Chrysoviruses in Magnaporthe oryzae
Abstract
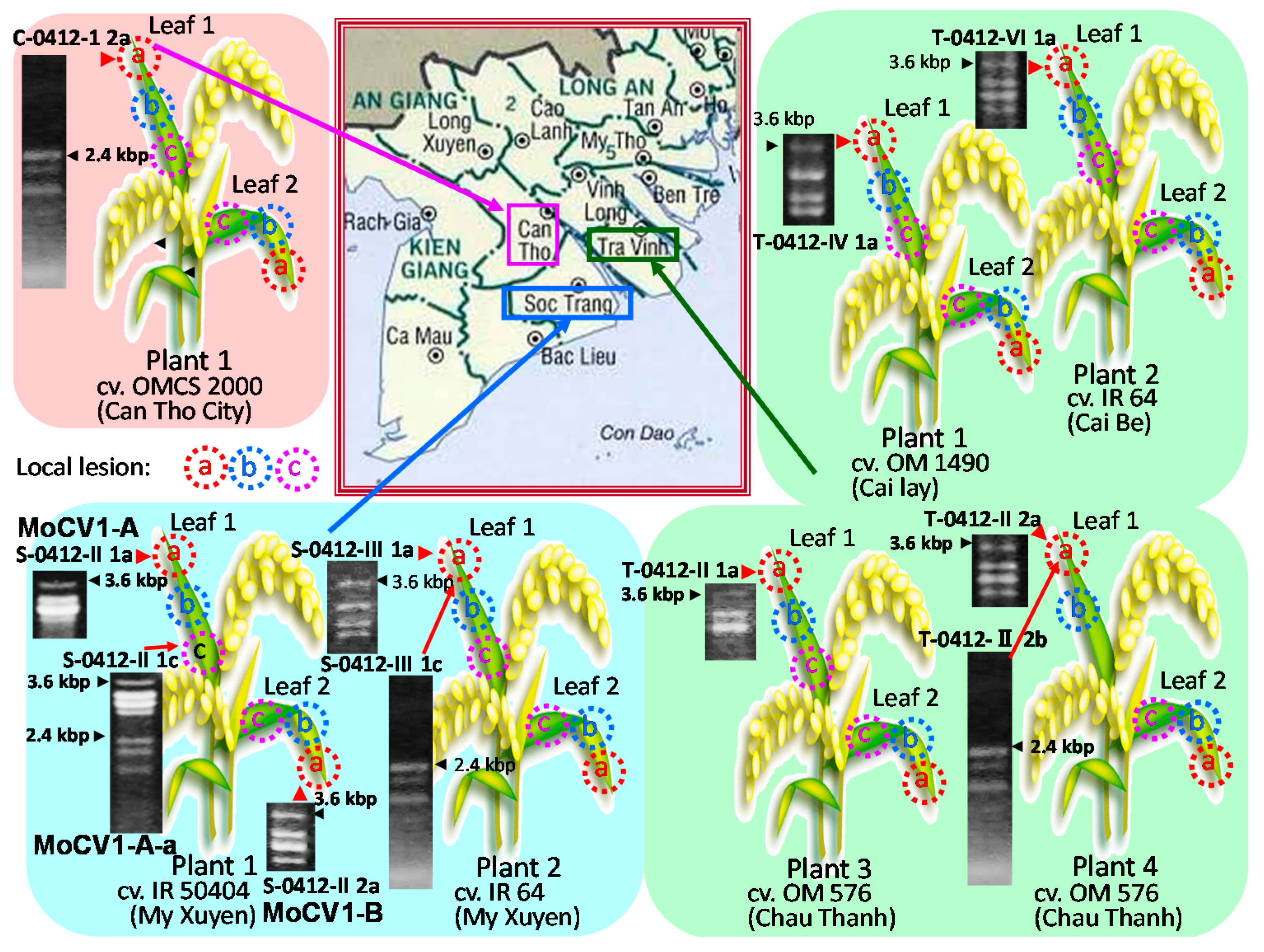
1. Introduction
2. Effects of Magnaporthe Chrysovirus on the Rice Blast Fungus
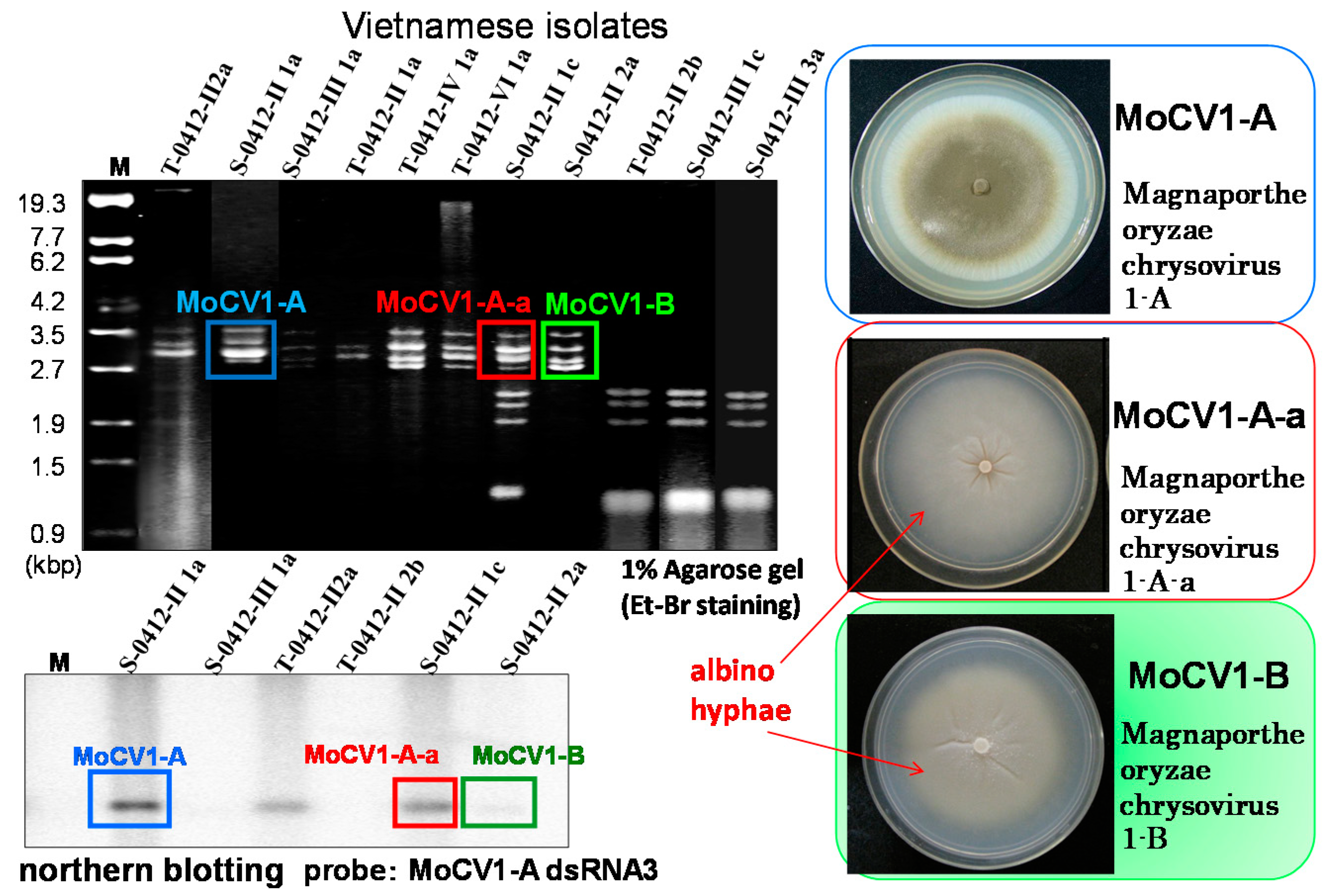
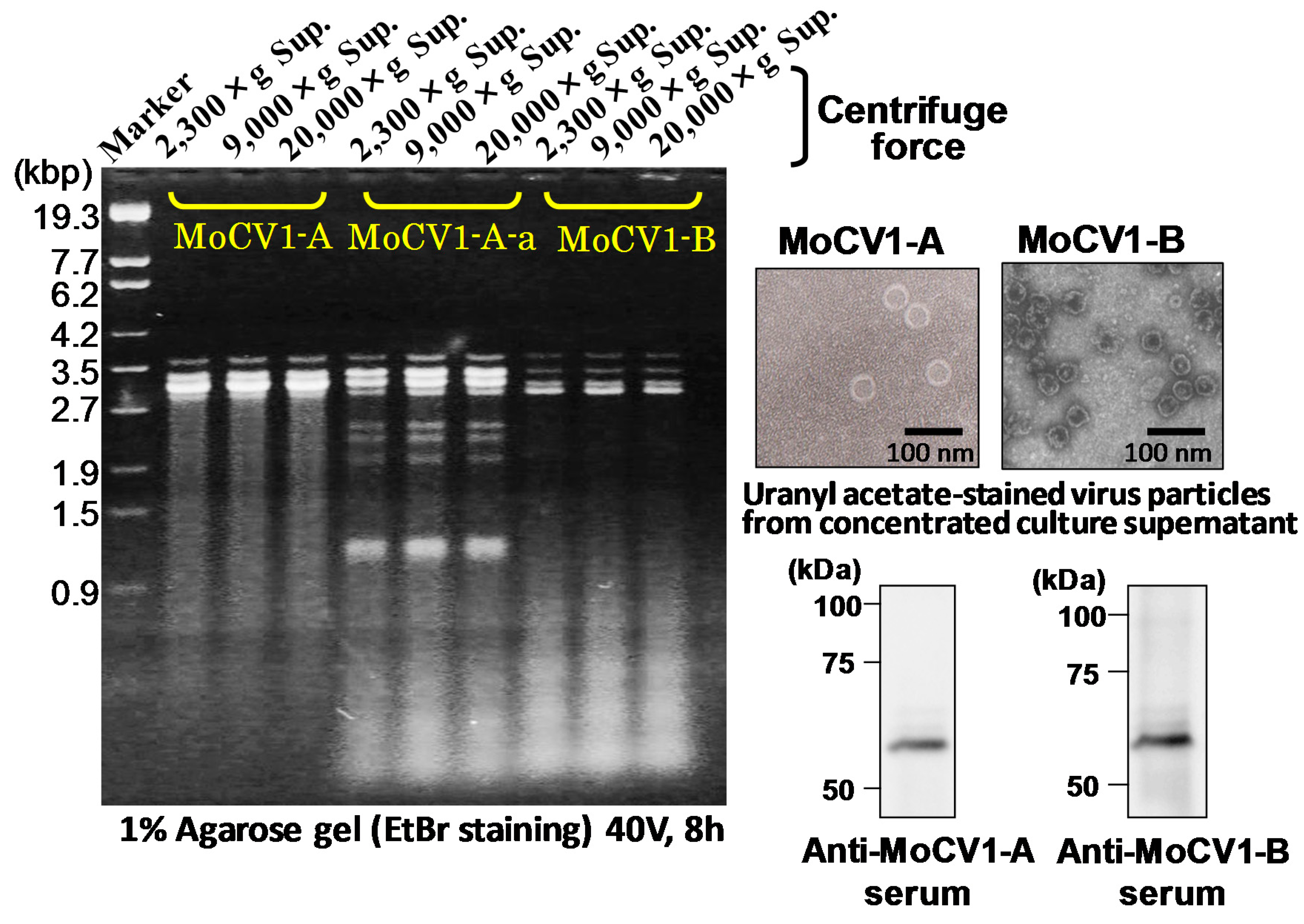
3. Molecular Properties of MoCV1-A and MoCV1-B
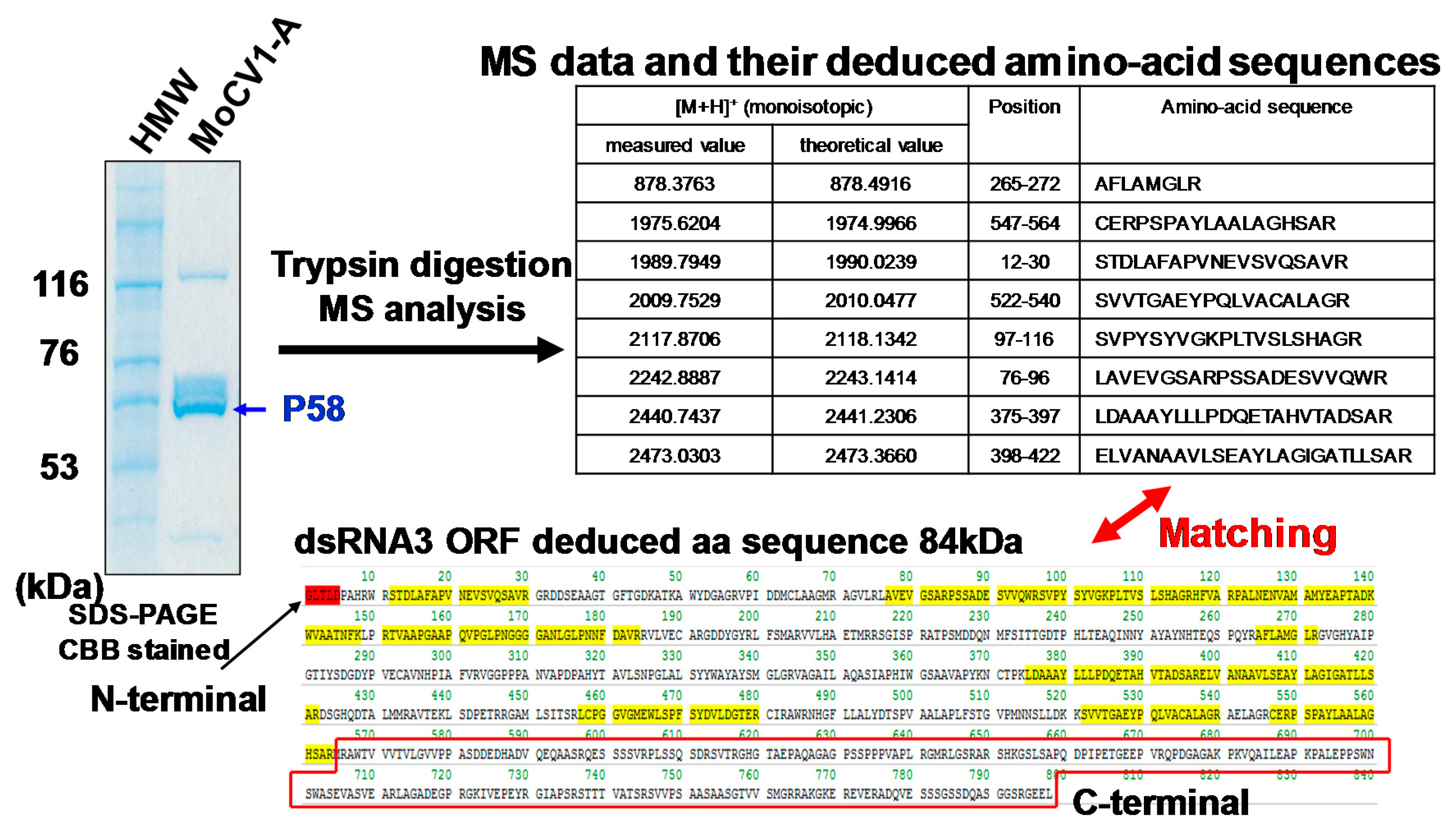
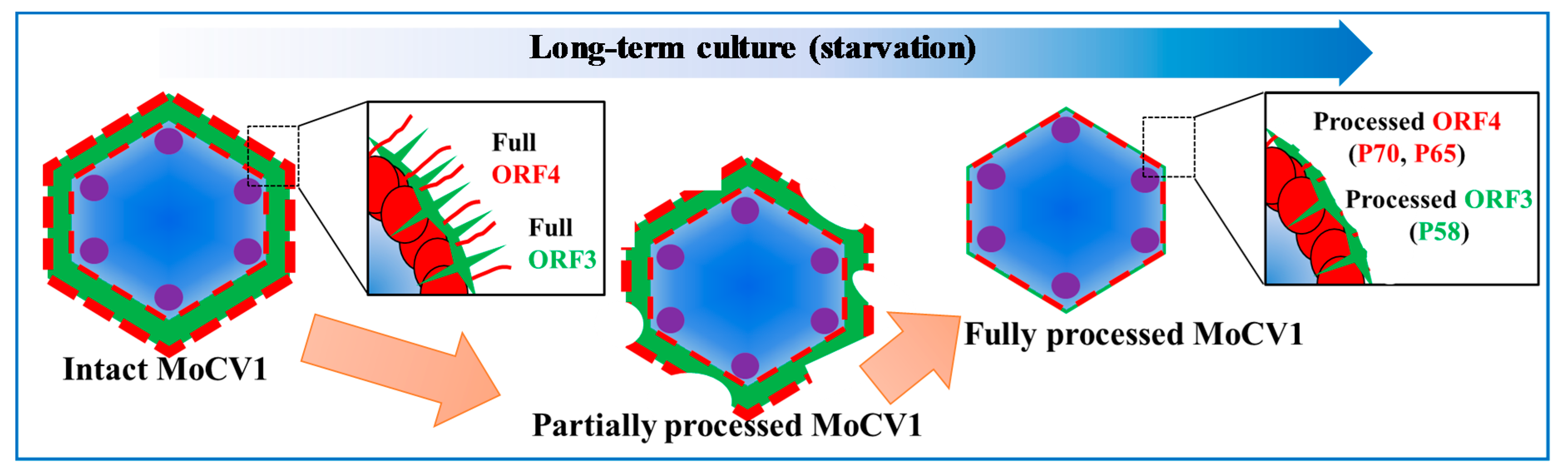
4. Virus Particles Containing dsRNAs and Multiform Structural Proteins
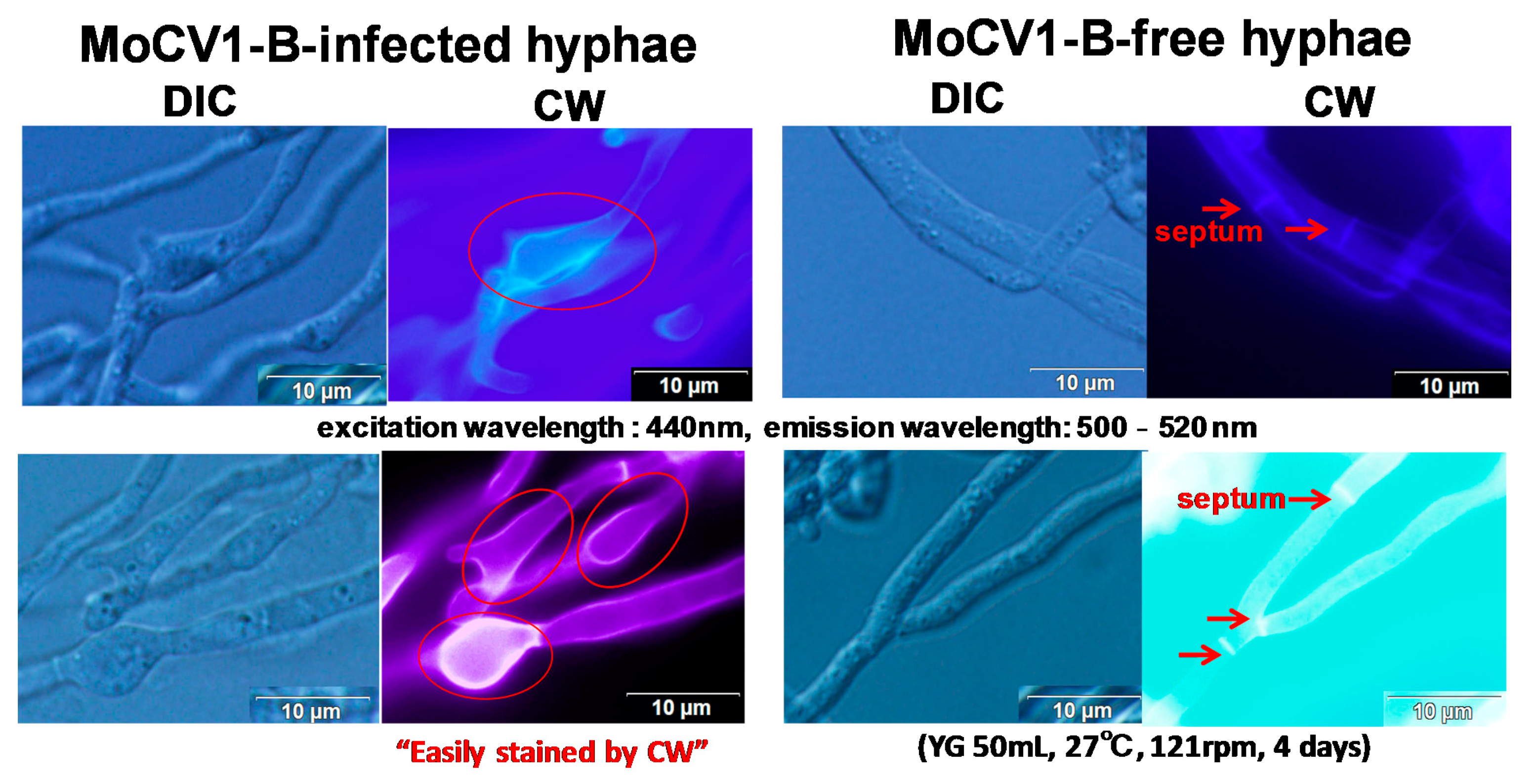
5. Release of Mycoviral dsRNA Genomes from the Mycelium of Mycovirus-Infected M. oryzae into the Culture Supernatant
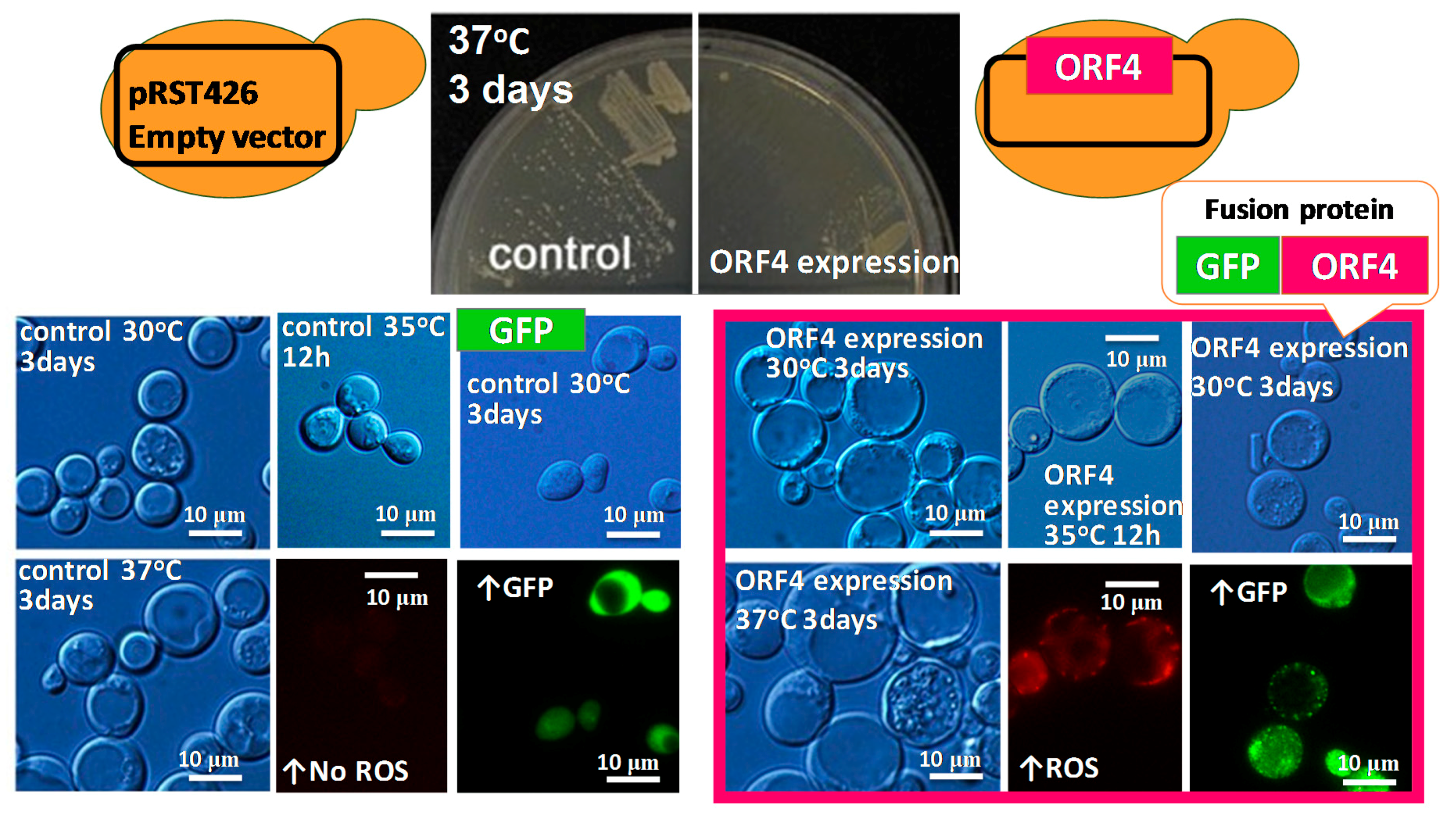
6. Heterologous Expression of Mycoviral Proteins Induced Cytological Damage in the Yeast, Saccharomyces cerevisiae
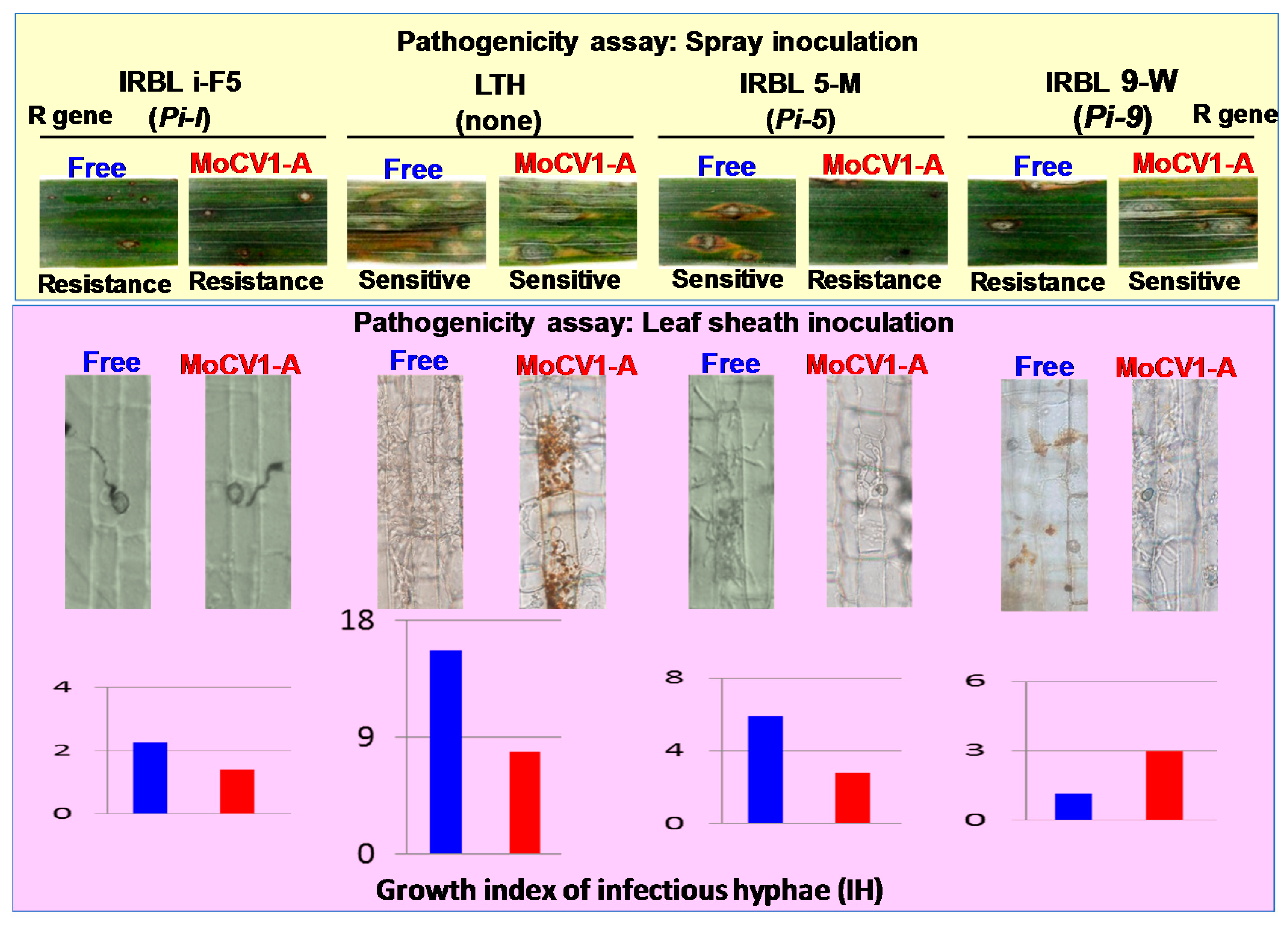
7. Influence of MoCV1-A on the Pathogenicity of Magnaporthe oryzae
8. Conclusions
Funding
Conflicts of Interest
References
- Ghabrial, S.A.; Castón, J.R.; Jiang, D.; Nibert, M.L.; Suzuki, N. 50-plus years of fungal viruses. Virology 2015, 479–480, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Hollings, M. Viruses associated with a die-back disease of cultivated mushroom. Nature 1962, 196, 962–965. [Google Scholar] [CrossRef]
- Wood, H.A.; Bozarth, R.F. Properties of virus-like particles of Penicillium chrysogenum: One double-stranded RNA molecule per particle. Virology 1972, 47, 604–609. [Google Scholar] [CrossRef]
- Herring, A.J.; Bevan, E.A. Virus-like particles associated with the double-stranded RNA species found in killer and sensitive strains of the yeast Saccharomyces cerevisiae. J. Gen. Virol. 1974, 22, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Doi, Y.; Yora, K. A polyhedral virus found in rice blast fungus, Pyricularia oryzae cavara. Ann. Phytopathol. Soc. Jpn. 1971, 37, 356–359. [Google Scholar] [CrossRef]
