Abstract
Chikungunya fever is a debilitating disease caused by Chikungunya virus (CHIKV) that can result in long-lasting arthralgias. The early diagnosis of CHIKV relies on PCR during the acute infection phase to allow differential diagnosis with other co-circulating arboviruses such as dengue and Zika. Alternatively, serology can support diagnosis and provide epidemiological information on current and past outbreaks. Many commercial serological ELISA assays are based on the inactivated whole CHIKV, but their sensitivity and specificity show great variability. We produced recombinant CHIKV E2 that is suitable for ELISA assays, which was used for the serodiagnosis of CHIKV infections occurring in an arbovirus endemic Mexican region within Michoacán state. A cross-sectional study was conducted in 2016–2017; sera was obtained from 15 healthy donors and 68 patients presenting undifferentiated febrile illness. Serum samples were screened by RT-PCR and by our in-house ELISA assay. Our results indicate that IgM and IgG anti-CHIKV E2 antibodies were detected with our ELISA assay with higher sensitivity than a commercially available CHIKV ELISA kit. Our simple and sensitive ELISA assay for the serodiagnosis of CHIKV infections can be applied to population-based seroprevalence surveys and has potential for monitoring vaccine immunogenicity in CHIKV vaccine clinical trials.
Keywords:
Chikungunya virus; envelope protein 2; diagnosis; ELISA; febrile patients; CHIKV antibodies; Mexico 1. Introduction
Chikungunya virus (CHIKV) is a re-emerging arbovirus that belongs to the Alphavirus genus of the Togaviridae family [1,2,3]. CHIKV is transmitted to humans by Aedes spp. mosquitoes [3,4,5]. The genome of CHIKV consists of a positive-sense, single-stranded RNA of approximately 11.8 kb, and contains structural genes that encode three structural proteins: envelope proteins 1 and 2 (E1 and E2) and a nucleocapsid protein along with two small peptides (E3 and 6K) [6,7,8]. The CHIKV E1 and E2 form spikes composed of triplets of E1–E2 heterodimers covering the viral surface [9,10]. Since its appearance in 2013 in the Caribbean, it has rapidly spread throughout the Americas, raising public health concerns [11,12,13]. CHIKV infections are characterized by an acute onset of fever associated with myalgia, headache, and severe debilitating arthropathy [3]. Vaccines or specific anti-viral therapies for CHIKV are not yet available; thus, rapid and simple methodologies to detect CHIKV infections are important for effective patient management and the control of future epidemics [12,14]. The primary laboratory test to diagnose CHIKV infection relies on the specific detection of CHIKV genomic sequences in serum collected up to five to six days after the onset of fever [15,16,17]. However, serological tests are required for the confirmation of CHIKV infection after the end of the viremia. Several commercial and in-house CHIKV serological assays that became available are based on whole virus antigens, and those reports indicate a variation of sensitivities and specificities [18,19,20,21,22].
Anti-CHIKV IgM antibodies appear as early as three days after infection, but their presence is usually detected only after three to four months [18,23]. In contrast, anti-CHIKV IgG antibodies remain detectable in convalescent individuals for many years; thus, anti-CHIKV IgG are able to provide insights into the seroepidemiology of CHIKV [20,24]. However, it is often difficult to differentiate acute from convalescent cases based on the detection of specific IgG alone in highly endemic areas. Despite this difficulty, a recent multi-country quality assurance study showed that the IgG detection of CHIKV infection may be more useful than IgM detection [25]. Therefore, we sought to develop an improved diagnostic tool to detect CHIKV-specific IgM and IgG based on a recombinant subunit protein that may be important in improving the sensitivity of serodiagnosis for clinical management and epidemiological surveillance. We have recently reported a recombinant CHIKV E2 protein in mammalian cells [26]. Based on this, we sought to detect and quantify anti-CHIKV E2 IgM and IgG E2 in human sera from CHIKV acute infections or convalescent patients. In order to validate our in-house ELISA, we have used a cohort of sera from infected patients tested by a reverse transcription-polymerase chain reaction (RT-PCR) along with the sera from healthy volunteers, and then compared these to a commercial ELISA assay. Results showed that our in-house E2 based IgM and IgG ELISA assays could detect anti-CHIKV IgM and IgG antibodies with similar or higher sensitivity than the commercial kit. The data presented here also provide valuable serological information regarding the current seroprevalence of CHIKV in the city of Lázaro Cárdenas, state of Michoacán, in México.
2. Materials and Methods
2.1. Study Design
A descriptive cross-sectional study was conducted in October 2016 and June–August 2017 at outpatient clinics at Lázaro Cárdenas, a port city in the state of Michoacán, México, where arboviruses are endemic. A total of 68 samples were collected and processed for anti-CHIKV IgM and IgG ELISA. Fifteen samples from healthy donors were collected in Morelia, the capital city in the state of Michoacán, where the circulation of CHIKV is not considered to be endemic.
