HIV-1 Latency and Latency Reversal: Does Subtype Matter?
Abstract
1. Introduction
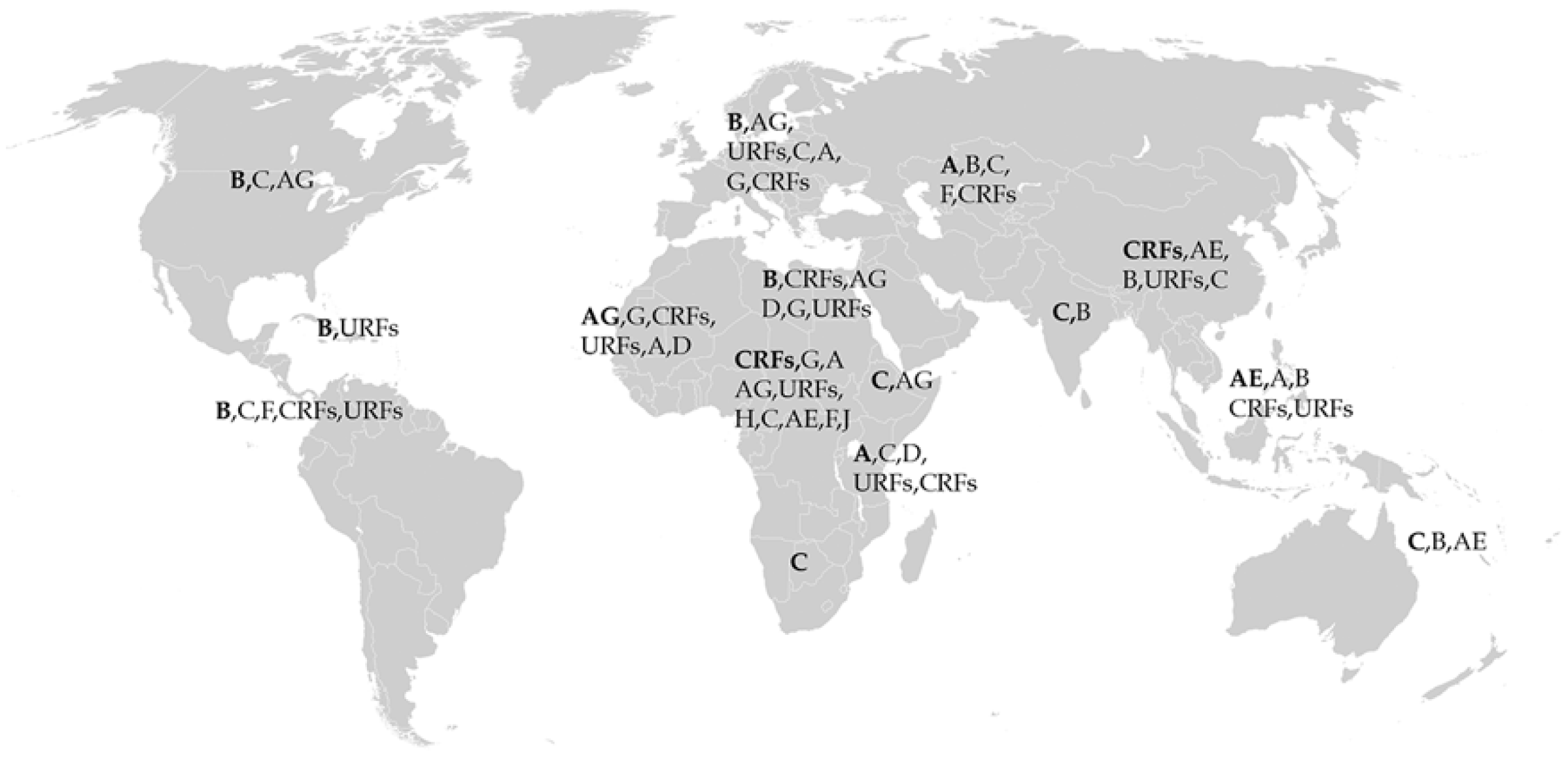
2. Role of Subtype in HIV-1 Pathogenesis
3. HIV-1 Coreceptor Usage and Tropism Switch
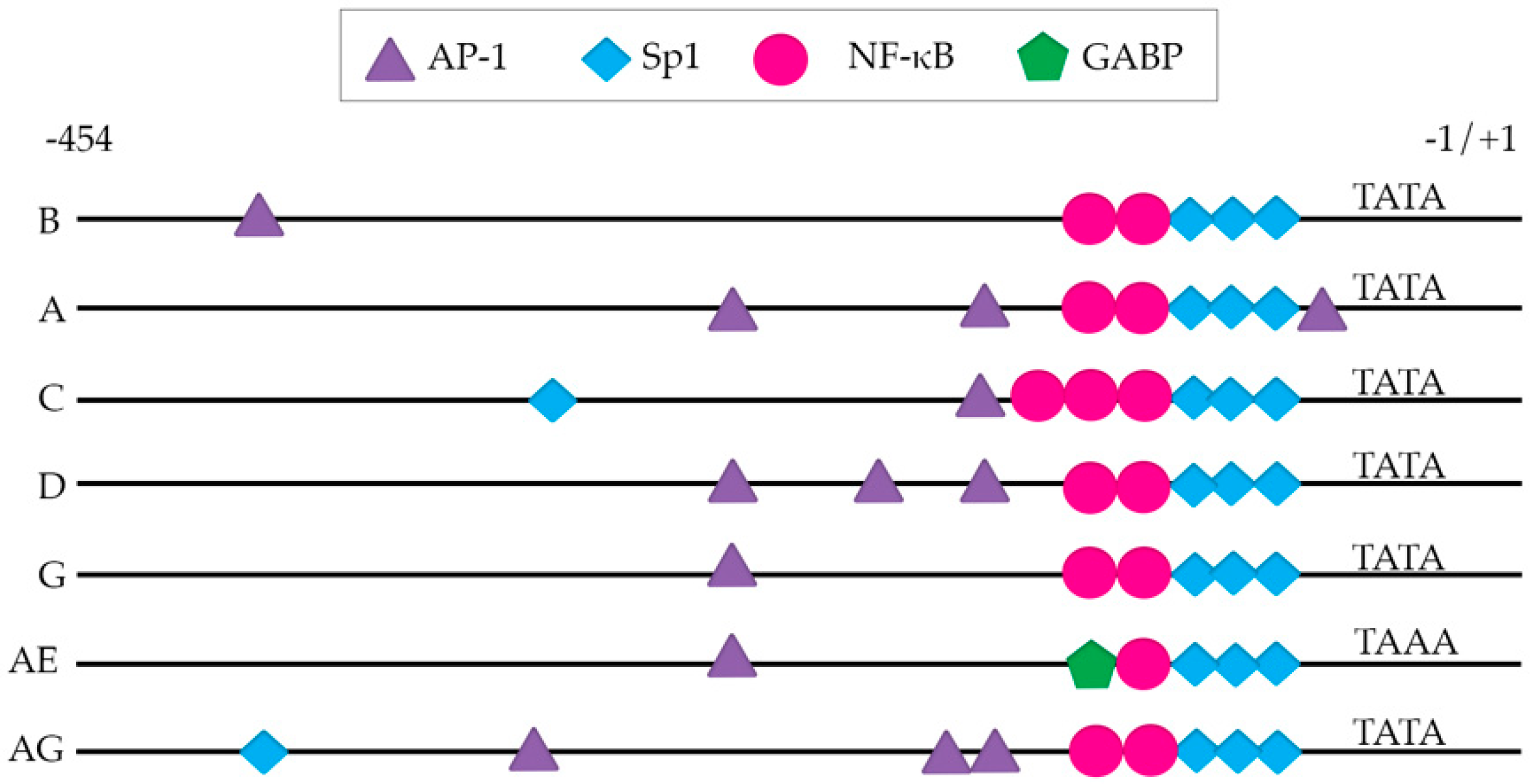
4. The Viral Long Terminal Repeat (LTR)
5. HIV-1 Viral Proteins and Latency
6. The Size of the Latent Reservoir
7. Latency Reversal—The “Shock”
7.1. Protein Kinase C (PKC) Agonists
7.2. Toll-Like Receptor Agonists
7.3. Non-Canonical NF-κB Agonists
7.4. STAT Modulators
7.5. Epigenetic Modifiers and PTEF-b Release Agents
7.6. Enhancing Viral Reactivation with Combination Treatments
8. Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chun, T.W.; Carruth, L.; Finzi, D.; Shen, X.; DiGiuseppe, J.A.; Taylor, H.; Hermankova, M.; Chadwick, K.; Margolick, J.; Quinn, T.C.; et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 1997, 387, 183–188. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D.; Hermankova, M.; Pierson, T.; Carruth, L.M.; Buck, C.; Chaisson, R.E.; Quinn, T.C.; Chadwick, K.; Margolick, J.; Brookmeyer, R.; et al. Identification of a Reservoir for HIV-1 in Patients on HIghly Active Antiretroviral Therapy. Science 1997, 278. [Google Scholar] [CrossRef] [PubMed]
- Siliciano, J.D.; Kajdas, J.; Finzi, D.; Quinn, T.C.; Chadwick, K.; Margolick, J.B.; Kovacs, C.; Gange, S.J.; Siliciano, R.F. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 2003, 9, 727–728. [Google Scholar] [CrossRef] [PubMed]
- Chun, T.W.; Stuyver, L.; Mizell, S.B.; Ehler, L.A.; Mican, J.A.; Baseler, M.; Lloyd, A.L.; Nowak, M.A.; Fauci, A.S. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 1997, 94, 13193–13197. [Google Scholar] [CrossRef]
- Siliciano, R.F.; Greene, W.C. HIV latency. Cold Spring Harb. Perspect. Med. 2011, 1, a007096. [Google Scholar] [CrossRef]
- Arts, E.J.; Hazuda, D.J. HIV-1 antiretroviral drug therapy. Cold Spring Harb. Perspect. Med. 2012, 2, a007161. [Google Scholar] [CrossRef]
- Sierra, S.; Kupfer, B.; Kaiser, R. Basics of the virology of HIV-1 and its replication. J Clin Virol 2005, 34, 233–244. [Google Scholar] [CrossRef]
- Ganor, Y.; Real, F.; Sennepin, A.; Dutertre, C.A.; Prevedel, L.; Xu, L.; Tudor, D.; Charmeteau, B.; Couedel-Courteille, A.; Marion, S.; et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat. Microbiol. 2019, 4, 633–644. [Google Scholar] [CrossRef]
- Chomont, N.; El-Far, M.; Ancuta, P.; Trautmann, L.; Procopio, F.A.; Yassine-Diab, B.; Boucher, G.; Boulassel, M.R.; Ghattas, G.; Brenchley, J.M.; et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 2009, 15, 893–900. [Google Scholar] [CrossRef]
- Brenchley, J.M.; Hill, B.J.; Ambrozak, D.R.; Price, D.A.; Guenaga, F.J.; Casazza, J.P.; Kuruppu, J.; Yazdani, J.; Migueles, S.A.; Connors, M.; et al. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: Implications for HIV pathogenesis. J. Virol. 2004, 78, 1160–1168. [Google Scholar] [CrossRef]
- McNamara, L.A.; Ganesh, J.A.; Collins, K.L. Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-kappaB activation. J. Virol. 2012, 86, 9337–9350. [Google Scholar] [CrossRef] [PubMed]
- Venanzi Rullo, E.; Cannon, L.; Pinzone, M.R.; Ceccarelli, M.; Nunnari, G.; O’Doherty, U. Genetic evidence that Naive T cells can contribute significantly to the HIV intact reservoir: Time to re-evaluate their role. Clin. Infect. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.K.; Yukl, S.A. Tissue reservoirs of HIV. Curr. Opin. HIV AIDS 2016, 11, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Barouch, D.H.; Deeks, S.G. Immunologic strategies for HIV-1 remission and eradication. Science 2014, 345, 169–174. [Google Scholar] [CrossRef]
- Spivak, A.M.; Planelles, V. HIV-1 Eradication: Early Trials (and Tribulations). Trends Mol. Med. 2016, 22, 10–27. [Google Scholar] [CrossRef]
- Macedo, A.B.; Novis, C.L.; Bosque, A. Targeting Cellular and Tissue HIV Reservoirs With Toll-Like Receptor Agonists. Front. Immunol. 2019, 10, 2450. [Google Scholar] [CrossRef]
- Dahabieh, M.S.; Battivelli, E.; Verdin, E. Understanding HIV latency: The road to an HIV cure. Annu. Rev. Med. 2015, 66, 407–421. [Google Scholar] [CrossRef]
- Hemelaar, J. Implications of HIV diversity for the HIV-1 pandemic. J. Infect. 2013, 66, 391–400. [Google Scholar] [CrossRef]
- Hsu, D.C.; O’Connell, R.J. Progress in HIV vaccine development. Hum. Vaccin. Immunother. 2017, 13, 1018–1030. [Google Scholar] [CrossRef]
- Taylor, B.S.; Sobieszczyk, M.E.; McCutchan, F.E.; Hammer, S.M. The Challenge of HIV-1 Subtype Diversity. N. Engl. J. Med. 2008, 358, 1590–1602. [Google Scholar] [CrossRef]
- Hemelaar, J.; Gouws, E.; Ghys, P.D.; Osmanov, S.; WHO-UNAIDS Network for HIV Isolation and Characterisation. Global trends in molecular epidemiology of HIV-1 during 2000-2007. AIDS 2011, 25, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, J. The origin and diversity of the HIV-1 pandemic. Trends Mol. Med. 2012, 18, 182–192. [Google Scholar] [CrossRef] [PubMed]
- Hemelaar, J.; Gouws, E.; Ghys, P.D.; Osmanov, S. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 2006, 20, W13–W23. [Google Scholar] [CrossRef] [PubMed]
- Pyne, M.T.; Hackett, J., Jr.; Holzmayer, V.; Hillyard, D.R. Large-scale analysis of the prevalence and geographic distribution of HIV-1 non-B variants in the United States. J. Clin. Microbiol. 2013, 51, 2662–2669. [Google Scholar] [CrossRef]
- Oster, A.M.; Switzer, W.M.; Hernandez, A.L.; Saduvala, N.; Wertheim, J.O.; Nwangwu-Ike, N.; Ocfemia, M.C.; Campbell, E.; Hall, H.I. Increasing HIV-1 subtype diversity in seven states, United States, 2006-2013. Ann. Epidemiol. 2017, 27, 244–251 e241. [Google Scholar] [CrossRef] [PubMed]
- Delgado, E.; Benito, S.; Montero, V.; Cuevas, M.T.; Fernandez-Garcia, A.; Sanchez-Martinez, M.; Garcia-Bodas, E.; Diez-Fuertes, F.; Gil, H.; Canada, J.; et al. Diverse Large HIV-1 Non-subtype B Clusters Are Spreading Among Men Who Have Sex With Men in Spain. Front. Microbiol. 2019, 10, 655. [Google Scholar] [CrossRef]
- Yamaguchi, J.; McArthur, C.; Vallari, A.; Sthreshley, L.; Cloherty, G.A.; Berg, M.G.; Rodgers, M.A. Complete genome sequence of CG-0018a-01 establishes HIV-1 subtype L. J. Acquir. Immune Defic. Syndr. 2019. [Google Scholar] [CrossRef]
- Alaeus, A.; Lidman, K.; Bjorkman, A.; Giesecke, J.; Albert, J. Similar rate of disease progression among individuals infected with HIV-1 genetic subtypes A-D. AIDS 1999, 13, 901–907. [Google Scholar] [CrossRef]