- Nuss, D.L. Hypovirulence: Mycoviruses at the fungal–plant interface. Nat. Rev. Microbiol. 2005, 3, 632–642. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Coutts, R.H.A. Mycoviruses in Aspergilli: A Comprehensive Review. Front. Microbiol. 2017, 8, 1699. [Google Scholar] [CrossRef]
- Dawe, A.L.; Nuss, D.L. Hypovirus molecular biology: From Koch’s postulates to host self-recognition genes that restrict virus transmission. Adv. Virus Res. 2013, 86, 109–147. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Dunn, S.E.; Li, H.; Xie, J.; Baker, T.S. Viruses of Helminthosporium (Cochlioblus) victoriae. Adv. Virus Res. 2013, 86, 289–325. [Google Scholar] [CrossRef]
- Drinnenberg, I.A.; Weinberg, D.E.; Xie, K.T.; Mower, J.P.; Wolfe, K.H.; Fink, G.R.; Bartel, D.P. RNAi in budding yeast. Science 2009, 326, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L. The fungal dimension of biodiversity: Magnitude, significance, and conservation. Mycol. Res. 1991, 95, 641–655. [Google Scholar] [CrossRef]
- Sato, T. Plant diseases and their pathogenic microbes in Japan. Microbiol. Cult. Coll. 2015, 29, 79–90. (In Japanese) [Google Scholar]
- Yokoi, T.; Yamashita, T.; Hibi, T. The nucleotide sequence and genome organization of Magnaporthe oryzae virus 1. Arch. Virol. 2007, 152, 2265–2269. [Google Scholar] [CrossRef]
- Maejima, K.; Himeno, M.; Komatsu, S.; Kakizawa, Y.; Yamaji, H.; Hamamoto, H.; Namba, S. Complete nucleotide sequence of a new double-stranded RNA virus from the rice blast fungus Magnaporthe oryzae. Arch. Virol. 2007, 153, 389–391. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; He, X.; Zhou, X.; Fang, S.; Deng, Q. Complete nucleotide sequence of Magnaporthe oryzae partitivirus 1. Arch. Virol. 2016, 161, 3295–3298. [Google Scholar] [CrossRef] [PubMed]
- Urayama, S.; Kato, S.; Suzuki, Y.; Aoki, N.; Le, M.T.; Arie, T.; Teraoka, T.; Fukuhara, T.; Moriyama, H. Mycoviruses related to chrysovirus affect vegetative growth in the rice blast fungus Magnaporthe oryzae. J. Gen. Virol. 2010, 91, 3085–3094. [Google Scholar] [CrossRef] [PubMed]