2.2. Sample Collection
Sera were obtained from febrile patients with signs or symptoms compatible with Chikungunya fever (fever, joint pain, or rash) in outpatient clinics at Lázaro Cárdenas at two different time points (seven samples in October 2016 and 61 samples in June–August 2017). These samples were tested with CHIKV RT-PCR to confirm the CHIKV infection. All the serum samples were initially kept at 4 °C, and after transportation to the laboratory, they were stored at −80 °C until further processing.
2.3. Real-Time Quantitative Reverse Transcription PCR (qRT-PCR)
Briefly, the extraction of viral RNA from serum samples was performed using a QIAamp Viral RNA Mini Kit (Qiagen, Germany) following the manufacturer’s instructions. A commercially available rtRT-PCR primer/probe kit (Path-CHIK-standard kits, Genesig, Primer design Ltd., Cambridge, UK) was used to detect and amplify Chikungunya RNA. All the serum samples were tested for Chikungunya RNA. The tests were performed using an Applied Biosystem step one real-time PCR machine (Applied Biosystem, CA, USA). A reaction mix with a final volume of 20 μL was prepared adding 10 μL of oasig One step PCR Master mix, 1 μL of CHIK primers/probe, 1 μL of internal extraction control primer/probe mix, 5 μL of viral RNA, and 3 μL of RNAse/DNAse free water. The cycling conditions for the RT-PCR consisted of a reverse transcription step of 55° for 30 min; enzyme activation for the PCR was performed at 95 °C for 2 min, followed by 50 cycles of denaturation 95 °C for 10 s, and data collection 60 °C for 60 s, as described by the manufacturer instructions. Fluorogenic data was collected through the FAM and VIC channels.
2.4. Production and Purification of Recombinant CHIKV E2 Protein
The production and purification of recombinant CHIKV E2 protein was carried out as described previously [26]. Briefly, the expression plasmid (500 μg) was individually transfected in HEK-293T cells using polyethyleneimine (PEI) in roller bottles (surface area of 2125 cm2) under standard cell culture conditions. Five days after transfection, cells were discarded and media was filtered through 0.22-μM disposable filters. The secreted proteins were purified from the supernatant by Niquel Sepharose affinity chromatography (HisTRAPTM, GE Healthcare Life Sciences, UK), using the Äkta Start chromatography system and eluted with 500 mM of imidazole. Finally, eluted proteins were dialyzed using a Slide-A-LyzerTM cassette (Fisher Scientific, UK) against 1× PBS.
2.5. Detection of Anti-CHIKV IgM and IgG by in-House ELISA
A total of 68 serum samples from febrile Mexican patients in an endemic area and 15 sera samples from healthy blood donors from a non-endemic area were tested in parallel. For the initial method development, seven patient sera from RT-PCR confirmed CHIKV infections were used for recombinant CHIKV E2-based IgG and IgM ELISA assay to measure the antibodies against E2 along with seven negative control sera from healthy volunteers. A CHIKV-positive control serum sample was obtained from a Mexican blood donor who was known to have confirmed CHIKV infection independent from this study. Briefly, human sera were diluted in Nunc Maxisorp Immuno ELISA plates coated with CHIKV E2 in PBS to a final concentration of 2 μg/mL. Following blocking, sera was incubated for 1 h; then, plates were washed six times with PBS/T (0.05%), and bound antibodies were detected by using a goat Anti-Human IgG–alkaline phosphatase-conjugated antibody (Sigma-Aldrich UK, A3187-.5ML) or a goat Anti-Human IgM (μ-chain specific)–alkaline phosphatase-conjugated antibody (Sigma, A3275-.5ML). Development was performed using 4-nitrophenylphosphate diluted in diethanolamine buffer, and absorbance values at OD405 were measured on a CLARIOstar instrument (BMG Labtech UK). Serum antibody endpoint titres for positive samples were defined by an absorbance value that was three standard deviations greater than the average OD405 of the negative controls.
2.6. Detection of Anti-CHIKV IgM and IgG by Commercial ELISA Kit
For the qualitative measurement of IgM and IgG class antibodies against CHIKV in human serum, an Anti-Chikungunya Virus IgM Human ELISA Kit (ab 177848) and an IgG Human ELISA Kit (ab 177835) from Abcam were used to test all the serum samples according to the manufacturer’s protocols. The results were calculated and interpreted as suggested in the protocol.
2.7. Maps and Geographical Data
Maps in Figure 1A–C were obtained using the QGIS 3.6 software Release 3.6 and the QGIS Geographic Information System, using the ESRI satellite database. Then, maps were layered with the OpenStretMap option. Geographical and climate conditions were obtained from historical databases available at the Mexican system for geographical information and the National meteorological service websites, respectively.
