The Natural Large Genomic Deletion Is Unrelated to the Increased Virulence of the Novel Genotype Fowl Adenovirus 4 Recently Emerged in China
Abstract
1. Introduction
2. Materials and Methods
2.1. Viruses and Cells
2.2. Total DNA Extraction and Genome Sequencing
2.3. Purified Viral DNA Preparation
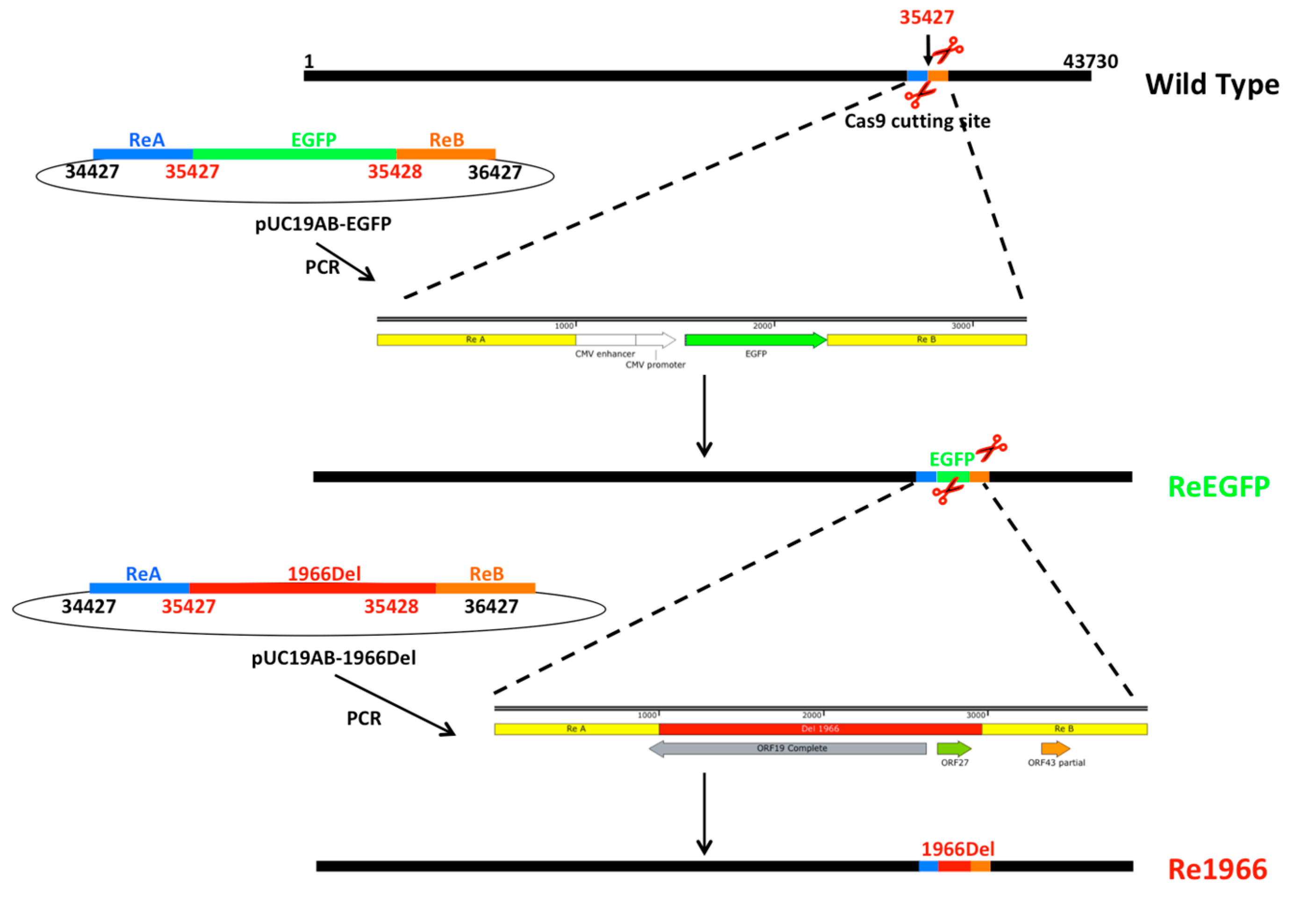
2.4. Donor Plasmid Construction
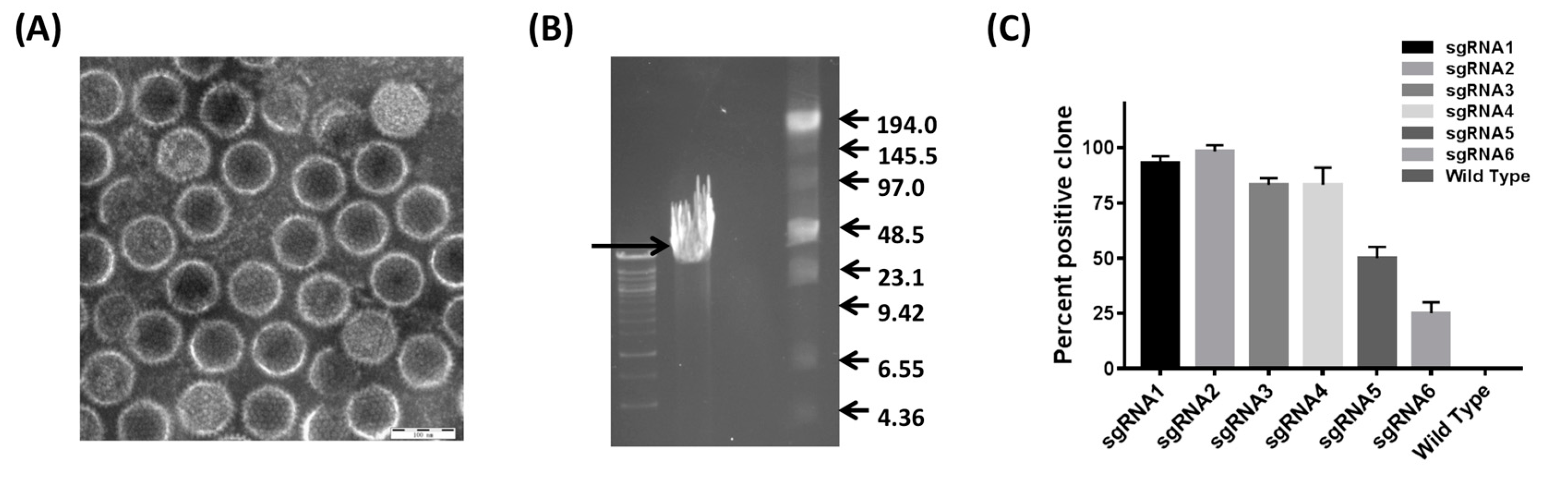
2.5. Generation of CRISPR/Cas9 sgRNA Plasmids
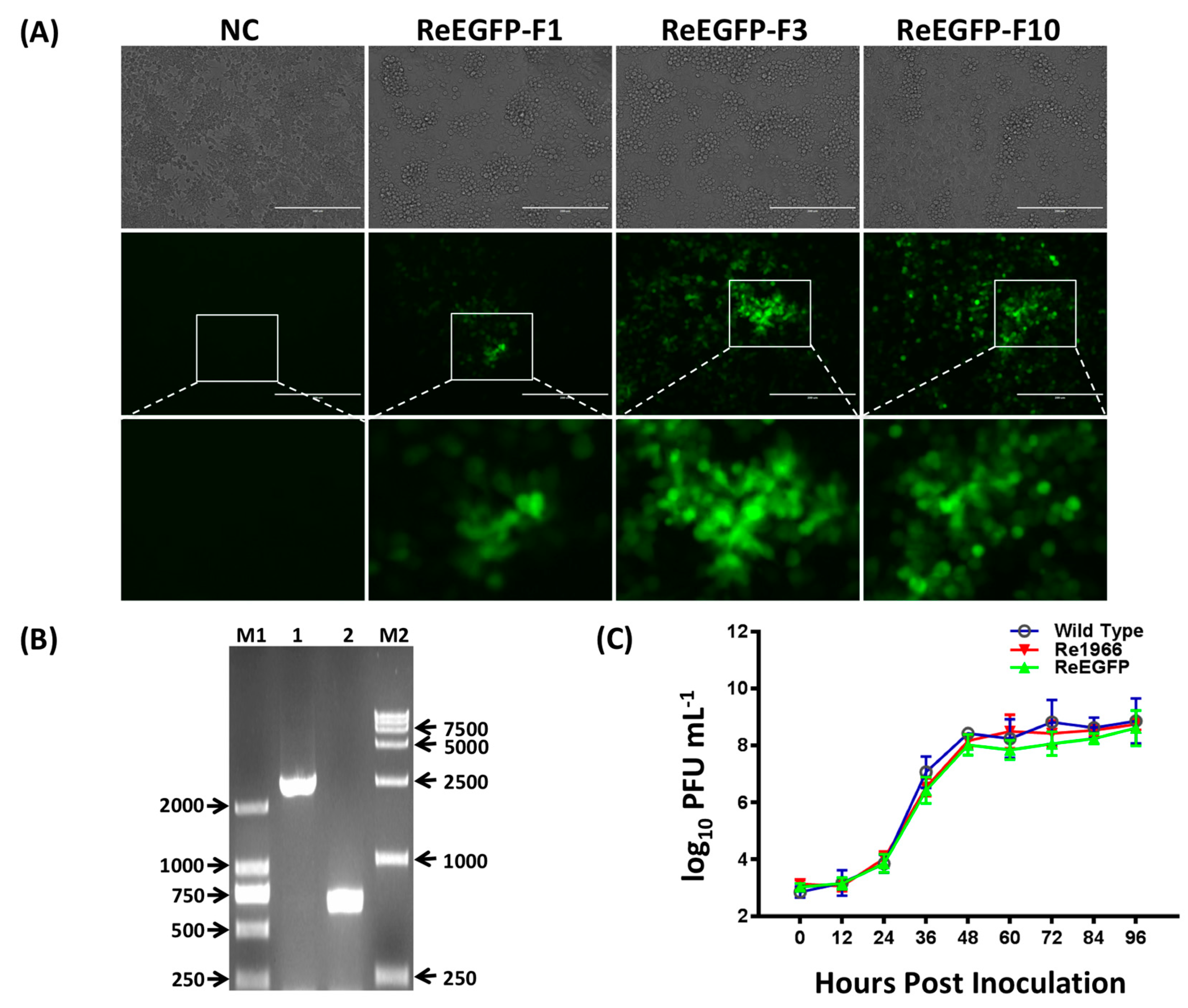
2.6. Cotransfection and Virus Rescue
2.7. Identification the Stability and Growth Properties of the Rescued Viruses
2.8. Pathogenicity Analyses of the Rescued Viruses
2.9. Ethics Statement
2.10. Statistical Analysis
3. Results
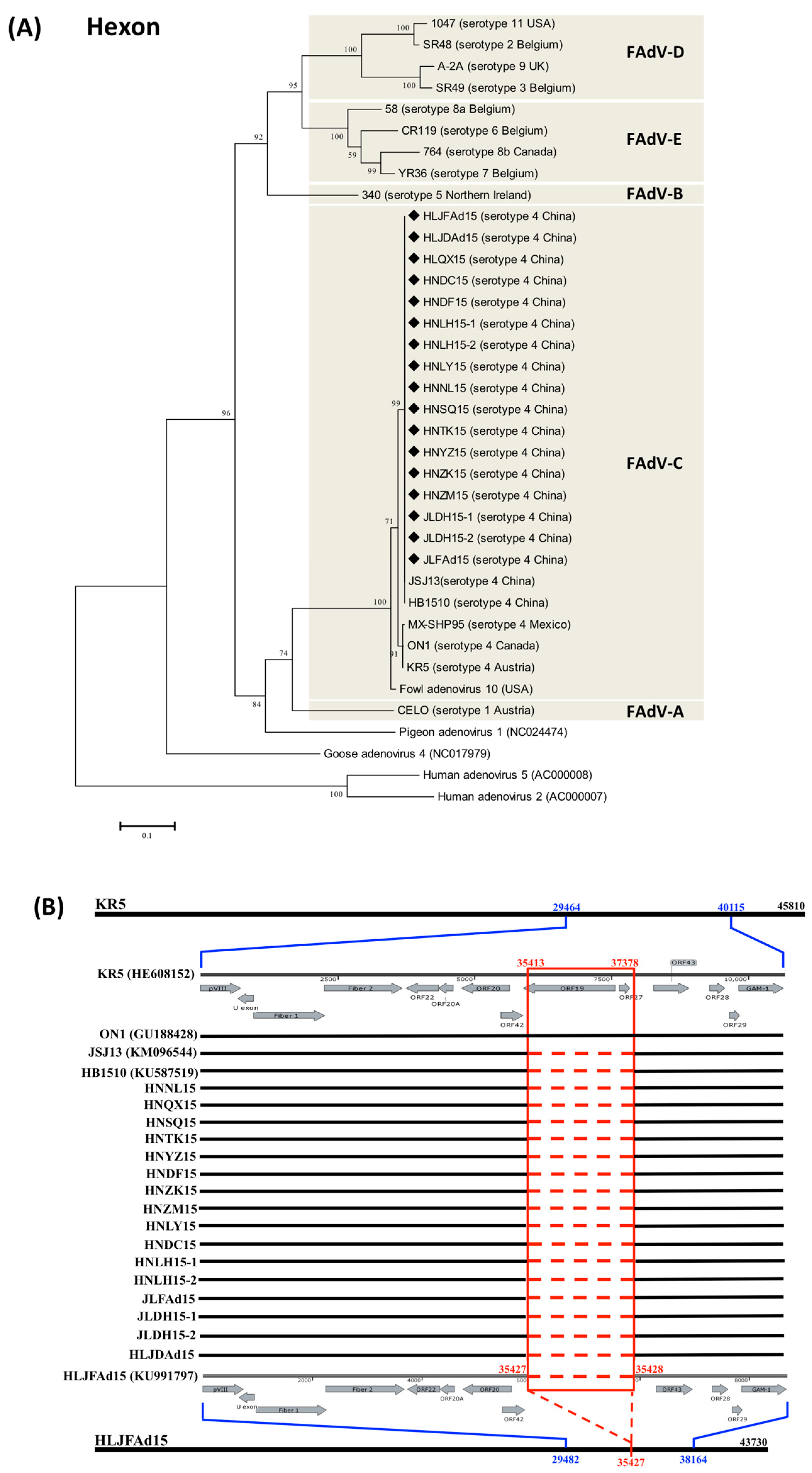
3.1. The Natural Large Genomic Deletion (1966Del) Specifically Characterized in the Novel Chinese FAdV-4 Strains
3.2. Development of the CRISPR/Cas9 Platform to Manipulate FAdV-4
3.3. Stability and Growth Properties of the Rescued Viruses
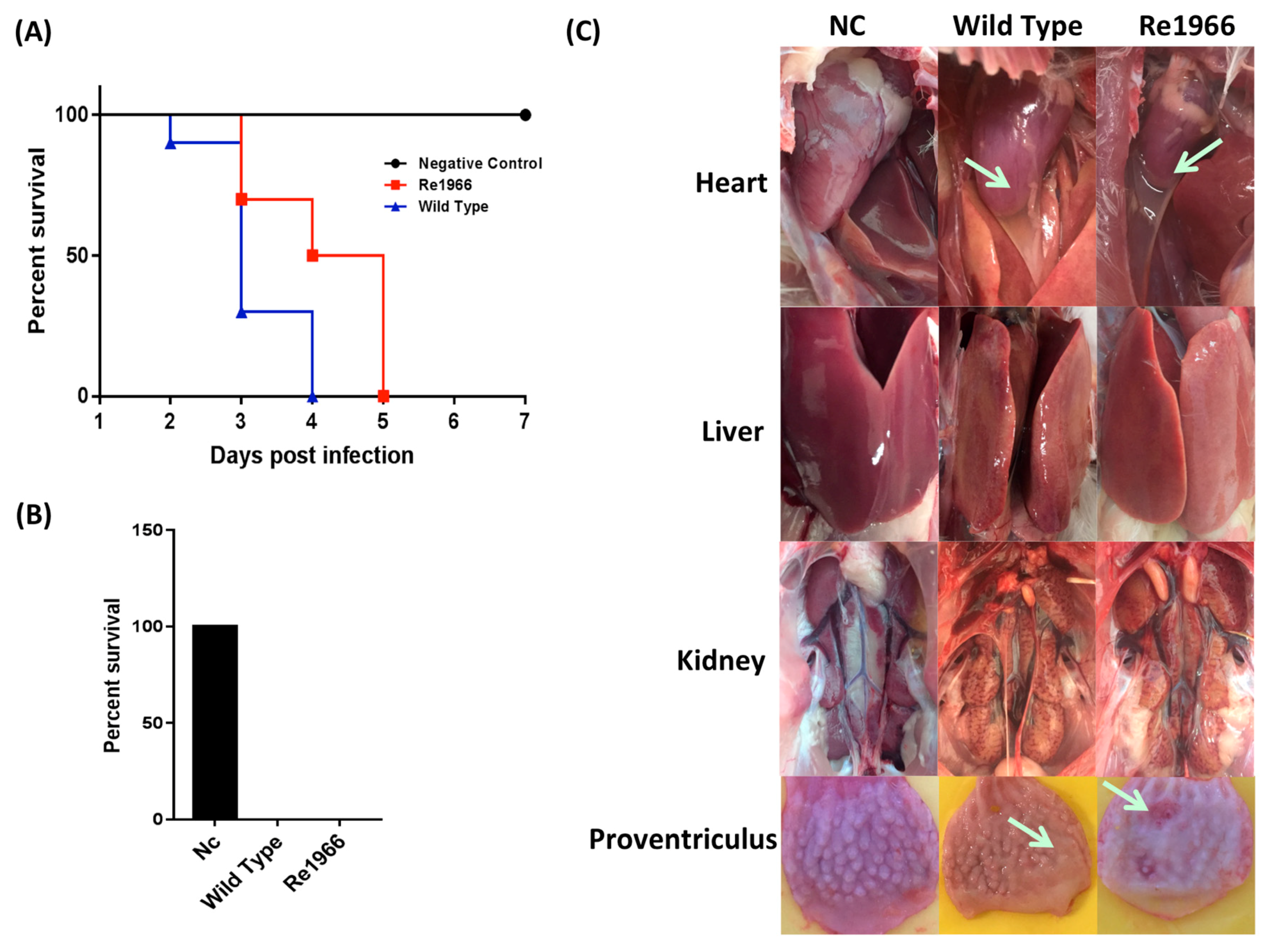
3.4. Pathogenicity of Re1966 and the Wild-Type Strain HLJFAd15 in SPF Chickens
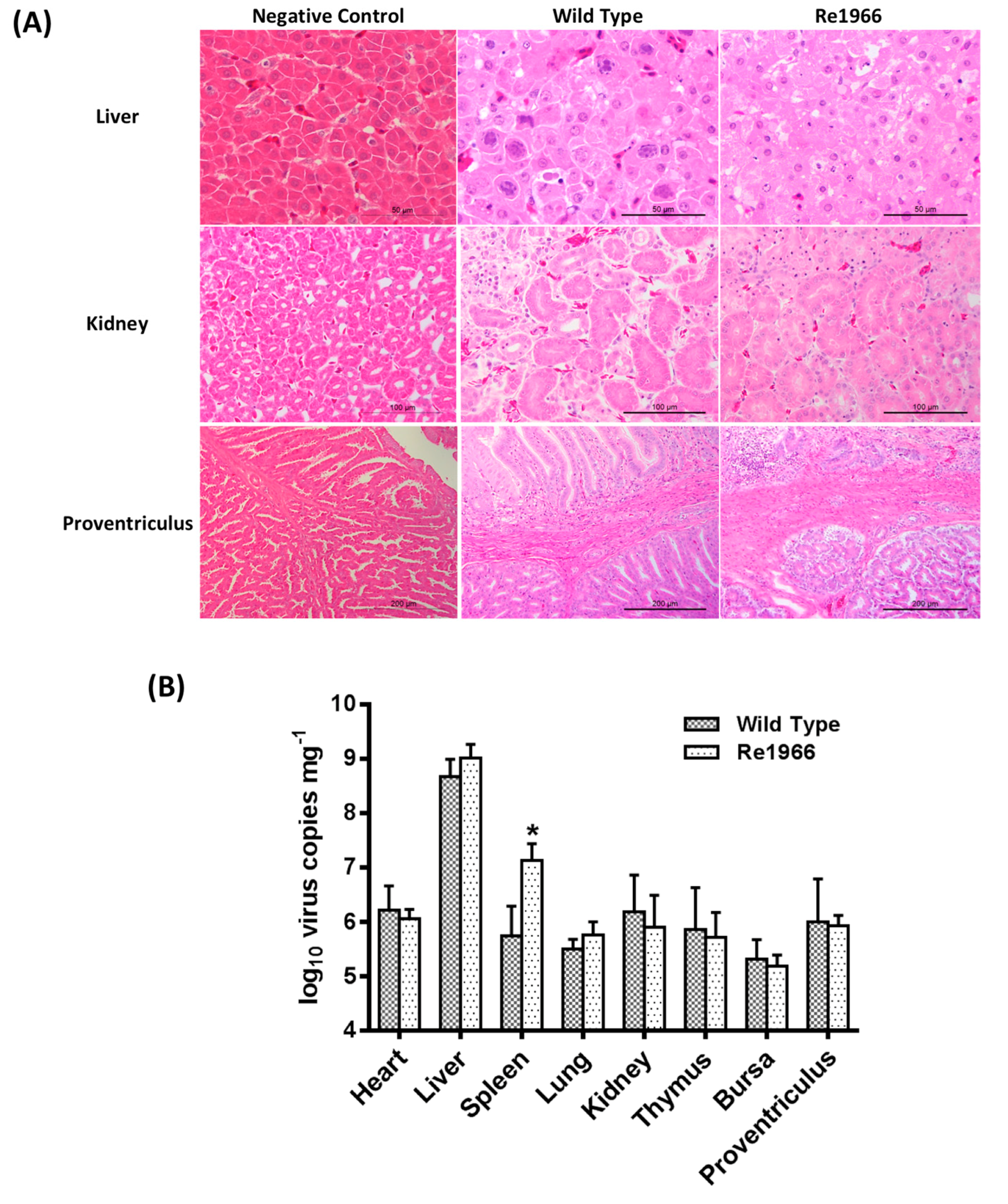
3.5. Histopathology Assay and Virus Distribution
4. Discussion
5. Conclusions
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hess, M. Detection and differentiation of avian adenoviruses: A review. Avian Pathol. 2000, 29, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.L.; Fuller, S.D.; Burnett, R.M. Difference imaging of adenovirus: Bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993, 12, 2589–2599. [Google Scholar] [CrossRef] [PubMed]
- Li, P.H.; Zheng, P.P.; Zhang, T.F.; Wen, G.Y.; Shao, H.B.; Luo, Q.P. Fowl adenovirus serotype 4: Epidemiology, pathogenesis, diagnostic detection, and vaccine strategies. Poult. Sci. 2017, 96, 2630–2640. [Google Scholar] [CrossRef] [PubMed]