- Urayama, S.; Sakoda, H.; Takai, R.; Katoh, Y.; Minh Le, T.; Fukuhara, T.; Arie, T.; Teraoka, T.; Moriyama, H. A dsRNA mycovirus, Magnaporthe oryzae chrysovirus 1-B, suppresses vegetative growth and development of the rice blast fungus. Virology 2014, 448, 265–273. [Google Scholar] [CrossRef]
- Ai, Y.P.; Zhong, J.; Chen, C.Y.; Zhu, H.J.; Gao, B.D. A novel single-stranded RNA virus isolated from the rice-pathogenic fungus Magnaporthe oryzae with similarity to members of the family Tombusviridae. Arch. Virol. 2016, 161, 725–729. [Google Scholar] [CrossRef]
- Illana, A.; Marconi, M.; Rodríguez-Romero, J.; Xu, P.; Dalmay, T.; Wilkinson, M.D.; Ayllón, M.; Sesma, A. Molecular characterization of a novel ssRNA ourmia-like virus from the rice blast fungus Magnaporthe oryzae. Arch. Virol. 2017, 162, 891–895. [Google Scholar] [CrossRef]
- Fujikawa, T.; Nishimura, M. Pathogenic fungi fleeing the plant’s immune system “Stealth strategy”. Shokubutu Boeki. 2010, 64, 740–744. (In Japanese) [Google Scholar]
- Okada, R.; Kiyota, E.; Moriyama, H.; Fukuhara, T.; Natsuaki, T. A simple and rapid method to purify viral dsRNA from plant and fungal tissue. J. Gen. Plant. Pathol. 2015, 81, 103–107. [Google Scholar] [CrossRef]
- Wheeler, M.H. Melanin biosynthesis in Verticillium dahliae: Dehydration and reduction reactions in cell-free homogenates. Exp. Mycol. 1982, 6, 171–179. [Google Scholar] [CrossRef]
- Hamada, T.; Asanagi, M.; Satozawa, T.; Araki, N.; Banba, S.; Higashimura, N.; Akase, T.; Hirase, K. Action mechanism of the novel rice blast fungicide tolprocarb distinct from that of conventional melamin biosynthesis inhibitors. J. Pestic. Sci. 2014, 39, 152–158. [Google Scholar] [CrossRef]