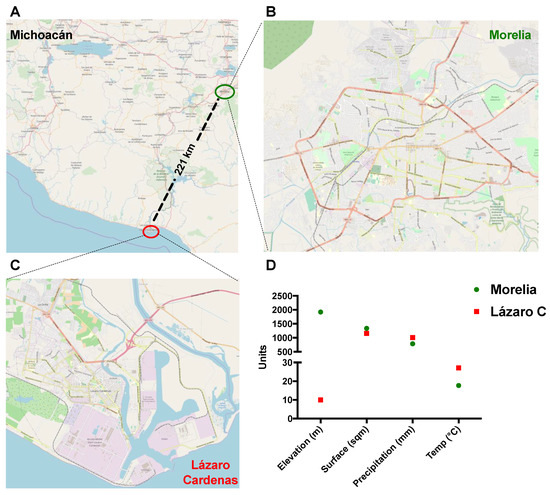
Figure 1.
The geographical location of study sites. (A–C). Map of Michoacan State in Mexico showing the locations of study sites (Lazaro Cardenas and Morelia) and illustration of the geographical differences between the two municipalities. (D). Graph showing the elevation, surface, precipitation, and temperature between the two municipalities.
2.8. Ethics Approval and Consent to Participate
The human sera statement was approved by the appropriate ethical review sub-committees, and written informed consent was obtained from all the study participants in Mexico (R-2016-785-104).
2.9. Data Analysis and Statistics
All the data analysis and statistics were performed using the Prism 7 software (GraphPad, Software, U.S.). p Values and R2 values reflect Pearson correlation tests for determining the correlation between the endpoint reciprocal titer values from ELISA assay and mean standard units (SU) from commercial kits.
3. Results
During October 2016 and June–August 2017, 68 febrile patients were recruited in this study from Lázaro Cárdenas, Michoacán, México (Figure 1A). Lázaro Cárdenas is a port city and an arbovirus endemic region (Figure 1C) with low altitude and higher temperature than Morelia (Figure 1B,D). By following the algorithm depicted in Figure 2, we performed qRT-PCR to all 68 samples. Seven out of 68 samples (10.3%) tested positive on RT-PCR confirming the acute CHIKV infection.
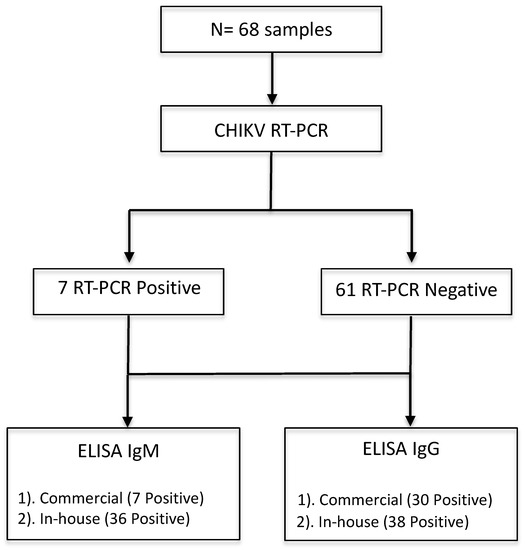
Figure 2.
Flow chart describing tests performed in samples using RT-PCR and two ELISA methodologies.
3.1. Development and Evaluation of CHIKV Recombinant E2 Based in-House ELISA
Next, we sought to test whether our recombinant CHIKV E2 could be used to detect anti- CHIKV E2 antibodies in human serum. We selected the seven CHIKV RT-PCR positive samples (blue), a positive CHIKV control (green), and seven control sera (red) from healthy volunteers to test the suitability of our recombinant CHIKV E2 proteins in IgG and IgM-based ELISA assay (Figure 3). The OD405 demonstrate that both IgG and IgM-based ELISA can reliably detect and differentiate the sera from CHIKV-infected patients from control sera (Figure 3A left and Figure 3B left), but OD405 values were higher for IgG-based ELISA than for IgM. Endpoint reciprocal titres were calculated and showed that all confirmed CHIKV patients have high mean titres of 3.4 and 2.7 on IgG and IgM ELISA, respectively (Figure 3A right and Figure 3B right). These results indicate that our purified CHIKV E2 recombinant protein can reliably detect anti-CHIKV E2 antibodies in infected patients, which may be relevant for the serodiagnosis of CHIKV infection.
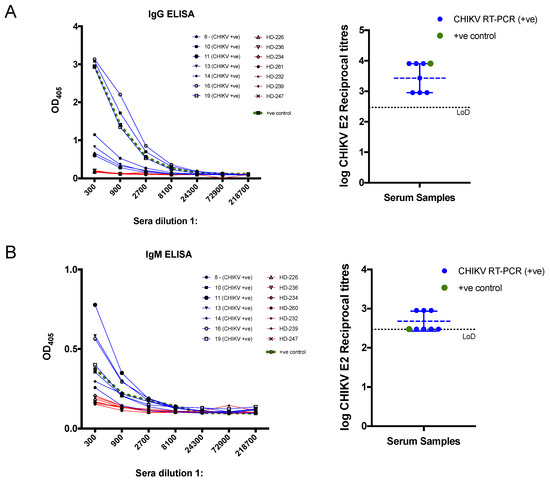
Figure 3.