- Schachner, A.; Matos, M.; Grafl, B.; Hess, M. Fowl adenovirus-induced diseases and strategies for their control–A review on the current global situation. Avian Pathol. 2018, 47, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Li, H.; Wang, D.; Wang, J.; Wang, Y.; Wang, X.; Li, J.; Liu, P.; Wang, J.; Xu, S. Characterization of fowl adenoviruses isolated between 2007 and 2014 in China. Vet. Microbiol. 2016, 197, 62–67. [Google Scholar]
- Absalon, A.E.; Morales-Garzon, A.; Vera-Hernandez, P.F.; Cortes-Espinosa, D.V.; Uribe-Ochoa, S.M.; Garcia, L.J.; Lucio-Decanini, E. Complete genome sequence of a non-pathogenic strain of Fowl Adenovirus serotype 11: Minimal genomic differences between pathogenic and non-pathogenic viruses. Virology 2017, 501, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Grafl, B.; Prokofieva, I.; Wernsdorf, P.; Steinborn, R.; Hess, M. Infection with an apathogenic fowl adenovirus serotype-1 strain (CELO) prevents adenoviral gizzard erosion in broilers. Vet. Microbiol. 2014, 172, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Marek, A.; Kajan, G.L.; Kosiol, C.; Benko, M.; Schachner, A.; Hess, M. Genetic diversity of species Fowl aviadenovirus D and Fowl aviadenovirus E. J. Gen. Virol. 2016, 97, 2323–2332. [Google Scholar] [CrossRef] [PubMed]
- Niczyporuk, J.S. Molecular characterisation of fowl adenovirus type 7 isolated from poultry associated with inclusion body hepatitis in Poland. Arch. Virol. 2017, 162, 1325–1333. [Google Scholar] [CrossRef] [PubMed]
- Khawaja, D.A.; Ahmad, S.; Rauf, A.M.; Zulfiqar, M.; Mahmood, S.M.I.; Mahmood, U.H.M. Isolation of an adeno virus from hydropericardium syndrome in broiler chicks. Pak. J. Vet. Res. 1988, 1, 2–17. [Google Scholar]