- Urayama, S.; Ohta, T.; Onozuka, N.; Sakoda, H.; Fukuhara, T.; Arie, T.; Teraoka, T.; Moriyama, H. Characterization of Magnaporthe oryzae chrysovirus 1 structural proteins and their expression in Saccharomyces cerevisiae. J. Virol. 2012, 86, 8287–8295. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Urayama, S.; Katoh, Y.; Fuji, S.; Hase, S.; Fukuhara, T.; Arie, T.; Teraoka, T.; Moriyama, H. Detection of Magnaporthe oryzae chrysovirus 1 in Japan and establishment of a rapid, sensitive and direct diagnostic method based on reverse transcription loop-mediated isothermal amplification. Arch. Virol. 2016, 161, 317–326. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Hu, Y.; Liu, L.; Wu, S.; Xie, J.; Cheng, J.; Fu, Y.; Zhang, G.; Ma, J.; Wang, Y.; et al. Genomic organization of a novel victorivirus from the rice blast fungus Magnaporthe oryzae. Arch. Virol. 2015, 160, 2907–2910. [Google Scholar] [CrossRef]
- Ghabrial, S.A.; Castón, J.R.; Coutts, R.H.A.; Hillman, B.I.; Jiang, D.; Kim, D.H.; Moriyama, H. ICTV Virus Taxonomy Profile: Chrysoviridae. J. Gen. Virol. 2018, 99, 19–20. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, J.; Wang, Y.; Hong, N.; Zhang, F.; Xu, W.; Wang, G. Hypovirulence of the phytopathogenic fungus Botryosphaeria dothidea: Association with a coinfecting chrysovirus and a partitivirus. J. Virol. 2014, 88, 7517–7527. [Google Scholar] [CrossRef]
- Nerva, L.; Ciuffo, M.; Vallino, M.; Margaria, P.; Varese, G.C.; Gnavi, G.; Turina, M. Multiple approaches for the detection and characterization of viral and plasmid symbionts from a collection of marine fungi. Virus Res. 2016, 219, 22–38. [Google Scholar] [CrossRef]
- Hammond, T.M.; Andrewski, M.D.; Roossinck, M.J.; Keller, N.P. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot. Cell. 2008, 7, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Van der Lende, T.R.; Duitman, E.H.; Gunnewijk, M.G.; Yu, L.; Wessels, J.G. Functional analysis of dsRNAs (L1, L3, L5, and M2) associated with isometric 34-nm virions of Agaricus bisporus (white button mushroom). Virology 1996, 217, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Complete genome sequence of a novel dsRNA mycovirus isolated from the phytopathogenic fungus Fusarium oxysporum f. sp. dianthi. Arch. Virol. 2015, 160, 2375–2379. [Google Scholar] [CrossRef] [PubMed]
- Lemus-Minor, C.G.; Canizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Fusarium oxysporum f. sp. dianthi virus 1 Accumulation Is Correlated with Changes in Virulence and Other Phenotypic Traits of Its Fungal Host. Phytopathology 2018, 108, 957–963. [Google Scholar] [CrossRef]
- Lemus-Minor, C.G.; Cañizares, M.C.; García-Pedrajas, M.D.; Pérez-Artés, E. Horizontal and vertical transmission of the hypovirulence-associated mycovirus Fusarium oxysporum f. sp. dianthi virus 1. Eur. J. Plant Pathol. 2018. [Google Scholar] [CrossRef]
- Darissa, O.; Willingmann, P.; Schafer, W.; Adam, G. A novel double-stranded RNA mycovirus from Fusarium graminearum: Nucleic acid sequence and genomic structure. Arch. Virol. 2011, 156, 647–658. [Google Scholar] [CrossRef]
- Herrero, N.; Zabalgogeazcoa, I. Mycoviruses infecting the endophytic and entomopathogenic fungus Tolypocladium cylindrosporum. Virus Res. 2011, 160, 409–413. [Google Scholar] [CrossRef]
- Okada, R.; Ichinose, S.; Takeshita, K.; Syun-ichi Urayama, S.; Fukuhara, T.; Komatsu, K.; Arie, T.; Ishihara, A.; Egusa, M.; Kodama, M.; et al. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 2018, 519, 23–32. [Google Scholar] [CrossRef]
- Aihara, M.; Urayama, S.I.; Le, M.T.; Katoh, Y.; Higashiura, T.; Fukuhara, T.; Arie, T.; Teraoka, T.; Komatsu, K.; Moriyama, H. Infection by Magnaporthe oryzae. chrysovirus 1 strain A triggers reduced virulence and pathogenic race conversion of it host fungus, Magnaporthe oryzae. J. Gen. Plant Pathol. 2018, 84, 92–103. [Google Scholar] [CrossRef]
