buchacher, A.; Predl, R.; Strutzenberger, K.; Steinfellner, W.; Trkola, A.; Purtscher, M.; Gruber, G.; Tauer, C.; Steindl, F.; Jungbauer, A.; et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and epstein-barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 1994, 10, 359–369. [Google Scholar] [CrossRef]
- Trkola, A.; Purtscher, M.; Muster, T.; Ballaun, C.; Buchacher, A.; Sullivan, N.; Srinivasan, K.; Sodroski, J.; Moore, J.P.; Katinger, H. Human monoclonal antibody 2g12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996, 70, 1100–1108. [Google Scholar]
- Mascola, J.R.; Lewis, M.G.; Stiegler, G.; Harris, D.; VanCott, T.C.; Hayes, D.; Louder, M.K.; Brown, C.R.; Sapan, C.V.; Frankel, S.S.; et al. Protection of macaques against pathogenic simian/human immunodeficiency virus 89.6pd by passive transfer of neutralizing antibodies. J. Virol. 1999, 73, 4009–4018. [Google Scholar]
- Etemad-Moghadam, B.; Sun, Y.; Nicholson, E.K.; Karlsson, G.B.; Schenten, D.; Sodroski, J. Determinants of neutralization resistance in the envelope glycoproteins of a simian-human immunodeficiency virus passaged in vivo. J. Virol. 1999, 73, 8873–8879. [Google Scholar] [PubMed]
- Crawford, J.M.; Earl, P.L.; Moss, B.; Reimann, K.A.; Wyand, M.S.; Manson, K.H.; Bilska, M.; Zhou, J.T.; Pauza, C.D.; Parren, P.W.H.I.; et al. Characterization of primary isolate-like variants of simian-human immunodeficiency virus. J. Virol. 1999, 73, 10199. [Google Scholar]
- Wyatt, R.; Moore, J.; Accola, M.; Desjardin, E.; Robinson, J.; Sodroski, J. Involvement of the v1/v2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 1995, 69, 5723–5733. [Google Scholar] [PubMed]
- Moore, J.P.; Thali, M.; Jameson, B.; Vignaux, F.; Lewis, G.; Poon, S.-W.; MacArthur, C.; Fung, M.; Sun, B.; Durda, P.J.; et al. Immunochemical analysis of the gpl20 surface glycoprotein of human immunodeficiency virus type 1: Probing the structure of the C4 and V4 domains and the interaction of the C4 domain with the v3 loop. J. Virol. 1993, 67, 12. [Google Scholar]
- Lee, W.S.; Prevost, J.; Richard, J.; van der Sluis, R.M.; Lewin, S.R.; Pazgier, M.; Finzi, A.; Parsons, M.S.; Kent, S.J. CD4- and time-dependent susceptibility of HIV-1-infected cells to ADCC. J. Virol. 2019, 93, e01901-18. [Google Scholar] [CrossRef] [PubMed]
- Buckner, C.M.; Kardava, L.; Zhang, X.; Gittens, K.; Justement, J.S.; Kovacs, C.; McDermott, A.B.; Li, Y.; Sajadi, M.M.; Chun, T.W.; et al. Maintenance of HIV-specific memory b-cell responses in elite controllers despite low viral burdens. J. Infect. Dis. 2016, 214, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Brocca-Cofano, E.; McKinnon, K.; Demberg, T.; Venzon, D.; Hidajat, R.; Xiao, P.; Daltabuit-Test, M.; Patterson, L.J.; Robert-Guroff, M. Vaccine-elicited siv and HIV envelope-specific iga and igg memory b cells in rhesus macaque peripheral blood correlate with functional antibody responses and reduced viremia. Vaccine 2011, 29, 3310–3319. [Google Scholar] [CrossRef]
- Lee, J.H.; Ozorowski, G.; Ward, A.B. Cryo-em structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 2016, 351, 1043–1048. [Google Scholar] [CrossRef]
- Hu, G.; Liu, J.; Taylor, K.A.; Roux, K.H. Structural comparison of HIV-1 envelope spikes with and without the V1/V2 loop. J. Virol. 2011, 85, 2741–2750. [Google Scholar] [CrossRef]
- Bartesaghi, A.; Merk, A.; Borgnia, M.J.; Milne, J.L.S.; Subramaniam, S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat. Struct. Mol. Boil. 2013, 20, 1352–1357. [Google Scholar] [CrossRef]
- Guzzo, C.; Zhang, P.; Liu, Q.; Kwon, A.L.; Uddin, F.; Wells, A.I.; Schmeisser, H.; Cimbro, R.; Huang, J.; Doria-Rose, N.; et al. Structural constraints at the trimer apex stabilize the HIV-1 envelope in a closed, antibody-protected conformation. mBio 2018, 9, e00955-18. [Google Scholar] [CrossRef] [PubMed]
- Brown, E.P.; Licht, A.F.; Dugast, A.-S.; Choi, I.; Bailey-Kellogg, C.; Alter, G.; Ackerman, M.E. High-throughput, multiplexed igg subclassing of antigen-specific antibodies from clinical samples. J. Immunol. Methods 2012, 386, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Veillette, M.; Coutu, M.; Richard, J.; Batraville, L.-A.; Désormeaux, A.; Roger, M.; Finzi, A. Conformational evaluation of HIV-1 trimeric envelope glycoproteins using a cell-based elisa assay. J. Vis. Exp. 2014, 14, 51995. [Google Scholar] [CrossRef] [PubMed]