ELISA assays to assess the reactivity of human sera. Chikungunya virus (CHIKV)-infected patients (blue) confirmed by RT-PCR, sera obtained from healthy donors (HD) from a non-endemic region (red) and a CHIKV-positive control serum (green). (A) The graph of CHIKV E2 IgG OD405 against sera dilutions (left) and the corresponding endpoint log reciprocal titres (right). (B) The graph of CHIKV E2 IgM OD405 (left) and the corresponding endpoint log reciprocal titres (right). The black dotted line indicates the limit of detection (LoD), which corresponds to 1:300 serum dilution on a log scale.
3.2. CHIKV Serological Diagnosis Using the Commercial ELISA Kits
After an initial screening of serum samples by RT-PCR, serological analysis was performed on all 68 serum samples along with 15 control samples from healthy blood donors obtained from Morelia, as shown in Figure 2. We tested all the serum samples by using commercial CHIKV IgM and IgG ELISA Kits for the serodiagnosis of CHIKV infection, and the mean standard units (SU) were calculated (Figure 4). Interestingly, among seven CHIKV RT-PCR positive samples, four out of seven CHIKV-infected patients (sample numbers 8, 10, 16, and 19) were identified by IgG commercial kit, while only one sample (sample number 10) tested positive as CHIKV infection. Overall, 30/68 (44.1%) samples that were both RT-PCR positive and negative were shown to be positive (mean SU > 11) on the commercial IgG ELISA, with one sample in the grey zone (inconclusive) (Figure 4 Top). On the other hand, 7/68 (10.3%) of samples tested positive (mean SU > 11) and four samples were inconclusive on the commercial IgG ELISA kit (Figure 4, bottom). All 15 serum samples from healthy volunteers yielded negative results by ELISA (Supplementary Table S1).
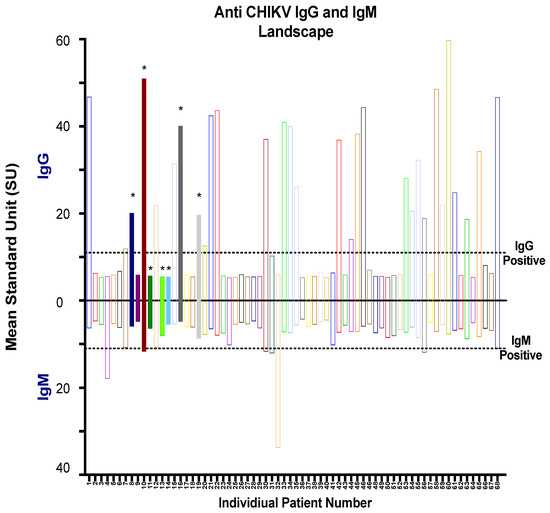
Figure 4.
Qualitative ELISA assays using the commercial kits to assess the reactivity of human serum samples. Mean standard unit (SU) for IgG (positive y-axis) and IgM (negative y-axis) are represented for all 68 serum samples. Positive samples are defined as >11 SU, and the negative samples are defined as <9. The SU of samples 9–11 are defined as in the grey zone (inconclusive results). RT-PCR positive samples are indicated by the (*) and color-filled bars.
3.3. CHIKV Serological Diagnosis Using the in-House ELISA
Next, we tested all the serum samples by using our in-house CHIKV IgM and IgG ELISA for the serodiagnosis of CHIKV infection. The graph in Figure 5A shows the OD405 values over sera dilutions in range of 300 to 218,700 for IgG-based ELISA, which reliably detected and differentiated the sera from CHIKV-infected patients from control sera and CHIKV seronegative sera. Endpoint reciprocal titres (Figure 5B) were calculated which showed that 57.4% and 54.4% of serum samples were positive for in-house IgG and IgM ELISA, respectively (Figure 5B top and bottom). Importantly, all the RT-PCR positive serum samples (8, 10, 11, 13, 14, 16, and 19) were shown to be positive in our in-house IgM and IgG ELISA (Table S1). This demonstrates that our in-house recombinant E2-based IgM and IgG may be more sensitive than the used commercial kit.
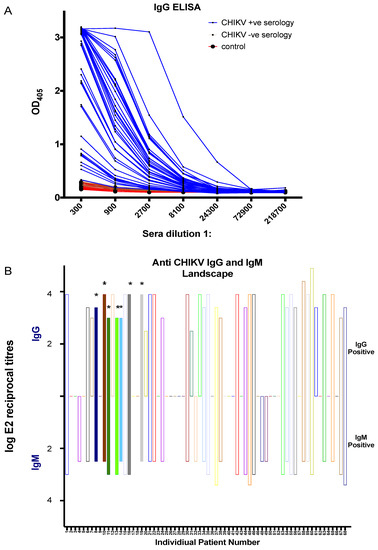
Figure 5.
In-house recombinant CHIKV envelope protein 2 (E2)-based ELISA. (A) IgG ELISA assay to assess the reactivity of all the human serum samples in this study. Positive CHIKV serology (blue), negative CHIKV serology (brown), and negative control serum samples from healthy volunteers (red). (B) Log reciprocal E2 endpoint titres for IgG (positive y-axis) and IgM (negative y-axis) are represented for all 68 serum samples. RT-PCR positive samples are indicated by the (*) and color-filled bars.