- Amornkul, P.N.; Tansuphasawadikul, S.; Limpakarnjanarat, K.; Likanonsakul, S.; Young, N.; Eampokalap, B.; Kaewkungwal, J.; Naiwatanakul, T.; Von Bargen, J.; Hu, D.J.; et al. Clinical disease associated with HIV-1 subtype B’ and E infection among 2104 patients in Thailand. AIDS 1999, 13, 1963–1969. [Google Scholar] [CrossRef]
- Galai, N.; Kalinkovich, A.; Burstein, R.; Vlahov, D.; Bentwich, Z. African HIV-1 subtype C and rate of progression among Ethiopian immigrants in Israel. Lancet 1997, 349, 180–181. [Google Scholar] [CrossRef]
- Kiwanuka, N.; Robb, M.; Laeyendecker, O.; Kigozi, G.; Wabwire-Mangen, F.; Makumbi, F.E.; Nalugoda, F.; Kagaayi, J.; Eller, M.; Eller, L.A.; et al. HIV-1 viral subtype differences in the rate of CD4+ T-cell decline among HIV seroincident antiretroviral naive persons in Rakai district, Uganda. J. Acquir. Immune. Defic. Syndr. 2010, 54, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Baeten, J.M.; Chohan, B.; Lavreys, L.; Chohan, V.; McClelland, R.S.; Certain, L.; Mandaliya, K.; Jaoko, W.; Overbaugh, J. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J. Infect. Dis. 2007, 195, 1177–1180. [Google Scholar] [CrossRef]
- Kaleebu, P.; French, N.; Mahe, C.; Yirrell, D.; Watera, C.; Lyagoba, F.; Nakiyingi, J.; Rutebemberwa, A.; Morgan, D.; Weber, J.; et al. Effect of human immunodeficiency virus (HIV) type 1 envelope subtypes A and D on disease progression in a large cohort of HIV-1-positive persons in Uganda. J. Infect. Dis. 2002, 185, 1244–1250. [Google Scholar] [CrossRef] [PubMed]
- Ssemwanga, D.; Nsubuga, R.N.; Mayanja, B.N.; Lyagoba, F.; Magambo, B.; Yirrell, D.; Van der Paal, L.; Grosskurth, H.; Kaleebu, P. Effect of HIV-1 subtypes on disease progression in rural Uganda: A prospective clinical cohort study. PLoS ONE 2013, 8, e71768. [Google Scholar] [CrossRef] [PubMed]
- Vasan, A.; Renjifo, B.; Hertzmark, E.; Chaplin, B.; Msamanga, G.; Essex, M.; Fawzi, W.; Hunter, D. Different rates of disease progression of HIV type 1 infection in Tanzania based on infecting subtype. Clin. Infect. Dis. 2006, 42, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Venner, C.M.; Nankya, I.; Kyeyune, F.; Demers, K.; Kwok, C.; Chen, P.L.; Rwambuya, S.; Munjoma, M.; Chipato, T.; Byamugisha, J.; et al. Infecting HIV-1 Subtype Predicts Disease Progression in Women of Sub-Saharan Africa. EBioMedicine 2016, 13, 305–314. [Google Scholar] [CrossRef]
- Amornkul, P.N.; Karita, E.; Kamali, A.; Rida, W.N.; Sanders, E.J.; Lakhi, S.; Price, M.A.; Kilembe, W.; Cormier, E.; Anzala, O.; et al. Disease progression by infecting HIV-1 subtype in a seroconverter cohort in sub-Saharan Africa. AIDS 2013, 27, 2775–2786. [Google Scholar] [CrossRef]
- Kanki, P.J.; Hamel, D.J.; Sankale, J.L.; Hsieh, C.; Thior, I.; Barin, F.; Woodcock, S.A.; Gueye-Ndiaye, A.; Zhang, E.; Montano, M.; et al. Human immunodeficiency virus type 1 subtypes differ in disease progression. J. Infect. Dis. 1999, 179, 68–73. [Google Scholar] [CrossRef]
- Berger, E.A.; Murphy, P.M.; Farber, J.M. Chemokine receptors as HIV-1 coreceptors: Roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 1999, 17, 657–700. [Google Scholar] [CrossRef]
- Connor, R.I.; Sheridan, K.E.; Ceradini, D.; Choe, S.; Landau, N.R. Change in coreceptor use correlates with disease progression in HIV-1--infected individuals. J. Exp. Med. 1997, 185, 621–628. [Google Scholar] [CrossRef]
- Moore, J.P.; Kitchen, S.G.; Pugach, P.; Zack, J.A. The CCR5 and CXCR4 Coreceptors— Central to Understanding the Transmission and Pathogenesis of Human Immunodeficiency VIrus Type 1 Infection. AIDS Res. Hum. Retroviruses 2004, 20, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Esbjornsson, J.; Mansson, F.; Martinez-Arias, W.; Vincic, E.; Biague, A.J.; da Silva, Z.J.; Fenyo, E.M.; Norrgren, H.; Medstrand, P. Frequent CXCR4 tropism of HIV-1 subtype A and CRF02_AG during late-stage disease--indication of an evolving epidemic in West Africa. Retrovirology 2010, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Beitari, S.; Wang, Y.; Liu, S.L.; Liang, C. HIV-1 Envelope Glycoprotein at the Interface of Host Restriction and Virus Evasion. Viruses 2019, 11, 311. [Google Scholar] [CrossRef] [PubMed]
- Lynch, R.M.; Shen, T.; Gnanakaran, S.; Derdeyn, C.A. Appreciating HIV type 1 diversity: Subtype differences in Env. AIDS Res. Hum. Retroviruses 2009, 25, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.C.; Tang, M.; Zhang, M.Y.; Majeed, S.; Montabana, E.; Stanfield, R.L.; Dimitrov, D.S.; Korber, B.; Sodroski, J.; Wilson, I.A.; et al. Structure of a V3-containing HIV-1 gp120 core. Science 2005, 310, 1025–1028. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.S.; Boyle, T.J.; Lyerly, H.K.; Cullen, B.R. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 1991, 253, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, N.G.; Seillier-Moiseiwitsch, F.; Ahn, J.; Walker, J.M.; Swanstrom, R. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 2002, 76, 3852–3864. [Google Scholar] [CrossRef]
- Alaeus, A. Significance of HIV-1 genetic subtypes. Scand. J. Infect. Dis. 2000, 32, 455–463. [Google Scholar] [CrossRef]
- Tscherning, C.; Alaeus, A.; Fredriksson, R.; Bjorndal, A.; Deng, H.; Littman, D.R.; Fenyo, E.M.; Albert, J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 1998, 241, 181–188. [Google Scholar] [CrossRef]
- Coetzer, M.; Cilliers, T.; Ping, L.H.; Swanstrom, R.; Morris, L. Genetic characteristics of the V3 region associated with CXCR4 usage in HIV-1 subtype C isolates. Virology 2006, 356, 95–105. [Google Scholar] [CrossRef]
- Felsovalyi, K.; Nadas, A.; Zolla-Pazner, S.; Cardozo, T. Distinct sequence patterns characterize the V3 region of HIV type 1 gp120 from subtypes A and C. AIDS Res. Hum. Retroviruses 2006, 22, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Eshleman, S.H.; Toma, J.; Fransen, S.; Stawiski, E.; Paxinos, E.E.; Whitcomb, J.M.; Young, A.M.; Donnell, D.; Mmiro, F.; et al. Coreceptor tropism in human immunodeficiency virus type 1 subtype D: High prevalence of CXCR4 tropism and heterogeneous composition of viral populations. J. Virol. 2007, 81, 7885–7893. [Google Scholar] [CrossRef] [PubMed]