- Toro, H.; Prusas, C.; Raue, R.; Cerda, L.; Geisse, C.; González, C.; Hess, M. Characterization of fowl adenoviruses from outbreaks of inclusion body hepatitis/hydropericardium syndrome in Chile. Avian Dis. 1999, 43, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Vera-Hernández, P.F.; Morales-Garzón, A.; Cortés-Espinosa, D.V.; Galiote-Flores, A.; García-Barrera, L.J.; Rodríguez-Galindo, E.T.; Toscano-Contreras, A.; Lucio-Decanini, E.; Absalón, A.E. Clinicopathological characterization and genomic sequence differences observed in a highly virulent fowl Aviadenovirus serotype 4. Avian Pathol. 2016, 45, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Byun, S.H.; Kim, M.J.; Kim, J.J.; Sung, H.W.; Mo, I.P. Outbreaks of hydropericardium syndrome and molecular characterization of Korean fowl adenoviral isolates. Avian Dis. 2008, 52, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Mittal, D.; Jindal, N.; Tiwari, A.K.; Khokhar, R.S. Characterization of fowl adenoviruses associated with hydropericardium syndrome and inclusion body hepatitis in broiler chickens. Virusdisease 2014, 25, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wan, W.; Gao, D.; Li, Y.; Yang, X.; Liu, H.; Yao, H.; Chen, L.; Wang, C.; Zhao, J. Genetic characterization of novel fowl aviadenovirus 4 isolates from outbreaks of hepatitis-hydropericardium syndrome in broiler chickens in China. Emerg. Microbes Infect. 2016, 5, e117. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Liang, G.; Zhang, J.; Wang, W.; Song, N.; Wang, P.; Zheng, W.; Xie, Q.; Shao, H.; Wan, Z. Outbreaks of serotype 4 fowl adenovirus with novel genotype, China. Emerg. Microbes Infect. 2016, 5, e50. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liu, L.; Gao, Y.; Liu, C.; Qi, X.; Zhang, Y.; Wang, Y.; Li, K.; Gao, L.; Wang, X. Characterization of a hypervirulent fowl adenovirus 4 with the novel genotype newly prevalent in China and establishment of reproduction infection model of hydropericardium syndrome in chickens. Poult. Sci. 2017, 96, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Liu, L.; Wang, Y.; Zhang, Y.; Qi, X.; Liu, C.; Gao, Y.; Wang, X.; Cui, H. The first whole genome sequence and pathogenicity characterization of a fowl adenovirus 4 isolated from ducks associated with inclusion body hepatitis and hydropericardium syndrome. Avian Pathol. 2017, 46, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Francois, A.; Eterradossi, N.; Delmas, B.; Payet, V.; Langlois, P. Construction of avian adenovirus CELO recombinants in cosmids. J. Virol. 2001, 75, 5288–5301. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Corredor, J.C.; Griffin, B.D.; Krell, P.J.; Nagy, E. Fowl Adenovirus 4 (FAdV-4)-Based Infectious Clone for Vaccine Vector Development and Viral Gene Function Studies. Viruses 2018, 10, 97. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.; Griffin, B.; de Jong, J.; Krell, P.J.; Nagy, E. Rapid generation of fowl adenovirus 9 vectors. J. Virol. Methods 2015, 223, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Suenaga, T.; Kohyama, M.; Hirayasu, K.; Arase, H. Engineering large viral DNA genomes using the CRISPR-Cas9 system. Microbiol. Immunol. 2014, 58, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Sun, L.; Gao, D.; Ding, C.; Li, Z.; Li, Y.; Cun, W.; Li, Q. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases. PLoS Pathog. 2014, 10, e1004090. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Sheng, C.; Wang, S.; Yang, L.; Liang, Y.; Huang, Y.; Liu, H.; Li, P.; Yang, C.; Yang, X. Removal of Integrated Hepatitis B Virus DNA Using CRISPR-Cas9. Front. Cell. Infect. Microbiol. 