- Bormann, J.; Heinze, C.; Blum, C.; Mentges, M.; Brockmann, A.; Alder, A.; Landt, S.K.; Josephson, B.; Indenbirken, D.; Spohn, M.; et al. Expression of a structural protein of the mycovirus FgV-ch9 negatively affects the transcript level of a novel symptom alleviation factor and causes virus-infection like symptoms in Fusarium graminearum. J. Virol. 2018. [Google Scholar] [CrossRef]
- Ejmal, M.A.; Holland, D.J.; MacDiarmid, R.M.; Pearson, M.N. The Effect of Aspergillus thermomutatus Chrysovirus 1 on the biology of three Aspergillus species. Viruses 2018, 10, 539. [Google Scholar] [CrossRef] [PubMed]
- Coustou, V.; Deleu, C.; Saupe, S.; Begueret, J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA 1997, 94, 9773–9778. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B. A new prion controls fungal cell fusion incompatibility. Proc. Natl. Acad. Sci. USA 1997, 94, 10012–10014. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Maddelein, M.L.; Taylor, K.; Moriyama, H. Prion of yeast and fungi: Proteins as genetic material. J. Biol. Chem. 1999, 274, 555–558. [Google Scholar] [CrossRef] [PubMed]
- Urayama, S.; Fukuhara, T.; Moriyama, H.; Toh, E.A.; Kawamoto, S. Heterologous expression of a gene of Magnaporthe oryzae chrysovirus 1 strain A disrupts growth of the human pathogenic fungus Cryptococcus neoformans. Microbiol. Immunol. 2014, 58, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Himeno, M.; Maejima, K.; Komatsu, K.; Ozeki, J.; Hashimoto, M.; Kagiwada, S.; Yamaji, Y.; Namba, S. Significantly low level of small RNA accumulation derived from an encapsidated mycovirus with dsRNA genome. Virology 2010, 396, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, K.; Katayama, Y.; Omatsu, T.; Mizutani, T.; Fukuhara, T.; Kodama, M.; Arie, T.; Teraoka, T.; Moriyama, H. Genome sequence of a novel victorivirus identified in the phytopathogenic fungus Alternaria arborescens. Arch. Virol. 2016, 161, 1701–1704. [Google Scholar] [CrossRef]