- Ataher, Q.; Portsmouth, S.; Napolitano, L.A.; Eng, S.; Greenacre, A.; Kambugu, A.; Wood, R.; Badal-Faesen, S.; Tressler, R. The epidemiology and clinical correlates of HIV-1 co-receptor tropism in non-subtype B infections from India, Uganda and South Africa. J. Int. AIDS Soc. 2012, 15, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Huang, Y.; He, T.; Cao, Y.; Ho, D.D. HIV-1 subtype and second-receptor use. Nature 1996, 383, 768. [Google Scholar] [CrossRef]
- Bednar, M.M.; Hauser, B.M.; Zhou, S.; Jacobson, J.M.; Eron, J.J., Jr.; Frank, I.; Swanstrom, R. Diversity and Tropism of HIV-1 Rebound Virus Populations in Plasma Level After Treatment Discontinuation. J. Infect. Dis. 2016, 214, 403–407. [Google Scholar] [CrossRef]
- Pardons, M.; Baxter, A.E.; Massanella, M.; Pagliuzza, A.; Fromentin, R.; Dufour, C.; Leyre, L.; Routy, J.P.; Kaufmann, D.E.; Chomont, N. Single-cell characterization and quantification of translation-competent viral reservoirs in treated and untreated HIV infection. PLoS Pathog. 2019, 15, e1007619. [Google Scholar] [CrossRef] [PubMed]
- Grau-Exposito, J.; Luque-Ballesteros, L.; Navarro, J.; Curran, A.; Burgos, J.; Ribera, E.; Torrella, A.; Planas, B.; Badia, R.; Martin-Castillo, M.; et al. Latency reversal agents affect differently the latent reservoir present in distinct CD4+ T subpopulations. PLoS Pathog. 2019, 15, e1007991. [Google Scholar] [CrossRef]
- Pereira, L.A.; Bentley, K.; Peeters, A.; Churchill, M.J.; Deacon, N.J. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 2000, 28, 663–668. [Google Scholar] [CrossRef]
- Kinoshita, S.; Su, L.; Amano, M.; Timmerman, L.A.; Kaneshima, H.; Nolan, G.P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 1997, 6, 235–244. [Google Scholar] [CrossRef]
- Bosque, A.; Planelles, V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 2009, 113, 58–65. [Google Scholar] [CrossRef]
- Williams, S.A.; Kwon, H.; Chen, L.F.; Greene, W.C. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 2007, 81, 6043–6056. [Google Scholar] [CrossRef] [PubMed]
- Taube, R.; Peterlin, M. Lost in transcription: Molecular mechanisms that control HIV latency. Viruses 2013, 5, 902–927. [Google Scholar] [CrossRef] [PubMed]
- Bosque, A.; Nilson, K.A.; Macedo, A.B.; Spivak, A.M.; Archin, N.M.; Van Wagoner, R.M.; Martins, L.J.; Novis, C.L.; Szaniawski, M.A.; Ireland, C.M.; et al. Benzotriazoles Reactivate Latent HIV-1 through Inactivation of STAT5 SUMOylation. Cell Rep. 2017, 18, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Pandelo Jose, D.; Bartholomeeusen, K.; da Cunha, R.D.; Abreu, C.M.; Glinski, J.; da Costa, T.B.; Bacchi Rabay, A.F.; Pianowski Filho, L.F.; Dudycz, L.W.; Ranga, U.; et al. Reactivation of latent HIV-1 by new semi-synthetic ingenol esters. Virology 2014, 462–463, 328–339. [Google Scholar] [CrossRef]
- Crotti, A.; Chiara, G.D.; Ghezzi, S.; Lupo, R.; Jeeninga, R.E.; Liboi, E.; Lievens, P.M.; Vicenzi, E.; Bovolenta, C.; Berkhout, B.; et al. Heterogeneity of signal transducer and activator of transcription binding sites in the long-terminal repeats of distinct HIV-1 subtypes. Open Virol. J. 2007, 1, 26–32. [Google Scholar] [CrossRef]
- Jeeninga, R.E.; Hoogenkamp, M.; Armand-Ugon, M.; de Baar, M.; Verhoef, K.; Berkhout, B. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 2000, 74, 3740–3751. [Google Scholar] [CrossRef]
- Rodriguez, M.A.; Shen, C.; Ratner, D.; Paranjape, R.S.; Kulkarni, S.S.; Chatterjee, R.; Gupta, P. Genetic and functional characterization of the LTR of HIV-1 subtypes A and C circulating in India. AIDS Res. Hum. Retroviruses 2007, 23, 1428–1433. [Google Scholar] [CrossRef]
- Roof, P.; Ricci, M.; Genin, P.; Montano, M.A.; Essex, M.; Wainberg, M.A.; Gatignol, A.; Hiscott, J. Differential regulation of HIV-1 clade-specific B, C, and E long terminal repeats by NF-kappaB and the Tat transactivator. Virology 2002, 296, 77–83. [Google Scholar] [CrossRef][Green Version]
- van Opijnen, T.; Jeeninga, R.E.; Boerlijst, M.C.; Pollakis, G.P.; Zetterberg, V.; Salminen, M.; Berkhout, B. Human immunodeficiency virus type 1 subtypes have a distinct long terminal repeat that determines the replication rate in a host-cell-specific manner. J. Virol. 2004, 78, 3675–3683. [Google Scholar] [CrossRef]
- Montano, M.A.; Novitsky, V.A.; Blackard, J.T.; Cho, N.L.; Katzenstein, D.A.; Essex, M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J. Virol. 1997, 71, 8657–8665. [Google Scholar]
- Burnett, J.C.; Lim, K.I.; Calafi, A.; Rossi, J.J.; Schaffer, D.V.; Arkin, A.P. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 2010, 84, 5958–5974. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Li, C.; Sang, F.; Li, Q.; Jiang, Z.Q.; Xu, L.R.; Guo, H.J.; Zhang, C.; Wang, J.H. The variances of Sp1 and NF-kappaB elements correlate with the greater capacity of Chinese HIV-1 B′-LTR for driving gene expression. Sci. Rep. 2016, 6, 34532. [Google Scholar] [CrossRef] [PubMed]
- Bachu, M.; Yalla, S.; Asokan, M.; Verma, A.; Neogi, U.; Sharma, S.; Murali, R.V.; Mukthey, A.B.; Bhatt, R.; Chatterjee, S.; et al. Multiple NF-kappaB sites in HIV-1 subtype C long terminal repeat confer superior magnitude of transcription and thereby the enhanced viral predominance. J. Biol. Chem. 2012, 287, 44714–44735. [Google Scholar] [CrossRef] [PubMed]
- Verhoef, K.; Sanders, R.W.; Fontaine, V.; Kitajima, S.; Berkhout, B. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-kappaB enhancer element into a GABP binding site. J. Virol. 1999, 73, 1331–1340. [Google Scholar] [PubMed]
- Williams, S.A.; Chen, L.F.; Kwon, H.; Ruiz-Jarabo, C.M.; Verdin, E.; Greene, W.C. NF-κB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional intiation. EMBO J. 2006, 24, 139–149. [Google Scholar] [CrossRef] [PubMed]
- van der Sluis, R.M.; Pollakis, G.; van Gerven, M.L.; Berkhout, B.; Jeeninga, R.E. Latency profiles of full length HIV-1 molecular clone variants with a subtype specific promoter. Retrovirology 2011, 8, 73. [Google Scholar] [CrossRef]
- Duverger, A.; Wolschendorf, F.; Zhang, M.; Wagner, F.; Hatcher, B.; Jones, J.; Cron, R.Q.; van der Sluis, R.M.; Jeeninga, R.E.; Berkhout, B.; et al. An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J. Virol. 2013, 87, 2264–2277. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Ooms, M.; Simon, V.; Sadowski, I. A doubly fluorescent HIV-1 reporter shows that the majority of integrated HIV-1 is latent shortly after infection. J. Virol. 2013, 87, 4716–4727. [Google Scholar] [CrossRef] [PubMed]
