Biodiversity of Streptococcus thermophilus Phages in Global Dairy Fermentations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Bacteriophage Isolation from Whey and Enumeration
2.3. Characterisation of Bacteriophage Isolates
2.3.1. Multiplex PCR typing
2.3.2. DNA Restriction Profiling
2.3.3. Host Range
2.4. Bacteriophage DNA Extraction
2.5. Genome Sequencing, Assembly, and Annotation
2.6. Genbank Accession Numbers
2.7. Comparative Genomic Analysis
2.8. Transmission Electron Microscopy
3. Results
3.1. Phage Screening of Whey Samples
3.2. Characterisation of Propagated Phage Isolates
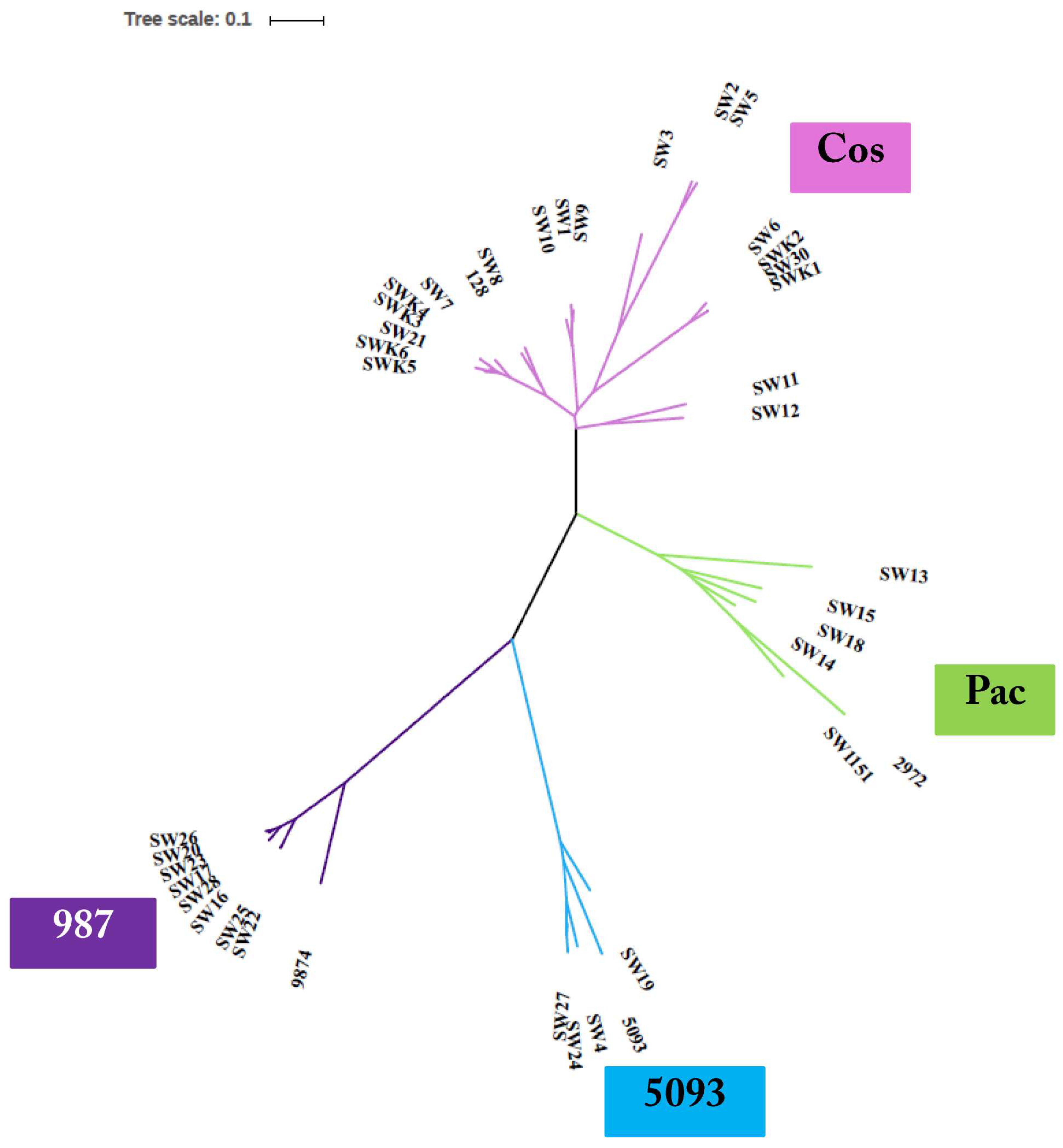
3.3. Phage Genome Sequencing
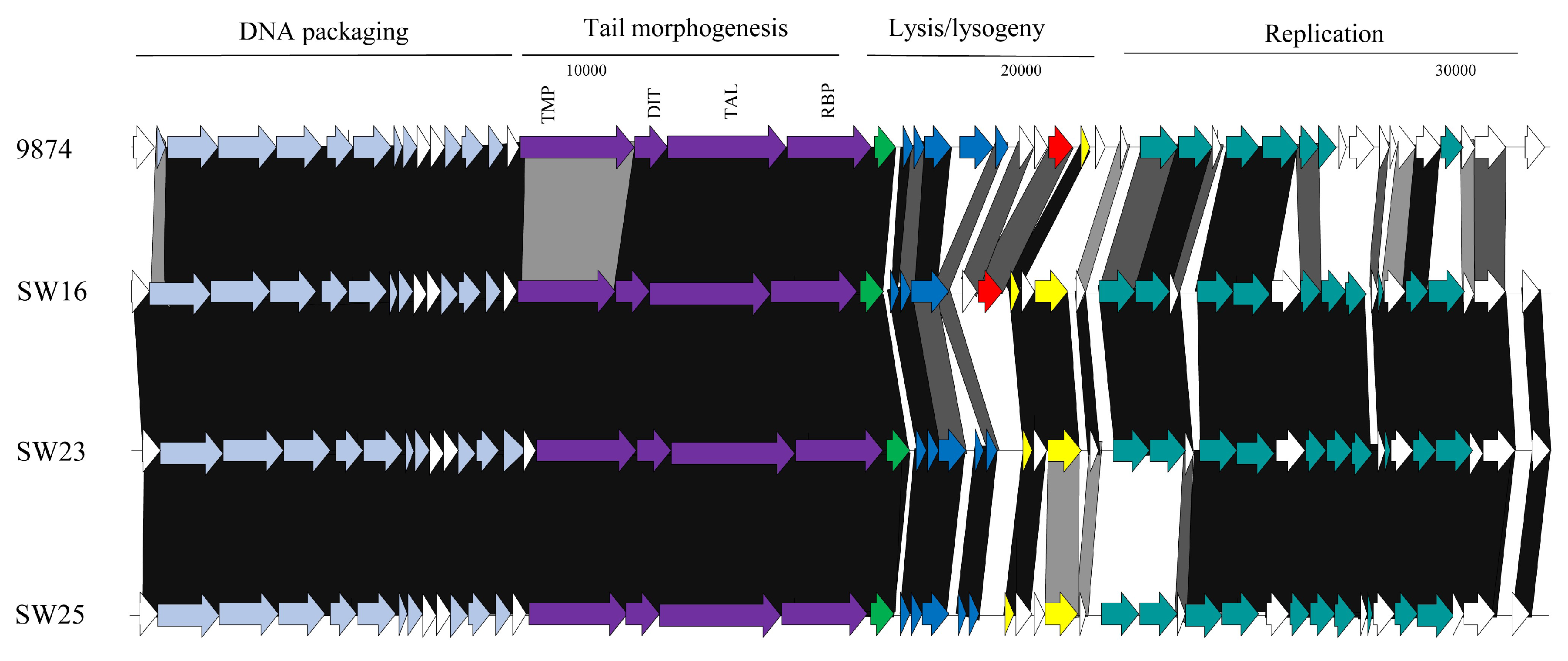
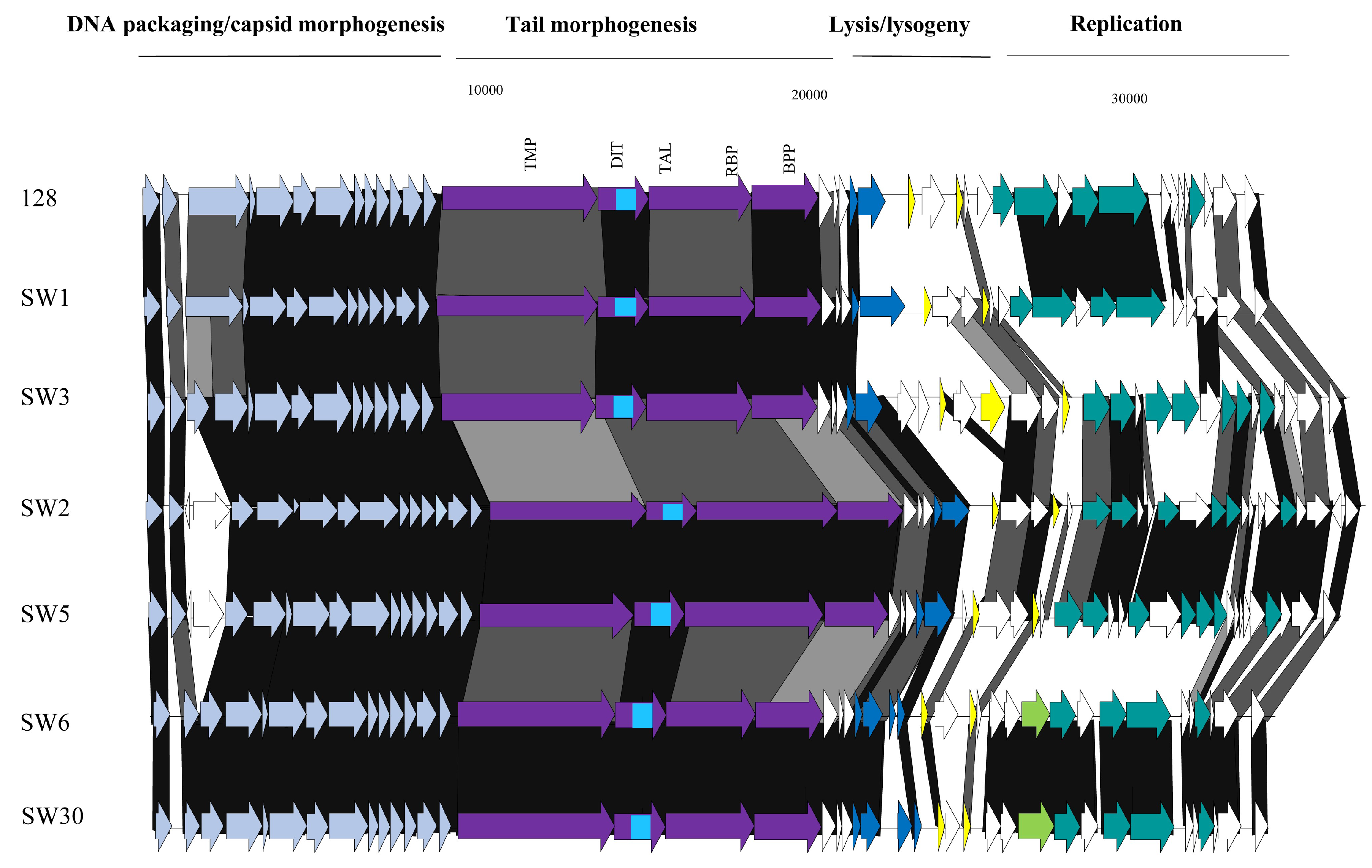
3.3.1. Genomic Diversity of the 987 Phages
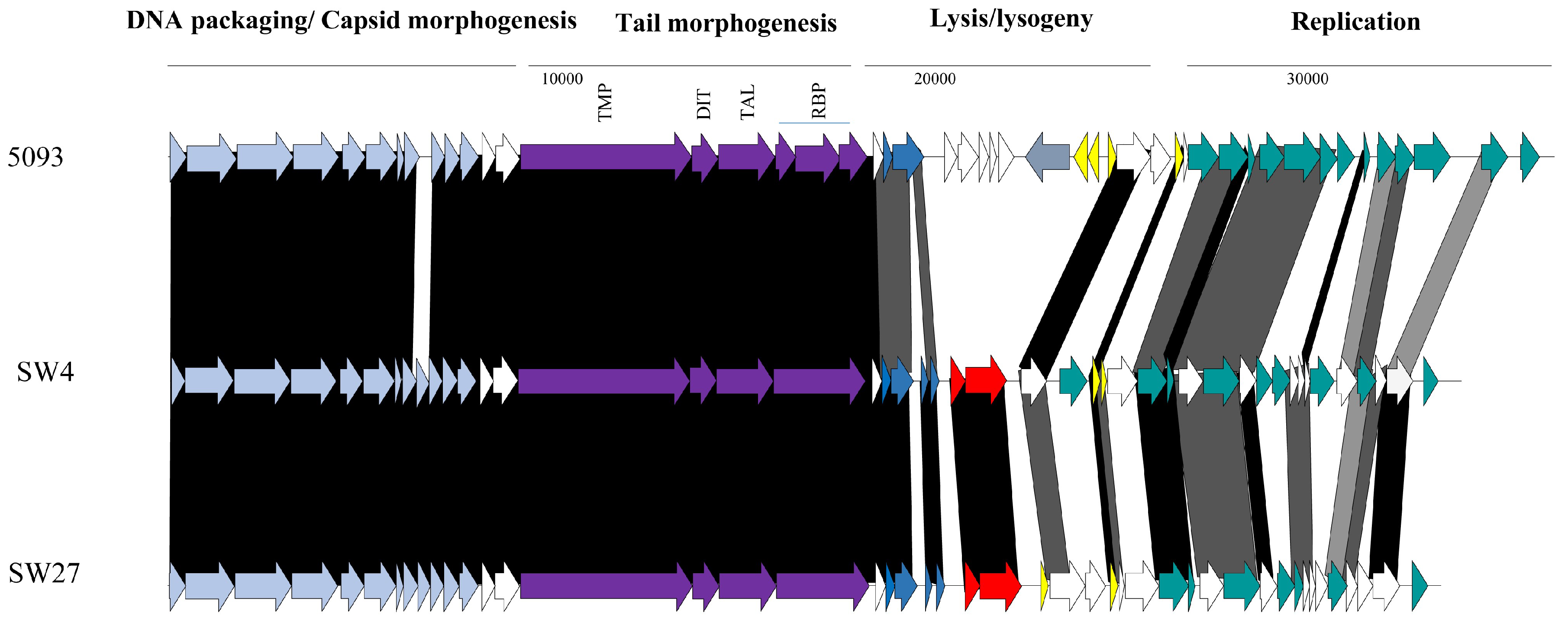
3.3.2. Genomic Diversity of the 5093 Phages
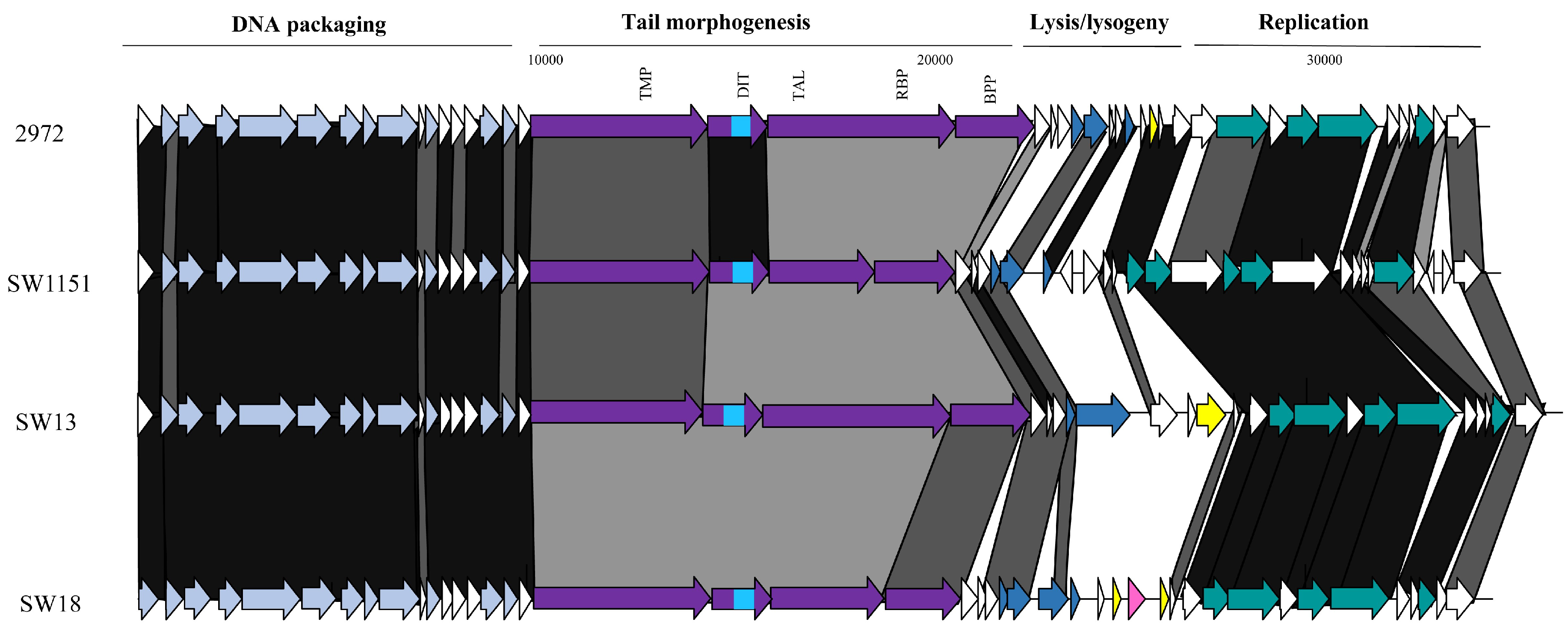
3.3.3. Genomic Diversity of the pac Phages
3.3.4. Genomic Diversity of the cos Phages
3.3.5. Phage-Encoded RBPs and Host Range
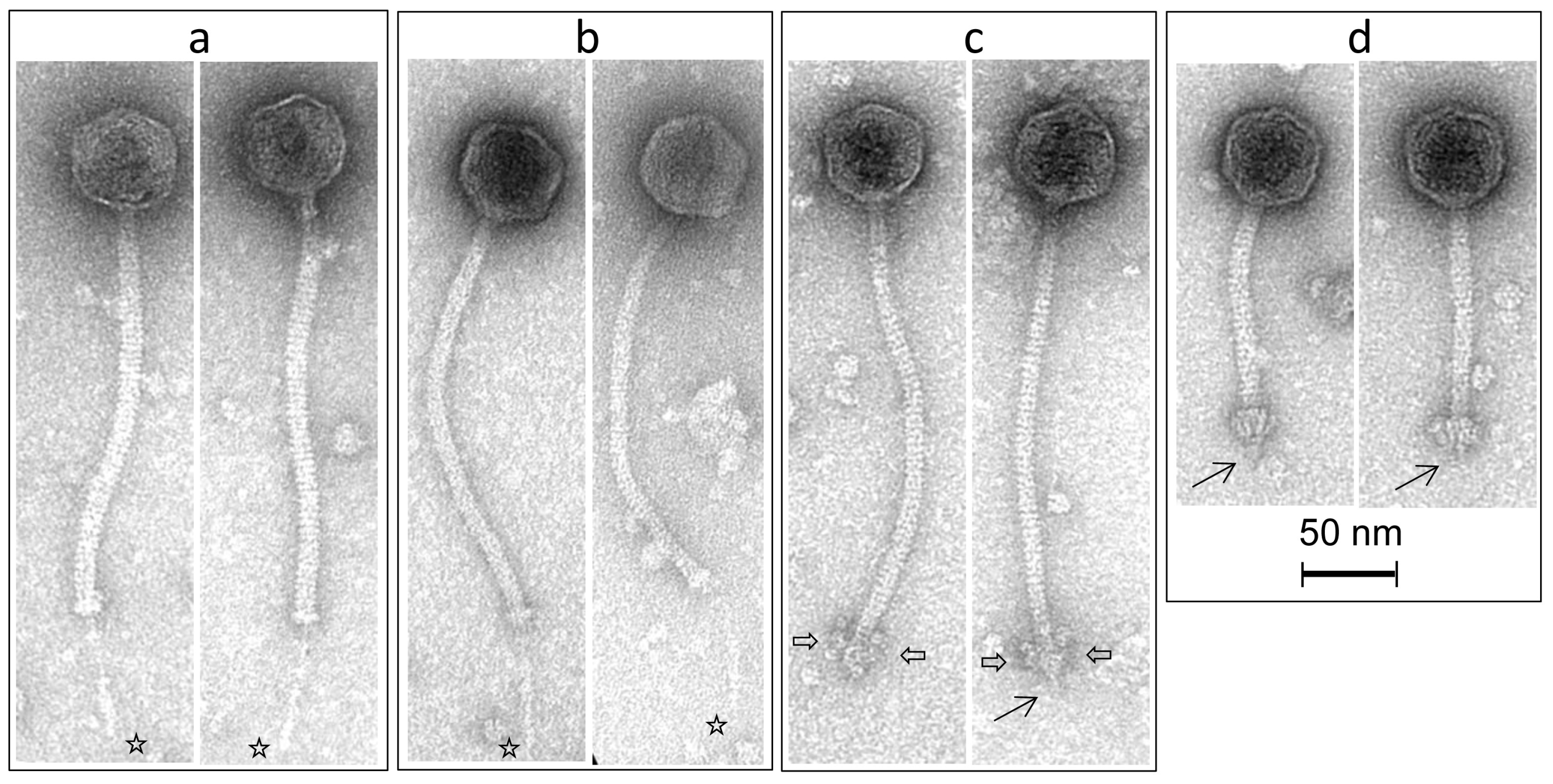
3.4. Morphological Analysis of Phage Isolates
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bolotin, A.; Quinquis, B.; Renault, P.; Sorokin, A.; Ehrlich, S.D.; Kulakauskas, S.; Lapidus, A.; Goltsman, E.; Mazur, M.; Pusch, G.D.; et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004, 22, 1554–1558. [Google Scholar] [CrossRef] [PubMed]
- Delorme, C.; Bartholini, C.; Bolotin, A.; Ehrlich, S.D.; Renault, P. Emergence of a cell wall protease in the Streptococcus thermophilus population. Appl. Environ. Microbiol. 2010, 76, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Le Marrec, C.; van Sinderen, D.; Walsh, L.; Stanley, E.; Vlegels, E.; Moineau, S.; Heinze, P.; Fitzgerald, G.; Fayard, B. Two groups of bacteriophages infecting Streptococcus thermophilus can be distinguished on the basis of mode of packaging and genetic determinants for major structural proteins. Appl. Environ. Microbiol. 1997, 63, 3246–3253. [Google Scholar] [PubMed]