- Aoki, N.; Moriyama, H.; Kodama, M.; Arie, T.; Teraoka, T.; Fukuhara, T. A novel mycovirus associated with four double-stranded RNAs affects host fungal growth in Alternaria alternata. Virus Res. 2009, 140, 179–187. [Google Scholar] [CrossRef]
- Icho, T.; Wickner, R.B. The double-stranded RNA genome of yeast virus L-A encodes its own putative RNA polymerase by fusing two open reading frames. J. Biol. Chem. 1989, 264, 6716–6723. [Google Scholar]
- Nagai, M.; Shimada, S.; Fujii, Y.; Moriyama, H.; Oba, M.; Katayama, Y.; Tsuchiaka, S.; Okazaki, S.; Omatsu, T.; Furuya, T.; et al. H2 genotypes of G4P[6], G5P[7], and G9[23] porcine rotaviruses show super-short RNA electropherotypes. Vet. Microbiol. 2015, 176, 250–256. [Google Scholar] [CrossRef]
- Urayama, S.; Kimura, Y.; Katoh, Y.; Ohta, T.; Onozuka, N.; Fukuhara, T.; Arie, T.; Teraoka, T.; Komatsu, K.; Moriyama, H. Suppressive effects of mycoviral proteins encoded by Magnaporthe oryzae chrysovirus 1 strain A on conidial germination of the rice blast fungus. Virus Res. 2016, 223, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Bhatti, M.F.; Jamal, A.; Petrou, M.A.; Cairns, T.C.; Bignell, E.M.; Coutts, R.H.A. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Ahn, I.P.; Lee, Y.H. A viral double-stranded RNA up regulates the fungal virulence of Nectria radicicola. Mol. Plant-Microbe Interact 2001, 14, 496–507. [Google Scholar] [CrossRef] [PubMed]
- Kanhayuwa, L.; Kotta-Loizou, I.; Özkan, S.; Gunning, A.P.; Coutts, R.H.A. A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 9100–9105. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Kiyosawa, S.; Yamaguchi, T.; Hirano, T.; Kobayashi, T.; Kushibuchi, K.; Watanabe, S. Proposal of a new method for differentiating races of Pyricularia oryzae Cavara in Japan. Ann. Phytopathol. Soc. Jpn. 1976, 42, 216–219. [Google Scholar] [CrossRef]