- Maldarelli, F.; Wu, X.; Su, L.; Simonetti, F.R.; Shao, W.; Hill, S.; Spindler, J.; Ferris, A.L.; Mellors, J.W.; Kearney, M.F.; et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 2014, 345, 179–183. [Google Scholar] [CrossRef]
- Wagner, T.A.; McLaughlin, S.; Garg, K.; Cheung, C.Y.; Larsen, B.B.; Styrchak, S.; Huang, H.C.; Edlefsen, P.T.; Mullins, J.I.; Frenkel, L.M. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 2014, 345, 570–573. [Google Scholar] [CrossRef]
- Garrido, C.; Geretti, A.M.; Zahonero, N.; Booth, C.; Strang, A.; Soriano, V.; De Mendoza, C. Integrase variability and susceptibility to HIV integrase inhibitors: Impact of subtypes, antiretroviral experience and duration of HIV infection. J Antimicrob. Chemother. 2010, 65, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Spira, S. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 2003, 51, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Ceccherini-Silberstein, F.; Malet, I.; D’Arrigo, R.; Antinori, A.; Marcelin, A.G.; Perno, C.F. Characterization and structural analysis of HIV-1 integrase conservation. AIDS Rev. 2009, 11, 17–29. [Google Scholar] [PubMed]
- Demeulemeester, J.; Vets, S.; Schrijvers, R.; Madlala, P.; De Maeyer, M.; De Rijck, J.; Ndung’u, T.; Debyser, Z.; Gijsbers, R. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host. Microbe 2014, 16, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.D.; Young, J.A. HIV-1: Fifteen proteins and an RNA. Annu. Rev. Biochem. 1998, 67, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Mehle, A.; Strack, B.; Ancuta, P.; Zhang, C.; McPike, M.; Gabuzda, D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004, 279, 7792–7798. [Google Scholar] [CrossRef]
- Lisovsky, I.; Schader, S.M.; Sloan, R.D.; Oliveira, M.; Coutsinos, D.; Bernard, N.F.; Wainberg, M.A. HIV-1 subtype variability in Vif derived from molecular clones affects APOBEC3G-mediated host restriction. Intervirology 2013, 56, 258–264. [Google Scholar] [CrossRef]
- Pollack, R.A.; Jones, R.B.; Pertea, M.; Bruner, K.M.; Martin, A.R.; Thomas, A.S.; Capoferri, A.A.; Beg, S.A.; Huang, S.H.; Karandish, S.; et al. Defective HIV-1 Proviruses Are Expressed and Can Be Recognized by Cytotoxic T Lymphocytes, which Shape the Proviral Landscape. Cell Host. Microbe 2017, 21, 494–506 e494. [Google Scholar] [CrossRef]
- Romani, B.; Kamali Jamil, R.; Hamidi-Fard, M.; Rahimi, P.; Momen, S.B.; Aghasadeghi, M.R.; Allahbakhshi, E. HIV-1 Vpr reactivates latent HIV-1 provirus by inducing depletion of class I HDACs on chromatin. Sci. Rep. 2016, 6, 31924. [Google Scholar] [CrossRef]
- Shen, A.; Siliciano, J.D.; Pierson, T.C.; Buck, C.B.; Siliciano, R.F. Establishment of latent HIV-1 infection of resting CD4(+) T lymphocytes does not require inactivation of Vpr. Virology 2000, 278, 227–233. [Google Scholar] [CrossRef]
- Bano, A.S.; Gupta, N.; Sood, V.; Banerjea, A.C. Vpr from HIV-1 subtypes B and C exhibit significant differences in their ability to transactivate LTR-mediated gene expression and also in their ability to promote apoptotic DNA ladder formation. Aids 2007, 21, 1832–1834. [Google Scholar] [CrossRef] [PubMed]
- Lindwasser, O.W.; Chaudhuri, R.; Bonifacino, J.S. Mechanisms of CD4 downregulation by the Nef and Vpu proteins of primate immunodeficiency viruses. Curr. Mol. Med. 2007, 7, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Langer, S.; Hammer, C.; Hopfensperger, K.; Klein, L.; Hotter, D.; De Jesus, P.D.; Herbert, K.M.; Pache, L.; Smith, N.; van der Merwe, J.A.; et al. HIV-1 Vpu is a potent transcriptional suppressor of NF-kappaB-elicited antiviral immune responses. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Jubier-Maurin, V.; Saragosti, S.; Perret, J.L.; Mpoudi, E.; Esu-Williams, E.; Mulanga, C.; Liegeois, F.; Ekwalanga, M.; Delaporte, E.; Peeters, M. Genetic characterization of the nef gene from human immunodeficiency virus type 1 group M strains representing genetic subtypes A, B, C, E, F, G, and H. AIDS Res. Hum. Retroviruses 1999, 15, 23–32. [Google Scholar] [CrossRef]
- Mann, J.K.; Byakwaga, H.; Kuang, X.T.; Le, A.Q.; Brumme, C.J.; Mwimanzi, P.; Omarjee, S.; Martin, E.; Lee, G.Q.; Baraki, B.; et al. Ability fo HIV-1 Nef to downregulate CD4 and HLA class I differs among viral subtypes. Retrovirology 2013, 10. [Google Scholar] [CrossRef]
- Mann, J.K.; Omarjee, S.; Khumalo, P.; Ndung’u, T. Genetic determinants of Nef-mediated CD4 and HLA class I down-regulation differences between HIV-1 subtypes B and C. Virol. J. 2015, 12, 200. [Google Scholar] [CrossRef]
- Fujinaga, K.; Zhong, Q.; Nakaya, T.; Kameoka, M.; Meguro, T.; Yamada, K.; Ikuta, K. Extracellular Nef protein regulates productive HIV-1 infection from latency. J. Immunol. 1995, 155, 5289–5298. [Google Scholar]
- Wolf, D.; Witte, V.; Clark, P.; Blume, K.; Lichtenheld, M.G.; Baur, A.S. HIV Nef enhances Tat-mediated viral transcription through a hnRNP-K-nucleated signaling complex. Cell Host. Microbe 2008, 4, 398–408. [Google Scholar] [CrossRef]
- Levy, D.N.; Refaeli, Y.; Weiner, D.B. Extracellular Vpr protein increases cellular permissiveness to human immunodeficiency virus replication and reactivates virus from latency. J. Virol. 1995, 69, 1243–1252. [Google Scholar]
- Mousseau, G.; Aneja, R.; Clementz, M.A.; Mediouni, S.; Lima, N.S.; Haregot, A.; Kessing, C.F.; Jablonski, J.A.; Thenin-Houssier, S.; Nagarsheth, N.; et al. Resistance to the Tat Inhibitor Didehydro-Cortistatin A Is Mediated by Heightened Basal HIV-1 Transcription. MBio 2019, 10. [Google Scholar] [CrossRef]
- Mousseau, G.; Clementz, M.A.; Bakeman, W.N.; Nagarsheth, N.; Cameron, M.; Shi, J.; Baran, P.; Fromentin, R.; Chomont, N.; Valente, S.T. An analog of the natural steroidal alkaloid cortistatin A potently suppresses Tat-dependent HIV transcription. Cell Host. Microbe 2012, 12, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Mousseau, G.; Kessing, C.F.; Fromentin, R.; Trautmann, L.; Chomont, N.; Valente, S.T. The Tat Inhibitor Didehydro-Cortistatin A Prevents HIV-1 Reactivation from Latency. MBio 2015, 6, e00465. [Google Scholar] [CrossRef] [PubMed]