- Desiere, F.; Lucchini, S.; Brussow, H. Comparative sequence analysis of the DNA packaging, head, and tail morphogenesis modules in the temperate cos-site Streptococcus thermophilus bacteriophage sfi21. Virology 1999, 260, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, S.; Desiere, F.; Brussow, H. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: Whole genome comparison of cos-site phages Sfi19 and Sfi21. Virology 1999, 260, 232–243. [Google Scholar] [CrossRef] [PubMed]
- Lucchini, S.; Desiere, F.; Brussow, H. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J. Virol. 1999, 73, 8647–8656. [Google Scholar] [PubMed]
- Achigar, R.; Magadan, A.H.; Tremblay, D.M.; Julia Pianzzola, M.; Moineau, S. Phage-host interactions in Streptococcus thermophilus: Genome analysis of phages isolated in Uruguay and ectopic spacer acquisition in CRISPR array. Sci. Rep. 2017, 7, 43438. [Google Scholar] [CrossRef] [PubMed]
- Lavelle, K.; Murphy, J.; Fitzgerald, B.; Lugli, G.A.; Zomer, A.; Neve, H.; Ventura, M.; Franz, C.M.; Cambillau, C.; van Sinderen, D.; et al. A decade of Streptococcus thermophilus phage evolution in an Irish dairy plant. Appl. Environ. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, B.; Mahony, J.; Hanemaaijer, L.; Neve, H.; Noben, J.P.; Lugli, G.A.; Ventura, M.; Kouwen, T.R.; van Sinderen, D. Global survey and genome exploration of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Front. Microbiol. 2017, 8, 1754. [Google Scholar] [CrossRef] [PubMed]
- Quiberoni, A.; Tremblay, D.; Ackermann, H.W.; Moineau, S.; Reinheimer, J.A. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J. Dairy Sci. 2006, 89, 3791–3799. [Google Scholar] [CrossRef]
- Mahony, J.; van Sinderen, D. Current taxonomy of phages infecting lactic acid bacteria. Front. Microbiol. 2014, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Carminati, D.; Giraffa, G. Evidence and characterization of temperate bacteriophage in Streptococcus salivarius subsp. thermophilus st18. J. Dairy Res. 1992, 59, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Fayard, B.; Haefliger, M.; Accolas, J.P. Interactions of temperate bacteriophages of Streptococcus salivarius subsp. thermophilus with lysogenic indicators affect phage DNA restriction patterns and host ranges. J. Dairy Res. 1993, 60, 385–399. [Google Scholar]