- Casazza, J.P.; Betts, M.R.; Picker, L.J.; Koup, R.A. Decay kinetics of human immunodeficiency virus-specific CD8+ T cells in peripheral blood after initiation of highly active antiretroviral therapy. J. Virol. 2001, 75, 6508–6516. [Google Scholar] [CrossRef] [PubMed]
- Lamine, A.; Caumont-Sarcos, A.; Chaix, M.-L.; Saez-Cirion, A.; Rouzioux, C.; Delfraissy, J.-F.; Pancino, G.; Lambotte, O. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS 2007, 21, 3. [Google Scholar] [CrossRef] [PubMed]
- Hatano, H.; Delwart, E.L.; Norris, P.J.; Lee, T.-H.; Dunn-Williams, J.; Hunt, P.W.; Hoh, R.; Stramer, S.L.; Linnen, J.M.; McCune, J.M.; et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 2009, 83, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Pereyra, F.; Palmer, S.; Miura, T.; Block, B.L.; Wiegand, A.; Rothchild, A.C.; Baker, B.; Rosenberg, R.; Cutrell, E.; Seaman, M.S.; et al. Persistent low-level viremia in HIV-1 elite controllers and relationship to immunologic parameters. J. Infect. Dis. 2009, 200, 984–990. [Google Scholar] [CrossRef]
- Burton, D.R.; Hangartner, L. Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu. Rev. Immunol. 2016, 34, 635–659. [Google Scholar] [CrossRef]
- West, A.P., Jr.; Scharf, L.; Scheid, J.F.; Klein, F.; Bjorkman, P.J.; Nussenzweig, M.C. Structural insights on the role of antibodies in HIV-1 vaccine and therapy. Cell 2014, 156, 633–648. [Google Scholar] [CrossRef]
- Rusert, P.; Kouyos, R.D.; Kadelka, C.; Ebner, H.; Schanz, M.; Huber, M.; Braun, D.L.; Hoze, N.; Scherrer, A.; Magnus, C.; et al. Determinants of HIV-1 broadly neutralizing antibody induction. Nat. Med. 2016, 22, 1260–1267. [Google Scholar] [CrossRef]
- Mouquet, H. Antibody b cell responses in HIV-1 infection. Trends Immunol. 2014, 35, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Diskin, R.; Scheid, J.F.; Gaebler, C.; Mouquet, H.; Georgiev, I.S.; Pancera, M.; Zhou, T.; Incesu, R.B.; Fu, B.Z.; et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell 2013, 153, 126–138. [Google Scholar] [CrossRef] [PubMed]
- Chung, A.W.; Kumar, M.P.; Arnold, K.B.; Yu, W.H.; Schoen, M.K.; Dunphy, L.J.; Suscovich, T.J.; Frahm, N.; Linde, C.; Mahan, A.E.; et al. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 2015, 163, 988–998. [Google Scholar] [CrossRef] [PubMed]
- Lewis, G.K. Role of fc-mediated antibody function in protective immunity against HIV-1. Immunology 2014, 142, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Lambotte, O.; Ferrari, G.; Moog, C.; Yates, N.L.; Liao, H.X.; Parks, R.J.; Hicks, C.B.; Owzar, K.; Tomaras, G.D.; Montefiori, D.C.; et al. Heterogeneous neutralizing antibody and antibody-dependent cell cytotoxicity responses in HIV-1 elite controllers. AIDS 2009, 23, 897–906. [Google Scholar] [CrossRef]
- Migueles, S.A.; Connors, M. Success and failure of the cellular immune response against HIV-1. Nat. Immunol. 2015, 16, 563–570. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kant, S.; Zhang, N.; Routy, J.-P.; Tremblay, C.; Thomas, R.; Szabo, J.; Côté, P.; Trottier, B.; LeBlanc, R.; Rouleau, D.; et al. Quantifying Anti-HIV Envelope-Specific Antibodies in Plasma from HIV Infected Individuals. Viruses 2019, 11, 487. https://doi.org/10.3390/v11060487
Kant S, Zhang N, Routy J-P, Tremblay C, Thomas R, Szabo J, Côté P, Trottier B, LeBlanc R, Rouleau D, et al. Quantifying Anti-HIV Envelope-Specific Antibodies in Plasma from HIV Infected Individuals. Viruses. 2019; 11(6):487. https://doi.org/10.3390/v11060487
Chicago/Turabian StyleKant, Sanket, Ningyu Zhang, Jean-Pierre Routy, Cécile Tremblay, Réjean Thomas, Jason Szabo, Pierre Côté, Benoit Trottier, Roger LeBlanc, Danielle Rouleau, and et al. 2019. "Quantifying Anti-HIV Envelope-Specific Antibodies in Plasma from HIV Infected Individuals" Viruses 11, no. 6: 487. https://doi.org/10.3390/v11060487
APA StyleKant, S., Zhang, N., Routy, J.-P., Tremblay, C., Thomas, R., Szabo, J., Côté, P., Trottier, B., LeBlanc, R., Rouleau, D., Harris, M., Dupuy, F. P., & Bernard, N. F. (2019). Quantifying Anti-HIV Envelope-Specific Antibodies in Plasma from HIV Infected Individuals. Viruses, 11(6), 487. https://doi.org/10.3390/v11060487