- Marsden, M.D.; Burke, B.P.; Zack, J.A. HIV latency is influenced by regions of the viral genome outside of the long terminal repeats and regulatory genes. Virology 2011, 417, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.T.; Allen, E.E.; Farmer, A.D.; Myers, G.L. Heterogeneity of HIV-1 and HIV-2. AIDS 1995, 9 (Suppl. A), S5–S18. [Google Scholar]
- Iordanskly, S.; Waltke, M.; Feng, Y.; Wood, C. Subtype-associated differences in HIV-1 reverse transcription affect the viral replication. Retrovirology 2010, 7, 85. [Google Scholar]
- Prodger, J.L.; Lai, J.; Reynolds, S.J.; Keruly, J.C.; Moore, R.D.; Kasule, J.; Kityamuweesi, T.; Buule, P.; Serwadda, D.; Nason, M.; et al. Reduced Frequency of Cells Latently Infected With Replication-Competent Human Immunodeficiency Virus-1 in Virally Suppressed Individuals Living in Rakai, Uganda. Clin. Infect. Dis. 2017, 65, 1308–1315. [Google Scholar] [CrossRef]
- Omondi, F.H.; Chandrarathna, S.; Mujib, S.; Brumme, C.J.; Jin, S.W.; Sudderuddin, H.; Miller, R.L.; Rahimi, A.; Laeyendecker, O.; Bonner, P.; et al. HIV Subtype and Nef-Mediated Immune Evasion Function Correlate with Viral Reservoir Size in Early-Treated Individuals. J. Virol. 2019, 93. [Google Scholar] [CrossRef]
- Bachmann, N.; von Siebenthal, C.; Vongrad, V.; Turk, T.; Neumann, K.; Beerenwinkel, N.; Bogojeska, J.; Fellay, J.; Roth, V.; Kok, Y.L.; et al. Determinants of HIV-1 reservoir size and long-term dynamics during suppressive ART. Nat. Commun. 2019, 10, 3193. [Google Scholar] [CrossRef]
- Ananworanich, J.; Dube, K.; Chomont, N. How does the timing of antiretroviral therapy initiation in acute infection affect HIV reservoirs? Curr. Opin. HIV AIDS 2015, 10, 18–28. [Google Scholar] [CrossRef]
- Archin, N.M.; Vaidya, N.K.; Kuruc, J.D.; Liberty, A.L.; Wiegand, A.; Kearney, M.F.; Cohen, M.S.; Coffin, J.M.; Bosch, R.J.; Gay, C.L.; et al. Immediate antiviral therapy appears to restrict resting CD4+ cell HIV-1 infection without accelerating the decay of latent infection. Proc. Natl. Acad. Sci. USA 2012, 109, 9523–9528. [Google Scholar] [CrossRef]
- Eriksson, S.; Graf, E.H.; Dahl, V.; Strain, M.C.; Yukl, S.A.; Lysenko, E.S.; Bosch, R.J.; Lai, J.; Chioma, S.; Emad, F.; et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013, 9, e1003174. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.C.; Shan, L.; Hosmane, N.N.; Wang, J.; Laskey, S.B.; Rosenbloom, D.I.; Lai, J.; Blankson, J.N.; Siliciano, J.D.; Siliciano, R.F. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013, 155, 540–551. [Google Scholar] [CrossRef] [PubMed]
- Bruner, K.M.; Murray, A.J.; Pollack, R.A.; Soliman, M.G.; Laskey, S.B.; Capoferri, A.A.; Lai, J.; Strain, M.C.; Lada, S.M.; Hoh, R.; et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat. Med. 2016, 22, 1043–1049. [Google Scholar] [CrossRef] [PubMed]
- Yue, F.Y.; Cohen, J.C.; Ho, M.; Rahman, A.; Liu, J.; Mujib, S.; Saiyed, A.; Hundal, S.; Khozin, A.; Bonner, P.; et al. HIV-Specific Granzyme B-Secreting but Not Gamma Interferon-Secreting T Cells Are Associated with Reduced Viral Reservoirs in Early HIV Infection. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Buzón, M.J.; Yang, Y.; Ouyang, Z.; Sun, H.; Seiss, K.; Rogich, J.; Le Gall, S.; Pereyra, F.; Rosenberg, E.S.; Yu, X.G.; et al. Susceptibility to CD8 T-cell-mediated killing influences the reservoir of latently HIV-1-infected CD4 T cells. J. Acquir. Immune. Defic. Syndr. 2014, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Vabret, N.; Bailly-Bechet, M.; Najburg, V.; Muller-Trutwin, M.; Verrier, B.; Tangy, F. The biased nucleotide composition of HIV-1 triggers type I interferon response and correlates with subtype D increased pathogenicity. PLoS ONE 2012, 7, e33502. [Google Scholar] [CrossRef]
- Kiguoya, M.W.; Mann, J.K.; Chopera, D.; Gounder, K.; Lee, G.Q.; Hunt, P.W.; Martin, J.N.; Ball, T.B.; Kimani, J.; Brumme, Z.L.; et al. Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Naidoo, L.; Mzobe, Z.; Jin, S.W.; Rajkoomar, E.; Reddy, T.; Brockman, M.A.; Brumme, Z.L.; Ndung’u, T.; Mann, J.K. Nef-mediated inhibition of NFAT following TCR stimulation differs between HIV-1 subtypes. Virology 2019, 531, 192–202. [Google Scholar] [CrossRef]
- Ojwach, D.B.A.; MacMillan, D.; Reddy, T.; Novitsky, V.; Brumme, Z.L.; Brockman, M.A.; Ndung’u, T.; Mann, J.K. Pol-Driven Replicative Capacity Impacts Disease Progression in HIV-1 Subtype C Infection. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Kim, Y.; Anderson, J.L.; Lewin, S.R. Getting the “Kill” into “Shock and Kill”: Strategies to Eliminate Latent HIV. Cell Host. Microbe 2018, 23, 14–26. [Google Scholar] [CrossRef]
- Spivak, A.M.; Planelles, V. Novel Latency Reversal Agents for HIV-1 Cure. Annu. Rev. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Margolis, D.M.; Archin, N.M. Proviral Latency, Persistent Human Immunodeficiency Virus Infection, and the Development of Latency Reversing Agents. J. Infect. Dis. 2017, 215, S111–S118. [Google Scholar] [CrossRef]
- van der Sluis, R.M.; Derking, R.; Breidel, S.; Speijer, D.; Berkhout, B.; Jeeninga, R.E. Interplay between viral Tat protein and c-Jun transcription factor in controlling LTR promoter activity in different human immunodeficiency virus type I subtypes. J. Gen. Virol. 2014, 95, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.; Rajagopalan, P.; Lotke, R.; Varghese, R.; Selvam, D.; Kundu, T.K.; Ranga, U. Functional Incompatibility between the Generic NF-kappaB Motif and a Subtype-Specific Sp1III Element Drives the Formation of the HIV-1 Subtype C Viral Promoter. J. Virol. 2016, 90, 7046–7065. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Dandekar, S. Targeting NF-kappaB signaling with protein kinase C agonists as an emerging strategy for combating HIV latency. AIDS Res. Hum. Retroviruses 2015, 31, 4–12. [Google Scholar] [CrossRef]
- Christian, F.; Smith, E.L.; Carmody, R.J. The Regulation of NF-kappaB Subunits by Phosphorylation. Cells 2016, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Hiscott, J.; Kwon, H.; Génin, P. Hostile takeovers: Viral appropriation of the NF-κB pathway. J. Clin. Investig. 2001, 107, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kulkosky, J.; Culnan, D.M.; Roman, J.; Dornadula, G.; Schnell, M.; Boyd, M.R.; Pomerantz, R.J. Prostratin: Activation of latent HIV-1 expression suggests a potential inductive adjuvant therapy for HAART. Blood 2001, 98, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.A.; Chen, L.F.; Kwon, H.; Fenard, D.; Bisgrove, D.; Verdin, E.; Greene, W.C. Prostratin antagonizes HIV latency by activating NF-kappaB. J. Biol. Chem. 2004, 279, 42008–42017. [Google Scholar] [CrossRef] [PubMed]