2017, 7, 91. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Furuse, Y.; Oshitani, H.; Kiyono, T. Highly Efficient CRISPR/Cas9-Mediated Cloning and Functional Characterization of Gastric Cancer-Derived Epstein-Barr Virus Strains. J. Virol. 2016, 90, 4383–4393. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.D.; Guo, J.C.; Wang, T.Y.; Zhao, K.; Liu, J.T.; Gao, J.C.; Tian, Z.J.; An, T.Q.; Cai, X.H. CRISPR/Cas9-mediated 2-sgRNA cleavage facilitates pseudorabies virus editing. FASEB J. 2018, 32, 4293–4301. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Zhang, Y.; Pedrera, M.; Chang, P.; Baigent, S.; Moffat, K.; Shen, Z.; Nair, V.; Yao, Y. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using CRISPR/Cas9 system. Vaccine 2018, 36, 716–722. [Google Scholar] [CrossRef] [PubMed]
- Parks, C.L.; Banerjee, S.; Spector, D.J. Organization of the transcriptional control region of the E1b gene of adenovirus type 5. J. Virol. 1988, 62, 54–67. [Google Scholar] [PubMed]
- Li, P.; Bellett, A.J.; Parish, C.R. Structural organization and polypeptide composition of the avian adenovirus core. J. Virol. 1984, 52, 638–649. [Google Scholar] [PubMed]
- Pan, Q.; Yang, Y.; Gao, Y.; Qi, X.; Liu, C.; Zhang, Y.; Cui, H.; Wang, X. An Inactivated Novel Genotype Fowl Adenovirus 4 Protects Chickens against the Hydropericardium Syndrome That Recently Emerged in China. Viruses 2017, 9, 216. [Google Scholar] [CrossRef] [PubMed]