- Mills, S.; Griffin, C.; O’Sullivan, O.; Coffey, A.; McAuliffe, O.E.; Meijer, W.C.; Serrano, L.M.; Ross, R.P. A new phage on the ‘mozzarella’ block: Bacteriophage 5093 shares a low level of homology with other Streptococcus thermophilus phages. Int. Dairy J. 2011, 21, 963–969. [Google Scholar] [CrossRef]
- McDonnell, B.; Mahony, J.; Neve, H.; Hanemaaijer, L.; Noben, J.P.; Kouwen, T.; van Sinderen, D. Identification and analysis of a novel group of bacteriophages infecting the lactic acid bacterium Streptococcus thermophilus. Appl. Environ. Microbiol. 2016, 82, 5153–5165. [Google Scholar] [CrossRef] [PubMed]
- Szymczak, P.; Janzen, T.; Neves, A.R.; Kot, W.; Hansen, L.H.; Lametsch, R.; Neve, H.; Franz, C.M.; Vogensen, F.K. Novel variants of Streptococcus thermophilus bacteriophages are indicative of genetic recombination among phages from different bacterial species. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed]
- Accolas, J.-P.; Spillmann, H. The morphology of six bacteriophages of Streptococcus thermophilus. J. Appl. Bacteriol. 1979, 47, 135–144. [Google Scholar] [CrossRef]
- Lillehaug, D. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 1997, 83, 85–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moineau, S.; Pandian, S.; Klaenhammer, T.R. Evolution of a lytic bacteriophage via DNA acquisition from the lactococcus lactis chromosome. Appl. Environ. Microbiol. 1994, 60, 1832–1841. [Google Scholar] [PubMed]
- Lugli, G.A.; Milani, C.; Mancabelli, L.; van Sinderen, D.; Ventura, M. Megannotator: A user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. Trnascan-se: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Borodovsky, M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999, 27, 3911–3920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped blast and psi-blast: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Soding, J.; Biegert, A.; Lupas, A.N. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005, 33, W244–W248. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, K.; Parkhill, J.; Crook, J.; Horsnell, T.; Rice, P.; Rajandream, M.A.; Barrell, B. Artemis: Sequence visualization and annotation. Bioinformatics 2000, 16, 944–945. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Deasy, T.; Mahony, J.; Neve, H.; Heller, K.J.; van Sinderen, D. Isolation of a virulent Lactobacillus brevis phage and its application in the control of beer spoilage. J. Food Prot. 2011, 74, 2157–2161. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Alqarni, M.; Stockdale, S.; Spinelli, S.; Feyereisen, M.; Cambillau, C.; van Sinderen, D. Functional and structural dissection of the tape measure protein of lactococcal phage TP901-1. Sci. Rep. 2016, 6, 36667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cumby, N.; Reimer, K.; Mengin-Lecreulx, D.; Davidson, A.R.; Maxwell, K.L. The phage tail tape measure protein, an inner membrane protein and a periplasmic chaperone play connected roles in the genome injection process of E. coli phage HK97. Mol. Microbiol. 2015, 96, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Piuri, M.; Hatfull, G.F. A peptidoglycan hydrolase motif within the mycobacteriophage tm4 tape measure protein promotes efficient infection of stationary phase cells. Mol. Microbiol. 2006, 62, 1569–1585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duplessis, M.; Levesque, C.M.; Moineau, S. Characterization of Streptococcus thermophilus host range phage mutants. Appl. Environ. Microbiol. 2006, 72, 3036–3041. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Oliveira, J.; Collins, B.; Hanemaaijer, L.; Lugli, G.A.; Neve, H.; Ventura, M.; Kouwen, T.R.; Cambillau, C.; van Sinderen, D. Genetic and functional characterisation of the lactococcal P335 phage-host interactions. BMC Genomics 2017, 18, 146. [Google Scholar] [CrossRef] [PubMed]
- Mahony, J.; Kot, W.; Murphy, J.; Ainsworth, S.; Neve, H.; Hansen, L.H.; Heller, K.J.; Sorensen, S.J.; Hammer, K.; Cambillau, C.; et al. Investigation of the relationship between lactococcal host cell wall polysaccharide genotype and 936 phage receptor binding protein phylogeny. Appl. Environ. Microbiol. 2013, 79, 4385–4392. [Google Scholar] [CrossRef] [PubMed]
- Dieterle, M.E.; Fina Martin, J.; Duran, R.; Nemirovsky, S.I.; Sanchez Rivas, C.; Bowman, C.; Russell, D.; Hatfull, G.F.; Cambillau, C.; Piuri, M. Characterization of prophages containing “evolved” Dit/Tal modules in the genome of Lactobacillus casei BL23. Appl. Microbiol. Biotechnol. 2016, 100, 9201–9215. [Google Scholar] [CrossRef] [PubMed]