3.4. Correlation of CHIKV Antibody Titres with the Commercial Kits
Next, we sought to correlate CHIKV antibody titres from our ELISA assay to the mean SU from the qualitative ELISA results from the commercial kits (Figure 6). Values of the reciprocal antibody titres and the mean SU for IgG showed a strong correlation (p < 0.0001 and R2 = 0.61) for the serologically positive patient samples that we tested (n = 39) (Figure 6A). On the other hand, no correlation was observed between our IgM ELISA and commercial IgM kit, which may be due to significant differences between the number of positive samples (7/68 versus 37/68) (Figure 6B). The results suggest that E2-specific antibodies could be measured by reciprocal endpoint titres in our in-house IgG ELISA assay, which correlates with the commercial IgG mean SU.
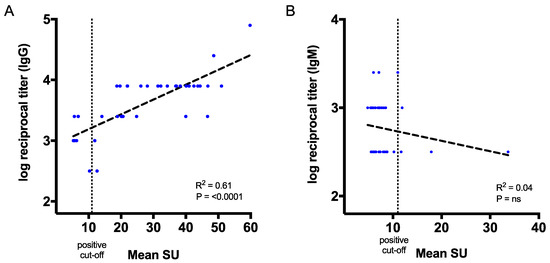
Figure 6.
Correlation between the endpoint titres from in-house ELISA assay and mean standard unit (SU) from the commercial CHIKV kit. (A) Correlation between the in-house IgG endpoint titres and the commercial IgG mean SU. (B) Correlation between the IgM endpoint titres and the commercial IgM mean SU. The p values and R2 values reflect Pearson correlation tests. A positive cut-off is defined as >11 SU.
4. Discussion
In this study, we aimed to test the potential for the clinical use of an in-house ELISA assay with an E2 CHIKV protein and compare it with an available commercial ELISA kit using samples of a cohort from a region of Mexico that is endemic for CHIKV. We have recently reported a recombinant CHIKV E2 protein produced in a mammalian expression system based on HEK293 cells [26]. In this study, we tested whether our recombinant CHIKV E2 could be used to detect anti-CHIKV antibodies in clinical samples. Our results suggest that both IgG-based and IgM-based ELISA can reliably detect anti-CHIKV E2 antibodies in both acute and convalescent patient sera. Moreover, our assays provided higher sensitivity than the commercial kit used in this study, in particular for IgM ELISA. Many commercially available anti-CHIKV ELISA or immunofluorescence assays use whole virus-derived antigens, which can have high production costs due to virus culture and purifications steps, thus decreasing affordability and limiting their use in CHIKV-endemic regions in developing countries [18,27,28,29,30]. A previous study reported that the Abcam ELISA kit, in particular, showed batch-to-batch variability, with some kits demonstrating lower sensitivity than others, and the kit had to be re-released [18]. This could account for low sensitivity (one out of seven) in detecting IgM antibodies in RT-PCR positive samples by the Abcam IgM kit. Alternatively, this may be due to the low level of IgM antibodies in the acute CHIKV serum samples, which were insufficient to be detected by whole virus antigen used in the kit, as the actual amount of E2/E1 glycoproteins in the whole virus antigen plus the capsid and the same concentration of purified recombinant E2 may differ significantly. Previous studies have evaluated the E. coli-based recombinant E1 and E2 for Indirect ELISA [28,31]. It was shown that recombinant CHIKV E2 had higher sensitivity and specificity than CHIKV E1-based ELISA, and the results were comparable to the native antigen-based ELISA [28]. Another study showed that IgG E2 ELISA provided ~90% sensitivity and 100% specificity in concordance with CHIKV-specific neutralization assay, whilst IgM to E2 showed similar sensitivity to a commercial immunochromatography for CHIKV IgG and IgM detection [31]. Other studies used the baculovirus-derived CHIKV E1 for ELISA [32,33]. Taken together, these studies provide strong pieces of evidence that recombinant protein-based ELISA may be safe and cost-effective, along with a high degree of sensitivity and specificity for setting up CHIKV diagnostics. We have been using a mammalian-based cell culture system to purify large quantities of viral recombinant proteins that could be used in diagnostic assays [26,34,35]. Protein expression in a mammalian system ensures the post-translational modifications such as N-glycosylation [35]. It was previously shown that insect cell-derived CHIKV VLPs mainly consisted of oligomannose glycans, while HEK293-derived VLPs contain the mixture of oligomannose, hybrid, and complex glycans [36].