- Kiyosawa, S. Pathogenic variations of Pyricularia oryzae and their use in genetic and breeding studies. SABRAO J. 1976, 8, 53–67. [Google Scholar]
- Fuke, K.; Takeshita, K.; Aoki, N.; Fukuhara, T.; Egusa, M.; Kodama, M.; Moriyama, H. The presence of double-stranded RNAs in Alternaria alternata Japanese pear pathotype is associated with morphological changes. J. Gen. Plant Pathol. 2011, 77, 248–252. [Google Scholar] [CrossRef]
- Akimitsu, K.; Tsuge, T.; Kodama, M.; Yamamoto, M.; Otani, H. Alternaria host-selective toxins: Determinant factors of plant disease. J. Gen. Plant Pathol. 2014, 80, 109–122. [Google Scholar] [CrossRef]
- Asai, S.; Shirasu, K. Plant cells under siege: Plant immune system versus pathogen effectors. Curr. Opin. Plant Biol. 2015, 28, 1–8. [Google Scholar] [CrossRef]
- Niu, Y.; Yuan, Y.; Mao, J.; Yang, Z.; Cao, Q.; Zhang, T.; Wang, S.; Liu, D. Characterization of two novel mycoviruses from Penicillium digitatum and the related fungicide resistance analysis. Sci. Rep. 2018, 8, 5513. [Google Scholar] [CrossRef]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moriyama, H.; Urayama, S.-i.; Higashiura, T.; Le, T.M.; Komatsu, K. Chrysoviruses in Magnaporthe oryzae. Viruses 2018, 10, 697. https://doi.org/10.3390/v10120697
Moriyama H, Urayama S-i, Higashiura T, Le TM, Komatsu K. Chrysoviruses in Magnaporthe oryzae. Viruses. 2018; 10(12):697. https://doi.org/10.3390/v10120697
Chicago/Turabian StyleMoriyama, Hiromitsu, Syun-ichi Urayama, Tomoya Higashiura, Tuong Minh Le, and Ken Komatsu. 2018. "Chrysoviruses in Magnaporthe oryzae" Viruses 10, no. 12: 697. https://doi.org/10.3390/v10120697
APA StyleMoriyama, H., Urayama, S.-i., Higashiura, T., Le, T. M., & Komatsu, K. (2018). Chrysoviruses in Magnaporthe oryzae. Viruses, 10(12), 697. https://doi.org/10.3390/v10120697