- Murphy, J.; Bottacini, F.; Mahony, J.; Kelleher, P.; Neve, H.; Zomer, A.; Nauta, A.; van Sinderen, D. Comparative genomics and functional analysis of the 936 group of lactococcal Siphoviridae phages. Sci. Rep. 2016, 6, 21345. [Google Scholar] [CrossRef] [PubMed]
- Duplessis, M.; Moineau, S. Identification of a genetic determinant responsible for host specificity in Streptococcus thermophilus bacteriophages. Mol. Microbiol. 2001, 41, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Levesque, C.; Duplessis, M.; Labonte, J.; Labrie, S.; Fremaux, C.; Tremblay, D.; Moineau, S. Genomic organization and molecular analysis of virulent bacteriophage 2972 infecting an exopolysaccharide-producing Streptococcus thermophilus strain. Appl. Environ. Microbiol. 2005, 71, 4057–4068. [Google Scholar] [CrossRef] [PubMed]

| S. thermophilus Strain | Strain Characteristics * | Phage Sensitive |
|---|---|---|
| STP1 | NS | + |
| UCCSt57 | NS | + |
| UCCSt13 | NS | + |
| UCCSt82 | NS | + |
| UCCSt63 | NS | + |
| UCCSt23 | NS | + |
| UCCSt83 | NS | + |
| UCCSt84 | NS | + |
| UCCSt85 | NS | + |
| UCCSt86 | NS | + |
| UCCSt87 | NS | + |
| UCCSt88 | NS | + |
| UCCSt89 | NS | + |
| UCCSt90 | NS | + |
| UCCSt91 | NS | + |
| UCCSt9 | NS | + |
| UCCSt97 | NS | + |
| UCCSt101 | NS | + |
| UCCSt53 | NS | + |
| STP2 | NS | - |
| UCCSt14 | NS | - |
| UCCSt61 | NS | - |
| UCCSt62 | NS | - |
| UCCSt65 | NS | - |
| UCCSt98 | NS | - |
| UCCSt67 | NS | - |
| UCCSt68 | NS | - |
| UCCSt55 | NS | - |
| UCCSt57 | NS | - |
| UCCSt28 | NS | - |
| UCCSt29 | NS | - |
| UCCSt64 | NS | - |
| UCCSt66 | NS | - |
| UCCSt73 | EPS+ | - |
| UCCSt69 | EPS+ | + |
| UCCSt54 | EPS+ | - |
| UCCSt99 | NS | - |
| UCCSt8 | NS | - |
| UCCSt21 | NS | - |
| UCCSt33 | NS | - |
| UCCSt22 | NS | - |
| UCCSt10 | NS | - |
| UCCSt92 | S | + |
| UCCSt51 | S | + |
| UCCSt93 | S | + |
| UCCSt12 | S | + |
| UCCSt94 | S | + |
| UCCSt50 | S | + |
| UCCSt95 | S | + |
| UCCSt96 | S | + |
| UCCSt31 | S/NS | + |
| UCCSt1 | S/NS | + |
| Phage | Whey Sample | Country of Origin | Host Strain | # Strains Infected † | Type | Genome Size (bp) | # ORFs | G&C% | Genbank Accession No. |