- Mehla, R.; Bivalkar-Mehla, S.; Zhang, R.; Handy, I.; Albrecht, H.; Giri, S.; Nagarkatti, P.; Nagarkatti, M.; Chauhan, A. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS ONE 2010, 5, e11160. [Google Scholar] [CrossRef] [PubMed]
- Spivak, A.M.; Bosque, A.; Balch, A.H.; Smyth, D.; Martins, L.; Planelles, V. Ex Vivo Bioactivity and HIV-1 Latency Reversal by Ingenol Dibenzoate and Panobinostat in Resting CD4(+) T Cells from Aviremic Patients. Antimicrob. Agents Chemother. 2015, 59, 5984–5991. [Google Scholar] [CrossRef] [PubMed]
- Martins, L.J.; Bonczkowski, P.; Spivak, A.M.; De Spiegelaere, W.; Novis, C.L.; DePaula-Silva, A.B.; Malatinkova, E.; Trypsteen, W.; Bosque, A.; Vanderkerckhove, L.; et al. Modeling HIV-1 Latency in Primary T Cells Using a Replication-Competent Virus. AIDS Res. Hum. Retroviruses 2016, 32, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Brogdon, J.; Ziani, W.; Wang, X.; Veazey, R.S.; Xu, H. In vitro effects of the small-molecule protein kinase C agonists on HIV latency reactivation. Sci. Rep. 2016, 6, 39032. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Maverakis, E.; Cheng, M.Y.; Elsheikh, M.M.; Deleage, C.; Mendez-Lagares, G.; Shimoda, M.; Yukl, S.A.; Hartigan-O’Connor, D.J.; Thompson, G.R., 3rd; et al. Disruption of latent HIV in vivo during the clearance of actinic keratosis by ingenol mebutate. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- Mifsud, E.J.; Tan, A.C.; Jackson, D.C. TLR Agonists as Modulators of the Innate Immune Response and Their Potential as Agents Against Infectious Disease. Front. Immunol. 2014, 5, 79. [Google Scholar] [CrossRef]
- Macedo, A.B.; Novis, C.L.; De Assis, C.M.; Sorensen, E.S.; Moszczynski, P.; Huang, S.-h.; Ren, Y.; Spivak, A.M.; Jones, R.B.; Planelles, V.; et al. Dual TLR2 and TLR7 agonists as HIV latency-reversing agents. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Novis, C.L.; Archin, N.M.; Buzon, M.J.; Verdin, E.; Round, J.L.; Lichterfeld, M.; Margolis, D.M.; Planelles, V.; Bosque, A. Reactivation of latent HIV-1 in central memory CD4(+) T cells through TLR-1/2 stimulation. Retrovirology 2013, 10, 119. [Google Scholar] [CrossRef]
- Offersen, R.; Nissen, S.K.; Rasmussen, T.A.; Ostergaard, L.; Denton, P.W.; Sogaard, O.S.; Tolstrup, M. A Novel Toll-Like Receptor 9 Agonist, MGN1703, Enhances HIV-1 Transcription and NK Cell-Mediated Inhibition of HIV-1-Infected Autologous CD4+ T Cells. J. Virol. 2016, 90, 4441–4453. [Google Scholar] [CrossRef]
- Tsai, A.; Irrinki, A.; Kaur, J.; Cihlar, T.; Kukolj, G.; Sloan, D.D.; Murry, J.P. Toll-Like Receptor 7 Agonist GS-9620 Induces HIV Expression and HIV-Specific Immunity in Cells from HIV-Infected Individuals on Suppressive Antiretroviral Therapy. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Pache, L.; Dutra, M.S.; Spivak, A.M.; Marlett, J.M.; Murry, J.P.; Hwang, Y.; Maestre, A.M.; Manganaro, L.; Vamos, M.; Teriete, P.; et al. BIRC2/cIAP1 Is a Negative Regulator of HIV-1 Transcription and Can Be Targeted by Smac Mimetics to Promote Reversal of Viral Latency. Cell Host. Microbe 2015, 18, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Sampey, G.C.; Irlbeck, D.M.; Browne, E.P.; Kanke, M.; McAllister, A.B.; Ferris, R.G.; Brehm, J.H.; Favre, D.; Routy, J.P.; Jones, C.D.; et al. The SMAC Mimetic AZD5582 is a Potent HIV Latency Reversing Agent. BioRxic Prepr. 2018. [Google Scholar] [CrossRef]
- Kuo, L.; Lawrence, D.; McDonald, D.; Refsland, E.; Bridges, S.; Smiley, S.; Tressler, R.L.; Beaubien, C.; Salzwedel, K. Highlights from the Fourth Biennial Strategies for an HIV Cure Meeting, 10-12 October 2018, Bethesda, MD, USA. J. Virus. Erad. 2019, 5, 50–59. [Google Scholar] [PubMed]
- Mbonye, U.; Karn, J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr. HIV Res. 2011, 9, 554–567. [Google Scholar]
- Alqahtani, A.; Choucair, K.; Ashraf, M.; Hammouda, D.M.; Alloghbi, A.; Khan, T.; Senzer, N.; Nemunaitis, J. Bromodomain and extra-terminal motif inhibitors: A review of preclinical and clinical advances in cancer therapy. Future Sci. OA 2019, 5, FSO372. [Google Scholar] [CrossRef]
- Bolden, J.E.; Peart, M.J.; Johnstone, R.W. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006, 5, 769–784. [Google Scholar] [CrossRef]
- Zhang, C.; Su, Z.Y.; Wang, L.; Shu, L.; Yang, Y.; Guo, Y.; Pung, D.; Bountra, C.; Kong, A.N. Epigenetic blockade of neoplastic transformation by bromodomain and extra-terminal (BET) domain protein inhibitor JQ-1. Biochem. Pharmacol. 2016, 117, 35–45. [Google Scholar] [CrossRef]
- Li, Z.; Guo, J.; Wu, Y.; Zhou, Q. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res. 2013, 41, 277–287. [Google Scholar] [CrossRef]
- Lu, P.; Shen, Y.; Yang, H.; Wang, Y.; Jiang, Z.; Yang, X.; Zhong, Y.; Pan, H.; Xu, J.; Lu, H.; et al. BET inhibitors RVX-208 and PFI-1 reactivate HIV-1 from latency. Sci. Rep. 2017, 7, 16646. [Google Scholar] [CrossRef]
- Bouchat, S.; Delacourt, N.; Kula, A.; Darcis, G.; Van Driessche, B.; Corazza, F.; Gatot, J.S.; Melard, A.; Vanhulle, C.; Kabeya, K.; et al. Sequential treatment with 5-aza-2′-deoxycytidine and deacetylase inhibitors reactivates HIV-1. EMBO Mol. Med. 2016, 8, 117–138. [Google Scholar] [CrossRef]
- Bouchat, S.; Gatot, J.S.; Kabeya, K.; Cardona, C.; Colin, L.; Herbein, G.; De Wit, S.; Clumeck, N.; Lambotte, O.; Rouzioux, C.; et al. Histone methyltransferase inhibitors induce HIV-1 recovery in resting CD4(+) T cells from HIV-1-infected HAART-treated patients. AIDS 2012, 26, 1473–1482. [Google Scholar] [CrossRef] [PubMed]
- Elliott, J.H.; Wightman, F.; Solomon, A.; Ghneim, K.; Ahlers, J.; Cameron, M.J.; Smith, M.Z.; Spelman, T.; McMahon, J.; Velayudham, P.; et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014, 10, e1004473. [Google Scholar] [CrossRef] [PubMed]
- Zaikos, T.D.; Painter, M.M.; Sebastian Kettinger, N.T.; Terry, V.H.; Collins, K.L. Class 1-Selective Histone Deacetylase (HDAC) Inhibitors Enhance HIV Latency Reversal while Preserving the Activity of HDAC Isoforms Necessary for Maximal HIV Gene Expression. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Imai, K.; Togami, H.; Okamoto, T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J. Biol. Chem. 2010, 285, 16538–16545. [Google Scholar] [CrossRef]