- Steer, P.A.; Kirkpatrick, N.C.; O’Rourke, D.; Noormohammadi, A.H. Classification of fowl adenovirus serotypes by use of high-resolution melting-curve analysis of the hexon gene region. J. Clin. Microbiol. 2009, 47, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Pan, Q.; Yang, Y.; Shi, Z.; Liu, L.; Gao, Y.; Qi, X.; Liu, C.; Zhang, Y.; Cui, H.; Wang, X. Different Dynamic Distribution in Chickens and Ducks of the Hypervirulent, Novel Genotype Fowl Adenovirus Serotype 4 Recently Emerged in China. Front. Microbiol. 2017, 8, 1005. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhong, Q.; Zhao, Y.; Hu, Y.X.; Zhang, G.Z. Pathogenicity and Complete Genome Characterization of Fowl Adenoviruses Isolated from Chickens Associated with Inclusion Body Hepatitis and Hydropericardium Syndrome in China. PLoS ONE 2015, 10, e0133073. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Yang, H.; Chen, W.; Cao, W.; Zhong, G.; Jiao, P.; Deng, G.; Yu, K.; Yang, C.; Bu, Z. A naturally occurring deletion in its NS gene contributes to the attenuation of an H5N1 swine influenza virus in chickens. J. Virol. 2008, 82, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Gao, Y.; Wang, Y.; Qin, L.; Qi, X.; Qu, Y.; Gao, H.; Wang, X. A 205-nucleotide deletion in the 3′ untranslated region of avian leukosis virus subgroup J, currently emergent in China, contributes to its pathogenicity. J. Virol. 2012, 86, 12849–12860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Zhang, J.; Zeng, J.; Yin, S.; Li, Y.; Zheng, L.; Guo, X.; Ge, X.; Yang, H. The 30-amino-acid deletion in the Nsp2 of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China is not related to its virulence. J. Virol. 2009, 83, 5156–5167. [Google Scholar] [CrossRef] [PubMed]
- Kamil, J.P.; Tischer, B.K.; Trapp, S.; Nair, V.K.; Osterrieder, N.; Kung, H.J. vLIP, a viral lipase homologue, is a virulence factor of Marek’s disease virus. J. Virol. 2005, 79, 6984–6996. [Google Scholar] [CrossRef] [PubMed]