|---|---|---|---|---|---|---|---|---|---|
| SW1 | 16 | Italy | UCCSt23 | 1 | cos | 34,821 | 38 | 38.85 | MH892352 |
| SW2 | 13 | Italy | UCCSt86 | 1 | cos | 37,222 | 47 | 38.81 | MH892353 |
| SW3 | 6 | Italy | UCCSt83 | 1 | cos | 37,181 | 45 | 38.64 | MH892354 |
| SW5 | 14 | Italy | UCCSt86 | 1 | cos | 36,836 | 47 | 38.78 | MH973661 |
| SW6 | 8 | Italy | UCCSt95 | 1 | cos | 34,276 | 42 | 38.64 | MH892351 |
| SW7 | 12 | Italy | UCCSt84 | 2 | cos | 35,917 | 42 | 39.11 | MH973662 |
| SW8 | 31 | Mexico | UCCSt88 | 1 | cos | 33,930 | 41 | 39.11 | MH892357 |
| SW9 | 17 | Italy | UCCSt23 | 2 | cos | 33,999 | 38 | 38.71 | MH892358 |
| SW10 | 32 | France | UCCSt23 | 1 | cos | 34,336 | 39 | 38.76 | MH892359 |
| SW21 | 11 | Italy | UCCSt82 | 3 | cos | 35,351 | 39 | 38.78 | MH892356 |
| SW30 | 35 | Austria | UCCSt95 | 1 | cos | 34,158 | 41 | 38.46 | MH892375 |
| SW12 | 29 | Turkey | UCCSt96 | 1 | cos | 35,382 | 43 | 38.75 | MH892361 |
| SW11 | 29 | Turkey | UCCSt92 | 1 | cos | 36,250 | 42 | 38.23 | MH892360 |
| SWK2 | 9 | Italy | UCCSt63 | 3 | cos | 36,594 | 42 | 39.30 | MH892378 |
| SWK6 | 10 | Italy | UCCSt63 | 3 | cos | 34,999 | 38 | 39.01 | MH892382 |
| SWK1 | 9 | Italy | UCCSt63 | 3 | cos | 36,089 | 43 | 39.17 | MH892377 |
| SWK3 | 31 | Mexico | UCCSt63 | 3 | cos | 35,443 | 41 | 38.72 | MH892379 |
| SWK5 | 21 | Italy | UCCSt63 | 2 | cos | 35,849 | 39 | 38.90 | MH892381 |
| SWK4 | 28 | Turkey | UCCSt63 | 3 | cos | 35,341 | 41 | 38.69 | MH892380 |
| SW31 * | 12 | Italy | UCCSt50 | 5 | pac | 36,587 | 41 | 39.24 | MH892383 |
| SW32 * | 14 | Italy | UCCSt50 | 5 | pac | 36,587 | 41 | 39.24 | MH892384 |
| SW33 * | 27 | Argentina | UCCSt50 | 5 | pac | 36,587 | 41 | 39.24 | MH892385 |
| SW13 * | 29 | Turkey | UCCSt50 | 5 | pac | 36,587 | 41 | 39.24 | MH892362 |
| SW14 | 9 | Italy | UCCSt96 | 2 | pac | 35,913 | 46 | 39.87 | MH892363 |
| SW15 | 12 | Italy | UCCSt96 | 1 | pac | 34,289 | 44 | 39.39 | MH892364 |
| SW18 | 8 | Italy | UCCSt93 | 2 | pac | 34,825 | 44 | 39.43 | MH892366 |
| SW1151 | 14 | Italy | UCCSt10 | 1 | pac | 35,056 | 45 | 39.60 | MH892376 |
| SW16 | 7 | Italy | UCCSt97 | 1 | 987 | 32,106 | 45 | 36.95 | MH892350 |