The in-house developed ELISA has higher sensitivity, and is also more cost-effective than using the commercial CHIKV assays. However, our E2-based recombinant ELISA assay would not detect anti-CHIKV E1 antibodies and some conformational epitopes targeting the E1/E2 heterodimers. We have recently reported the formation of virus-like articles by expressing the full structural cassette of CHIKV [26]; it will be interesting to test those as captured antigens in the ELISA assay to compare with our in-house E2-based ELISA and commercial ELISA kit to address the recognition of both anti-E1 and anti-E2 antibodies conformational epitopes. In order to validate our ELISA assay further, it will be useful to test negative controls such as serum samples from ZIKV or DENV-infected individuals. In addition, it will be very informative to determine PRNT titres for positive serology serum samples [37] that can support a sensitivity and specificity assessment of our in-house ELISA assay in comparison to PRNT as well as correlating antibody and PRNT titres. It will also be interesting to explore the cross-reactivity toward the antibodies against other alphaviruses, as they could be co-circulating in CHIKV endemic areas and determine the specificity of our ELISA assay [38,39]. We have detected ~50% seropositive using our in-house IgM and IgG ELISA for the febrile patients in Lázaro Cárdenas. As CHIKV co-circulates with other arboviruses in various endemic regions, there is often difficulty in differentiating between acute and previous CHIKV infections when their symptomatology is similar. Nonetheless, our ELISA assay could be helpful for the serodiagnosis of CHIKV in acute patients in the endemic regions of Mexico where there is no availability of specialized laboratories for RT-PCR, which may be important for effective patient management. In conclusion, our in-house CHIKV-specific ELISA can be a useful tool for the serodiagnosis of CHIKV infection, for seroprevalence studies in endemic regions to contribute to the management of the potential future epidemics, and for the monitoring of vaccine immunogenicity during CHIKV vaccine clinical trials.
Supplementary Materials
The following are available online at https://www.mdpi.com/1999-4915/11/5/407/s1, Table S1: Comparative evaluation of recombinant E2 based in-house ELISA with respect to the commercial kit for all the serum samples used in this study.
Author Contributions
Conceived and designed the experiments: Y.C.K., A.R.-S., C.L.-C., and Y.C.K. performed the production and purification of recombinant proteins. M.A.M. and M.E.V.-S. contributed human sera, reagents, and materials. K.G.H.-F., C.A.D.-A., and H.V.-C. performed RT-PCR assay. Y.C.K. performed ELISA assays. N.G.-L., A.C.-M., and M.E.V.-S. assisted with ELISA. Y.C.K. and C.L.-C. performed data analysis. Y.C.K. wrote the original manuscript. Y.C.K., C.L.-C., M.E.V.-S., and A.R.-S. reviewed and edited the manuscript. M.E.V.-S., H.V.-C., and A.R.-S. are grant holders.
Funding
A.R.-S. is a Jenner Investigator and an Oxford Martin Fellow. This report is work commissioned by Innovate UK and the UK Department of Health and Social Care (projects No. 972216 and 971557). N.G.-L. and A.C.-M. are recipients of CONACYT fellowships. This work was supported by grants from CONACYT Fonteras de la Ciencia 2015-02-1192, Ciencia Básica SEP-CONACyT 256235, and Fosiss-CONACyT 233697 to H.V.-C. The views expressed in this publication are those of the author(s) and not necessarily those of Innovate UK or the Department of Health and Social Care.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Strauss, J.H.; Strauss, E.G. The alphaviruses: Gene expression, replication, and evolution. Microbiol. Mol. Biol. Rev. 1994, 58, 491–562. [Google Scholar]
- Khan, A.H.; Morita, K.; del Carmen Parquet, M.; Hasebe, F.; Mathenge, E.G.M.; Igarashi, A. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J. Gen. Virol. 2002, 83, 3075–3084. [Google Scholar] [CrossRef] [PubMed]
- Pialoux, G.; Gaüzère, B.-A.; Jauréguiberry, S.; Strobel, M. Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 2007, 7, 319–327. [Google Scholar] [CrossRef]
- Mason, P.J.; Haddow, A.J. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952-53; an additional note on Chikungunya virus isolations and serum antibodies. Trans. R. Soc. Trop. Med. Hyg. 1957, 51, 238–240. [Google Scholar] [CrossRef]