- Banerjee, C.; Archin, N.; Michaels, D.; Belkina, A.C.; Denis, G.V.; Bradner, J.; Sebastiani, P.; Margolis, D.M.; Montano, M. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J. Leukoc. Biol. 2012, 92, 1147–1154. [Google Scholar] [CrossRef]
- Bartholomeeusen, K.; Xiang, Y.; Fujinaga, K.; Peterlin, B.M. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J. Biol. Chem. 2012, 287, 36609–36616. [Google Scholar] [CrossRef]
- Archin, N.M.; Liberty, A.L.; Kashuba, A.D.; Choudhary, S.K.; Kuruc, J.D.; Crooks, A.M.; Parker, D.C.; Anderson, E.M.; Kearney, M.F.; Strain, M.C.; et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 2012, 487, 482–485. [Google Scholar] [CrossRef]
- Sagot-Lerolle, N.; Lamine, A.; Chaix, M.L.; Boufassa, F.; Costagliola, D.; Goujard, C.; Paller, C.; Delfraissy, J.F.; Lambotte, O. Prolonged valproic acid treatment does not reduce the size of the latent HIV reservoir. AIDS 2008, 22, 1125–1129. [Google Scholar] [CrossRef]
- Blazkova, J.; Murray, D.; Justement, J.S.; Funk, E.K.; Nelson, A.K.; Moir, S.; Chun, T.W.; Fauci, A.S. Paucity of HIV DNA methylation in latently infected, resting CD4+ T cells from infected individuals receiving antiretroviral therapy. J. Virol. 2012, 86, 5390–5392. [Google Scholar] [CrossRef]
- Blazkova, J.; Trejbalova, K.; Gondois-Rey, F.; Halfon, P.; Philibert, P.; Guiguen, A.; Verdin, E.; Olive, D.; Van Lint, C.; Hejnar, J.; et al. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009, 5, e1000554. [Google Scholar] [CrossRef]
- Yukl, S.A.; Kaiser, P.; Kim, P.; Telwatte, S.; Joshi, S.K.; Vu, M.; Lampiris, H.; Wong, J.K. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci. Transl. Med. 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Price, D.H. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 2000, 20, 2629–2634. [Google Scholar] [CrossRef] [PubMed]
- Peterlin, B.M.; Price, D.H. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006, 23, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Bisgrove, D.A.; Mahmoudi, T.; Henklein, P.; Verdin, E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl. Acad. Sci. USA 2007, 104, 13690–13695. [Google Scholar] [CrossRef]
- Bullen, C.K.; Laird, G.M.; Durand, C.M.; Siliciano, J.D.; Siliciano, R.F. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 2014, 20, 425–429. [Google Scholar] [CrossRef]
- Laird, G.M.; Bullen, C.K.; Rosenbloom, D.I.; Martin, A.R.; Hill, A.L.; Durand, C.M.; Siliciano, J.D.; Siliciano, R.F. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015, 125, 1901–1912. [Google Scholar] [CrossRef]
- Bui, J.K.; Cyktor, J.C.; Fyne, E.; Campellone, S.; Mason, S.W.; Mellors, J.W. Blockade of the PD-1 axis alone is not sufficient to activate HIV-1 virion production from CD4+ T cells of individuals on suppressive ART. PLoS ONE 2019, 14, e0211112. [Google Scholar] [CrossRef]
- Fromentin, R.; DaFonseca, S.; Costiniuk, C.T.; El-Far, M.; Procopio, F.A.; Hecht, F.M.; Hoh, R.; Deeks, S.G.; Hazuda, D.J.; Lewin, S.R.; et al. PD-1 blockade potentiates HIV latency reversal ex vivo in CD4(+) T cells from ART-suppressed individuals. Nat. Commun. 2019, 10, 814. [Google Scholar] [CrossRef]
| Geographical Location | Study Population | Study Size | Subtypes Included | Subtyping Method | End Points | Results | Reference |
|---|---|---|---|---|---|---|---|
| Israel | Ethiopian immigrants, non-Ethiopian Israeli men | 168 (77 subtype C Ethiopian immigrants, 91 subtype B non-Ethiopian Israeli men) | C, B | V3 env peptide immunoassay, direct sequencing of V3 env region PCR | CD4 and CD8 counts | No difference in rates of CD4 decline between both groups | [30] |
| Senegal | Seronegative registered female sex workers | 1683 seronegative enrolled, 81 seroconverted, 54 samples were subtyped | A, C, D, G | C2-V3 env region | AIDS-free survival, defined by <200 CD4 cells/mm3 | Non-A subtypes were 8 times more likely to develop AIDS than A subtypes | [38] |
| Thailand | HIV-1 positive inpatients | 2104 subtyped individuals | B’, E | V3 loop env-based peptide EIA | Total CD4 counts | No difference in immuno-suppression between subtypes | [29] |
| Sweden | HIV-1 infected outpatients | 98 individuals (49 ethnic Swedes, 39 ethnic Africans) | A, B, C, D | V3 env sequencing | CD4 count, CD4 decline, | No association in disease progression or CD4 decline and subtype | [28] |
| Uganda | HIV-1 infected adults | 1045 either A or D subtype individuals | A, D | Peptide serology, HMA | Progression to death, CD4 cell count | Subtype D associated with faster progression to death than subtype A | [33] |
| Tanzania | HIV-1 seropositive pregnant mothers | 428 samples where subtype was determined | A, C, D, Recombinants | C2-C5 env region and 3’ p24/5’-p7 region of gag | Progression to death, WHO stage 4 clinical disease, CD4 cell count | Subtype D associated with the fastest progression to death, WHO stage 4 of illness, CD4 <200 cells/mm3 than subtype A or C | [35] |
| Kenya | HIV-1 seronegative commercial female sex workers | 145 women | A, C, D | V1-V3 loops of env, HMA, sequencing and phylogenetic analysis | Mortality, CD4 counts | Subtype D associated with higher mortality and faster CD4 decline | [32] |
| Uganda | HIV-1 seroconverters | 312 individuals | A, D, Recombinants, multiple | Multiregion hybridization assay | CD4 decline | Subtype D associated with faster CD4 decline than subtype A | [31] |
| Uganda | HIV-1 incident ART-naïve individuals | 292 individuals | A, D, A/D, C, other recombinants | Partial gag, env, pol sequencing | CD4 ≤250 cells/mm3, WHO clinical stage 4 AIDS, death before and after ART introduction | Subtype D associated with faster disease progression than subtype A | [34] |
| Kenya, Rwanda, South Africa, Uganda, Zambia | Adult and youths with documented HIV-1 infection | 579 individuals were subtyped | A, C, D | pol sequencing | CD4 count <350 cells/µL, viral load of 1x105 copies/mL, clinical AIDS | Subtype C progressed faster than subtype A, subtype D progressed faster than subtype A | [37] |
| Sub-Saharan Africa (Uganda, Zimbabwe) | Newly infected HIV-1 women | 303 women | A, C, D | PR, RT, and C2-V3 env region | CD4 decline | Subtype D was associated with faster CD4 decline, followed by subtype A, then subtype C | [36] |