| SW17 | 8 | Italy | UCCSt97 | 1 | 987 | 32,005 | 46 | 36.84 | MH892365 |
| SW20 | 9 | Italy | UCCSt97 | 1 | 987 | 32,567 | 46 | 36.81 | MH892368 |
| SW22 | 13 | Italy | UCCSt97 | 1 | 987 | 31,738 | 45 | 37.35 | MH892369 |
| SW23 | 13 | Italy | UCCSt97 | 1 | 987 | 31,422 | 45 | 37.03 | MH892370 |
| SW25 | 14 | Italy | UCCSt97 | 1 | 987 | 32,013 | 46 | 37.20 | MH892371 |
| SW26 | 15 | Italy | UCCSt97 | 1 | 987 | 31,283 | 44 | 36.98 | MH892372 |
| SW28 | 21 | Italy | UCCSt97 | 1 | 987 | 32,093 | 45 | 36.88 | MH892374 |
| SW4 | 26 | Argentina | UCCSt89 | 1 | 5093 | 34,556 | 47 | 38.15 | MH892355 |
| SW19 | 9 | Italy | UCCSt89 | 1 | 5093 | 35,153 | 46 | 38.10 | MH892367 |
| SW24 | 14 | Italy | UCCSt89 | 1 | 5093 | 32,092 | 42 | 38.31 | MH973663 |
| SW27 | 21 | Italy | UCCSt89 | 1 | 5093 | 33,953 | 45 | 38.25 | MH892373 |
| Whey Sample Number | Country of Origin | Factory # | Starter Used ᴥ | Sampling Year | Phage Positive | Types of Phages (# Propagated Isolates) | # Strains Infected | Phage Titre Range (pfu/mL) |
|---|---|---|---|---|---|---|---|---|
| 1 | Italy | 1 | DS1 | 2017 | No | - | 0 | 0 |
| 2 | Italy | 1 | DS1 | 2017 | No | - | 0 | 0 |
| 3 | Italy | 1 | DS1 | 2017 | No | - | 0 | 0 |
| 4 | Italy | 2 | DS2 | 2017 | No | - | 0 | 0 |
| 5 | Italy | 2 | DS2 | 2017 | No | - | 0 | 0 |
| 6 | Italy | 3 | DS3 | 2017 | Yes | cos (1) | 1 | 102 |
| 7 | Italy | 4 | DS4 | 2017 | Yes | 987 (1) | 1 | 102 |
| 8 | Italy | 5 | DS5 | 2017 | Yes | cos (3), pac (2), 987 (1) | 3 | 102 |
| 9 | Italy | 6 | DS6 | 2017 | Yes | cos (16), pac (3), 5093 (1), 987 (1) | 9 | 102–106 |
| 10 | Italy | 7 | DS7 | 2017 | Yes | cos (1) | 1 | 102 |
| 11 | Italy | 7 | DS7 | 2017 | Yes | cos (1) | 1 | 102 |
| 12 | Italy | 8 | DS8 | 2017 | Yes | cos (8), pac (1) | 4 | 104–106 |
| 13 | Italy | 8 | DS8 | 2017 | Yes | cos (14), pac (1), 987 (2) | 8 | 104–109 |
| 14 | Italy | 9 | DS9 | 2017 | Yes | cos (11), pac (2), 5093 (1), 987 (1) | 8 | 103–107 |
| 15 | Italy | 9 | DS9 | 2017 | Yes | cos (15), 987 (1) | 8 | 103–107 |
| 16 | Italy | 10 | DS10 | 2016 | Yes | cos (3) | 2 | 102–108 |
| 17 | Italy | 10 | DS10 | 2016 | Yes | cos (12) | 3 | 103–105 |
| 18 | Italy | 11 | DS11 | 2016 | No | - | 0 | 0 |
| 19 | Italy | 12 | DS12 | 2017 | No | - | 0 | 0 |