- Tulman, E.R.; Afonso, C.L.; Lu, Z.; Zsak, L.; Rock, D.L.; Kutish, G.F. The genome of a very virulent Marek’s disease virus. J. Virol. 2000, 74, 7980–7988. [Google Scholar] [CrossRef] [PubMed]
- Corredor, J.C.; Garceac, A.; Krell, P.J.; Nagy, E. Sequence comparison of the right end of fowl adenovirus genomes. Virus Genes 2008, 36, 331–344. [Google Scholar] [CrossRef] [PubMed]
- Griffin, B.D.; Nagy, E. Coding potential and transcript analysis of fowl adenovirus 4: Insight into upstream ORFs as common sequence features in adenoviral transcripts. J. Gen. Virol. 2011, 92, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
| No. | Name | Location | Host | Age | Clinical Signs a | Mortality |
|---|---|---|---|---|---|---|
| 1 | HLJFAd15 | Zhaodong, Heilongjiang | Layers | 180 | HHS, IBH | >50% |
| 2 | HLJDAd15 | Shuangcherng, Heilongjiang | Ducks | 45 | IBH | 15% |
| 3 | JLFAd15 | Jiutai, Jilin | Layers | 72 | HHS, IBH | 18% |
| 4 | JLDH-1 | Dehui, Jilin | Broiler breeders | 160 | HHS, IBH | 15% |
| 5 | JLDH-2 | Dehui, Jilin | Broiler breeders | 160 | HHS, IBH | 15% |
| 6 | HNNL15 | Ningling, Henan | Layers | 46 | HHS, IBH | 61% |
| 7 | HNQX15 | Qixian, Henan | Layers | 45 | HHS, IBH | 43% |
| 8 | HNSQ15 | Shenqiu, Henan | Layers | 90 | HHS, IBH | 32% |
| 9 | HNTK15 | Taikang, Henan | Layers | 55 | HHS, IBH | 62% |
| 10 | HNYZ15 | Yuzhou, Henan | Layers | 52 | HHS, IBH | 42% |
| 11 | HNDF15 | Dengfeng, Henan | Layers | 46 | HHS, IBH | 46% |
| 12 | HNZK15 | Zhoukou, Henan | Layers | 34 | HHS, IBH | 52% |
| 13 | HNZM15 | Zhongmu, Henan | Layers | 64 | HHS, IBH | 36% |
| 14 | HNLY15 | Linying, Henan | Layers | 55 | HHS, IBH | 56% |
| 15 | HNDC15 | Dancheng, Henan | Layers | 63 | HHS, IBH | 48% |
| 16 | HNLH15-1 | Luohe, Henan | Layers | 44 | HHS, IBH | 57% |