| Model System | Viruses Used | Subtype Assessed | Experimental Methods | Conclusions | Reference |
|---|---|---|---|---|---|
| C33A, HeLa, COS, U87, U373, SupT1 | Subtype-specific LTR in a subtype B LAI background, subtype-specific LTR-luciferase reporter plasmid | A, C1, C2, D, E, F, G, G’’ | Transfection of subtype-specific LTR- luciferase constructs to measure basal LTR activity and LTR activity in response to subtype B tat protein and TNF-α stimulation | Correlation between number of NF-κB sites and TNF-α response, subtype C had the greatest response Basal LTR activity in non-B subtypes significantly higher than subtype B, no difference in response to subtype B Tat transactivation Subtype-E LTR -LAI virus replicated better than subtype B-LAI | [66] |
| U937, Jurkat | LTR-luciferase reporter | B, C, E | Transfection of subtype-specific LTR-luciferase construct and addition of subtype B, C, E Tat to measure Tat transactivation, with or without TSA | Clade E Tat has the most transactivation activity Duplicated NF-κB sites in subtype C do not compete for NF-κB binding Basal activity of LTRs varied by subtype | [68] |
| SupT1, MT2 | Subtype-specific LTR in a subtype B LAI background | A, B, C, D, E, F, G | Used CA-p24 ELISA to measure viral fitness/replication | Subtype-specific LTR impacts viral replication, viral fitness influenced by cellular environment | [69] |
| Jurkat, Jurkat Tat-T | Subtype-specific LTR luciferase reporter plasmid | A, C | Used transfection of LTR-luciferase in presence or absence of subtype B tat, to measure transcriptional activity | No difference in transcriptional activity between subtypes A and C | [67] |
| 293T/U937 | Subtype-specific LTR-luciferase construct | A, B, C, D, E, F, G, G’’ | LTR transactivation studied by Co-transfection of subtype-specific LTR-luciferase construct and active pSTAT5 | Potency of LTR transactivation by active STAT5 differs between subtype | [65] |
| Jurkat, Primary CD4 T cells | LTR-GFP-IRES-Tat (LGIT) virus with subtype-specific LTRs | A, B, A2, A/G, B/C, C’, C, B/F, D, F, H | Flow cytometry to determine LGIT-virus infected cells and latency reactivation with LRAs | Varying degrees of sensitivity to reactivation by single agent or combination of LRAs across subtypes | [71] |
| Jurkat, SUPT1 | Subtype-specific LTR in a subtype B LAI background | A, B, C1, C2, D, AE, F, G, AG | Flow cytometry and ELISA for CAp24 for assessing reactivation | No differences except subtype AE and G were less prone to become latent, subtype AE had a significantly different response to LRA Vorinostat | [76] |
| Jurkat | Double-labeled (Red mCherry protein, Green eGFP protein) with subtype-specific LTR in a subtype B LAI background | A, B, C, D, F, G, AE | Flow cytometry to measure mCherry(red) expression and eGFP(green) expression. mCherry+eGFP- cells are latently infected | Difference in degree of silent infections across subtypes, as well as sensitivity to PMA/ionomycin-mediated reactivation. | [77] |
| J2574 | Subtype-specific LTR in a subtype B LAI background | A, B, C1, D, E, F, G | Flow cytometry: difference in %GFP between untreated infected population and PMA-treated infected population is latent population | AP-1 binding site in LTR is important for establishment of latency. Subtype E promoter lacks this site and has reduced ability to establish latency, Subtype A and subtype C exhibited greater latency establishment. LRAs reactivated similarly in subtype A compared to subtype B. | [78] |
| HEK293T, Primary CD4 T cells | Subtype-specific LTR in a HIV-eGFP/VSV-G single cycle infectious virus, LTR driven luciferase reporter and LTR-driven dual luciferase/renilla reporter | AE, B, B’, C, BC | measurement of luciferase/renilla as HIV-1 gene expression, measurement of GFP positive cells as a measure of viral reactivation | Blockade of Sp1 or NF-κB sites using dead Cas9 suppresses viral reactivation. HIV-1 B’ LTR had the greatest transcriptional activity when stimulated with TNF-α | [72] |
| Cohort Size | Cohort Type | Subtype | Measure of Viral Reservoir | Conclusions | Reference |
|---|---|---|---|---|---|
| Ugandan cohort: 70 Baltimore cohort: 51 | ART-treated, virally suppressed adults | A, D, AD, B | QVOA | Non-B individuals had a reduced frequency of latently infected cells, the mechanism underlying this observation could not be determined | [106] |
| 30 | ART-naïve men with acute/early HIV-1 infection | B, G, AE | QVOA | Subtype-specific Nef-mediated HLA downregulation correlates with reservoir size; HIV subtype is a statistically significant multivariable correlate of reservoir size. | [107] |
| 1057 | ART-treated long term suppressed individuals | B, AE, AG, A, C, D, F, G, Numerous recombinant forms | Total HIV-1 DNA | HIV-1 non-B subtype was associated with faster decay of reservoir in multivariable analysis; HIV-1 non-B subtype was only significantly associated with a smaller reservoir in a univariable model, trend observed in multivariable analysis | [108] |
| Model System | LRAs Assessed | Subtypes Tested | Reference |
|---|---|---|---|
| LTR-GFP-IRES-Tat (LGIT) virus-infected Jurkat cells and primary CD4 T cells | SAHA Prostratin HMBA, TSA, Valproic acid | A, A2, B, C, C, D, F, AG, BC, BF, H | [71] |
| Subtype-specific LTR in a subtype B LAI virus background infected SUPT1 or Jurkat | Vorinostat | A, B, C1, C2, D, AE, F, G, AG | [76] |
| LTR-luciferase reporter construct and tat transfection into U937, Jurkat cells | TSA | B, C, E | [68] |
| Subtype specific LTR in a subtype B LAI virus-infected J2574 cells | TSA SAHA HMBA | A | [78] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarabia, I.; Bosque, A. HIV-1 Latency and Latency Reversal: Does Subtype Matter? Viruses 2019, 11, 1104. https://doi.org/10.3390/v11121104
Sarabia I, Bosque A. HIV-1 Latency and Latency Reversal: Does Subtype Matter? Viruses. 2019; 11(12):1104. https://doi.org/10.3390/v11121104
Chicago/Turabian StyleSarabia, Indra, and Alberto Bosque. 2019. "HIV-1 Latency and Latency Reversal: Does Subtype Matter?" Viruses 11, no. 12: 1104. https://doi.org/10.3390/v11121104
APA StyleSarabia, I., & Bosque, A. (2019). HIV-1 Latency and Latency Reversal: Does Subtype Matter? Viruses, 11(12), 1104. https://doi.org/10.3390/v11121104