| 20 | Italy | 13 | DS13 | 2017 | Yes | cos (2) | 1 | 107 |
| 21 | Italy | 14 | DS14 | 2016 | Yes | cos (9), 5093 (2), 987 (2) | 8 | 102–106 |
| 22 | Spain | 15 | DS15 | 2017 | Yes | cos (4) | 3 | 106–107 |
| 23 | Spain | 15 | DS15 | 2017 | Yes | cos (5) | 3 | 105–107 |
| 24 | Spain | 16 | DS16 | 2017 | No | - | 0 | 0 |
| 25 | Israel | 17 | DS17 | 2017 | Yes | cos (3) | 2 | 102–104 |
| 26 | Argentina | 18 | DS18 | 2017 | Yes | cos (2), 5093 (1) | 2 | 104–109 |
| 27 | Argentina | 19 | DS19 | 2017 | Yes | cos (4) pac (1) | 2 | 102–103 |
| 28 | Turkey | 20 | DS20 | 2016 | Yes | cos (16) | 6 | 103–107 |
| 29 | Turkey | 21 | DS21 | 2016 | Yes | cos (6), pac (1) | 7 | 102–104 |
| 30 | Russia | 22 | DS22 | 2017 | Yes | cos (1) | 0 | 108 |
| 31 | Mexico | 23 | DS23 | 2016 | Yes | cos (2) | 1 | 102 |
| 32 | France | 24 | DS24 | 2016 | Yes | cos (3) | 2 | 103–107 |
| 33 | Egypt | 25 | DS25 | 2016 | No | - | 0 | 0 |
| 34 | Congo | 26 | DS26 | 2016 | No | - | 0 | 0 |
| 35 | Austria | 27 | DS27 | 2017 | Yes | cos (5) | 1 | 103 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lavelle, K.; Martinez, I.; Neve, H.; Lugli, G.A.; Franz, C.M.A.P.; Ventura, M.; Bello, F.D.; Sinderen, D.V.; Mahony, J. Biodiversity of Streptococcus thermophilus Phages in Global Dairy Fermentations. Viruses 2018, 10, 577. https://doi.org/10.3390/v10100577
Lavelle K, Martinez I, Neve H, Lugli GA, Franz CMAP, Ventura M, Bello FD, Sinderen DV, Mahony J. Biodiversity of Streptococcus thermophilus Phages in Global Dairy Fermentations. Viruses. 2018; 10(10):577. https://doi.org/10.3390/v10100577
Chicago/Turabian StyleLavelle, Katherine, Ines Martinez, Horst Neve, Gabriele A. Lugli, Charles M. A. P. Franz, Marco Ventura, Fabio Dal Bello, Douwe Van Sinderen, and Jennifer Mahony. 2018. "Biodiversity of Streptococcus thermophilus Phages in Global Dairy Fermentations" Viruses 10, no. 10: 577. https://doi.org/10.3390/v10100577
APA StyleLavelle, K., Martinez, I., Neve, H., Lugli, G. A., Franz, C. M. A. P., Ventura, M., Bello, F. D., Sinderen, D. V., & Mahony, J. (2018). Biodiversity of Streptococcus thermophilus Phages in Global Dairy Fermentations. Viruses, 10(10), 577. https://doi.org/10.3390/v10100577