| 17 | HNLH15-2 | Luohe, Henan | Layers | 52 | HHS, IBH | 41% |
| Name | Sequences (5′-3′) | Name | Sequences (5′-3′) |
|---|---|---|---|
| sgRNA-1 | CACCGATTAGGTCGCGCATTCCGAT | sgRNA-4 | CACCGTATATGGCGGATGTTCGGAT |
| AAACATCGGAATGCGCGACCTAATC | AAACATCCGAACATCCGCCATATAC | ||
| sgRNA-2 | CACCGCGGATCGGAGTTTATTCGCC | sgRNA-5 | CACCGCTCAGAGGCTCCTTCTCGAG |
| AAACGGCGAATAAACTCCGATCCGC | AAACCTCGAGAAGGAGCCTCTGAGC | ||
| sgRNA-3 | CACCGAATGCGCGACCTAATATCAC | sgRNA-6 | CACCGTTACGAAAGCTGTGTTTATA |
| AAACGTGATATTAGGTCGCGCATTC | AAACTATAAACACAGCTTTCGTAAC |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, Q.; Wang, J.; Gao, Y.; Cui, H.; Liu, C.; Qi, X.; Zhang, Y.; Wang, Y.; Wang, X. The Natural Large Genomic Deletion Is Unrelated to the Increased Virulence of the Novel Genotype Fowl Adenovirus 4 Recently Emerged in China. Viruses 2018, 10, 494. https://doi.org/10.3390/v10090494
Pan Q, Wang J, Gao Y, Cui H, Liu C, Qi X, Zhang Y, Wang Y, Wang X. The Natural Large Genomic Deletion Is Unrelated to the Increased Virulence of the Novel Genotype Fowl Adenovirus 4 Recently Emerged in China. Viruses. 2018; 10(9):494. https://doi.org/10.3390/v10090494
Chicago/Turabian StylePan, Qing, Jing Wang, Yulong Gao, Hongyu Cui, Changjun Liu, Xiaole Qi, Yanping Zhang, Yongqiang Wang, and Xiaomei Wang. 2018. "The Natural Large Genomic Deletion Is Unrelated to the Increased Virulence of the Novel Genotype Fowl Adenovirus 4 Recently Emerged in China" Viruses 10, no. 9: 494. https://doi.org/10.3390/v10090494
APA StylePan, Q., Wang, J., Gao, Y., Cui, H., Liu, C., Qi, X., Zhang, Y., Wang, Y., & Wang, X. (2018). The Natural Large Genomic Deletion Is Unrelated to the Increased Virulence of the Novel Genotype Fowl Adenovirus 4 Recently Emerged in China. Viruses, 10(9), 494. https://doi.org/10.3390/v10090494