- Bodenmann, P.; Genton, B. Chikungunya: An epidemic in real time. Lancet 2006, 368, 258. [Google Scholar] [CrossRef]
- Simizu, B.; Yamamoto, K.; Hashimoto, K.; Ogata, T. Structural proteins of Chikungunya virus. J. Virol. 1984, 51, 254–258. [Google Scholar]
- Powers, A.M.; Brault, A.C.; Shirako, Y.; Strauss, E.G.; Kang, W.; Strauss, J.H.; Weaver, S.C. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 2001, 75, 10118–10131. [Google Scholar] [CrossRef] [PubMed]
- Higashi, N.; Matsumoto, A.; Tabata, K.; Nagatomo, Y. Electron microscope study of development of Chikungunya virus in green monkey kidney stable (VERO) cells. Virology 1967, 33, 55–69. [Google Scholar] [CrossRef]
- Cheng, R.H.; Kuhn, R.J.; Olson, N.H.; Rossmann, M.G.; Choi, H.K.; Smith, T.J.; Baker, T.S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 1995, 80, 621–630. [Google Scholar] [CrossRef]
- Voss, J.E.; Vaney, M.-C.; Duquerroy, S.; Vonrhein, C.; Girard-Blanc, C.; Crublet, E.; Thompson, A.; Bricogne, G.; Rey, F.A. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 2010, 468, 709–712. [Google Scholar] [CrossRef]
- Leparc-Goffart, I.; Nougairede, A.; Cassadou, S.; Prat, C.; Lamballerie, X. de Chikungunya in the Americas. Lancet 2014, 383, 514. [Google Scholar] [CrossRef]
- Weaver, S.C. Arrival of Chikungunya Virus in the New World: Prospects for Spread and Impact on Public Health. PLoS Negl. Trop. Dis. 2014, 8, e2921. [Google Scholar] [CrossRef]
- Fischer, M.; Staples, J.E. Notes from the field: Chikungunya virus spreads in the Americas-Caribbean and South America 2013–2014. MMWR 2014, 63, 500–501. [Google Scholar]
- Weaver, S.C.; Osorio, J.E.; Livengood, J.A.; Chen, R.; Stinchcomb, D.T. Chikungunya virus and prospects for a vaccine. Expert Rev. Vaccines 2012, 11, 1087–1101. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.; Ravi, V.; Desai, A.; Parida, M.; Powers, A.M.; Johnson, B.W. Utility of IgM ELISA, TaqMan real-time PCR, reverse transcription PCR, and RT-LAMP assay for the diagnosis of Chikungunya fever. J. Med. Virol. 2012, 84, 1771–1778. [Google Scholar] [CrossRef]
- Hasebe, F.; Parquet, M.C.; Pandey, B.D.; Mathenge, E.G.M.; Morita, K.; Balasubramaniam, V.; Saat, Z.; Yusop, A.; Sinniah, M.; Natkunam, S.; et al. Combined detection and genotyping of Chikungunya virus by a specific reverse transcription-polymerase chain reaction. J. Med. Virol. 2002, 67, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Telles, J.-N.; Le Roux, K.; Grivard, P.; Vernet, G.; Michault, A. Evaluation of real-time nucleic acid sequence-based amplification for detection of Chikungunya virus in clinical samples. J. Med. Microbiol. 2009, 58, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.W.; Russell, B.J.; Goodman, C.H. Laboratory Diagnosis of Chikungunya Virus Infections and Commercial Sources for Diagnostic Assays. J. Infect. Dis. 2016, 214, S471–S474. [Google Scholar] [CrossRef]
- Kosasih, H.; Widjaja, S.; Surya, E.; Hadiwijaya, S.H.; Butarbutar, D.P.; Jaya, U.A.; Alisjahbana, B.; Williams, M. Evaluation of two IgM rapid immunochromatographic tests during circulation of Asian lineage Chikungunya virus. Southeast Asian J. Trop. Med. Public Health 2012, 43, 55–61. [Google Scholar] [PubMed]
- Azami, N.A.M.; Salleh, S.A.; Shah, S.A.; Neoh, H.; Othman, Z.; Zakaria, S.Z.S.; Jamal, R. Emergence of chikungunya seropositivity in healthy Malaysian adults residing in outbreak-free locations: Chikungunya seroprevalence results from the Malaysian Cohort. BMC Infect. Dis. 2013, 13, 67. [Google Scholar] [CrossRef]
- Yap, G.; Pok, K.-Y.; Lai, Y.-L.; Hapuarachchi, H.-C.; Chow, A.; Leo, Y.-S.; Tan, L.-K.; Ng, L.-C. Evaluation of Chikungunya Diagnostic Assays: Differences in Sensitivity of Serology Assays in Two Independent Outbreaks. PLoS Negl. Trop. Dis. 2010, 4, e753. [Google Scholar] [CrossRef] [PubMed]
- Rianthavorn, P.; Wuttirattanakowit, N.; Prianantathavorn, K.; Limpaphayom, N.; Theamboonlers, A.; Poovorawan, Y. Evaluation of a rapid assay for detection of IgM antibodies to chikungunya. Southeast Asian J. Trop. Med. Public Health 2010, 41, 92–96. [Google Scholar] [PubMed]
- Pierro, A.; Rossini, G.; Gaibani, P.; Finarelli, A.C.; Moro, M.L.; Landini, M.P.; Sambri, V. Persistence of anti-chikungunya virus-specific antibodies in a cohort of patients followed from the acute phase of infection after the 2007 outbreak in Italy. New Microbes New Infect. 2015, 7, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.P.; Suresh, A.; Vanamail, P.; Sabesan, S.; Krishnamoorthy, K.G.; Mathew, J.; Jose, V.T.; Jambulingam, P. Chikungunya virus outbreak in Kerala, India, 2007: A seroprevalence study. Mem. Inst. Oswaldo Cruz 2011, 106, 912–916. [Google Scholar] [CrossRef]
- Niedrig, M.; Zeller, H.; Schuffenecker, I.; Drosten, C.; Emmerich, P.; Rumer, L.; Donoso-Mantke, O. International diagnostic accuracy study for the serological detection of chikungunya virus infection. Clin. Microbiol. Infect. 2009, 15, 880–884. [Google Scholar] [CrossRef] [PubMed]
- López-Camacho, C.; Chan Kim, Y.; Blight, J.; Lazaro Moreli, M.; Montoya-Diaz, E.; Huiskonen, J.T.; Mareike Kümmerer, B.; Reyes-Sandoval, A. Assessment of Immunogenicity and Neutralisation Efficacy of Viral-Vectored Vaccines Against Chikungunya Virus. Viruses 2019, 11, 322. [Google Scholar] [CrossRef]
- Prat, C.M.; Flusin, O.; Panella, A.; Tenebray, B.; Lanciotti, R.; Leparc-Goffart, I. Evaluation of Commercially Available Serologic Diagnostic Tests for Chikungunya Virus. Emerg. Infect. Dis. 2014, 20, 2129–2132. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Dhanwani, R.; Kumar, J.S.; Rao, P.V.L.; Parida, M. Comparative evaluation of the diagnostic potential of recombinant envelope proteins and native cell culture purified viral antigens of Chikungunya virus. J. Med. Virol. 2014, 86, 1169–1175. [Google Scholar] [CrossRef]
- Hierholzer, J.C.; Killington, R.A.; Stokes, A. Preparation of antigens. In Virology Methods Manual; Elsevier: Amsterdam, The Netherlands, 1996. [Google Scholar]
- Erasmus, J.H.; Needham, J.; Raychaudhuri, S.; Diamond, M.S.; Beasley, D.W.C.; Morkowski, S.; Salje, H.; Salas, I.F.; Kim, D.Y.; Frolov, I.; et al. Utilization of an Eilat Virus-Based Chimera for Serological Detection of Chikungunya Infection. PLoS Negl. Trop. Dis. 2015, 9, e0004119. [Google Scholar] [CrossRef]
- Fumagalli, M.J.; de Souza, W.M.; Espósito, D.L.A.; Silva, A.; Romeiro, M.F.; Martinez, E.Z.; da Fonseca, B.A.L.; Figueiredo, L.T.M. Enzyme-linked immunosorbent assay using recombinant envelope protein 2 antigen for diagnosis of Chikungunya virus. Virol. J. 2018, 15, 112. [Google Scholar] [CrossRef] [PubMed]
- Cho, B.; Jeon, B.-Y.; Kim, J.; Noh, J.; Kim, J.; Park, M.; Park, S. Expression and Evaluation of Chikungunya Virus E1 and E2 Envelope Proteins for Serodiagnosis of Chikungunya Virus Infection. Yonsei Med. J. 2008, 49, 828–835. [Google Scholar] [CrossRef]
- Kumar, P.; Pok, K.-Y.; Tan, L.-K.; Angela, C.; Leo, Y.-S.; Ng, L.-C. Development and evaluation of baculovirus-expressed Chikungunya virus E1 envelope proteins for serodiagnosis of Chikungunya infection. J. Virol. Methods 2014, 206, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Lopez-Camacho, C.; Nettleship, J.E.; Rahman, N.; Hill, M.L.; Silva-Reyes, L.; Ortiz-Martinez, G.; Figueroa-Aguilar, G.; Mar, M.A.; Vivanco-Cid, H.; et al. Optimization of Zika virus envelope protein production for ELISA and correlation of antibody titers with virus neutralization in Mexican patients from an arbovirus endemic region. Virol. J. 2018, 15, 193. [Google Scholar] [CrossRef]
- Nettleship, J.E.; Rahman-Huq, N.; Owens, R.J. The production of glycoproteins by transient expression in Mammalian cells. In High Throughput Protein Expression and Purification; Methods in Molecular Biology; Springer: Berlin, Germany, 2009; pp. 245–263. [Google Scholar]
- Lancaster, C.; Pristatsky, P.; Hoang, V.M.; Casimiro, D.R.; Schwartz, R.M.; Rustandi, R.; Ha, S. Characterization of N-glycosylation profiles from mammalian and insect cell derived chikungunya VLP. J. Chromatogr. B 2016, 1032, 218–223. [Google Scholar] [CrossRef] [PubMed]
- Azami, N.A.M.; Moi, M.L.; Takasaki, T. Neutralization Assay for Chikungunya Virus Infection: Plaque Reduction Neutralization Test. In Chikungunya Virus; Methods in Molecular Biology; Springer: Berlin, Germany, 2016; pp. 273–282. [Google Scholar]
- Weaver, S.C.; Winegar, R.; Manger, I.D.; Forrester, N.L. Alphaviruses: Population genetics and determinants of emergence. Antivir. Res. 2012, 94, 242–257. [Google Scholar] [CrossRef] [PubMed]
- Navarro, J.-C.; Carrera, J.-P.; Liria, J.; Auguste, A.J.; Weaver, S.C. Alphaviruses in Latin America and the introduction of chikungunya virus. In Human Virology in Latin America; Springer: Berlin, Germany, 2017; pp. 169–192. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).