Abstract
The exo-xis region of lambdoid bacteriophage genomes contains several established and potential genes that are evolutionarily conserved, but not essential for phage propagation under laboratory conditions. Nevertheless, deletion or overexpression of either the whole exo-xis region and important regulatory elements can significantly influence the regulation of phage development. This report defines specific roles for orf60a and orf61 in bacteriophage λ and Φ24B, a specific Shiga toxin-converting phage with clinical relevance. We observed that mutant phages bearing deletions of orf60a and orf61 impaired two central aspects of phage development: the lysis-versus-lysogenization decision and prophage induction. These effects were more pronounced for phage Φ24B than for λ. Surprisingly, adsorption of phage Φ24B on Escherichia coli host cells was less efficient in the absence of either orf60a or orf61. We conclude that these open reading frames (ORFs) play important, but not essential, roles in the regulation of lambdoid phage development. Although phages can propagate without these ORFs in nutrient media, we suggest that they may be involved in the regulatory network, ensuring optimization of phage development under various environmental conditions.
1. Introduction
Bacteriophage λ, and other bacteriophages of the lambdoid class that share a similar genome organization and life cycle, have contributed to many important discoveries in molecular biology, from the regulation of gene expression and DNA replication to macromolecular interactions and novel biological structures [,,]. Some lambdoid phages carry genes encoding toxins that function as virulence factors for several species of pathogenic bacteria. Examples of such bacteria are Shiga toxin-producing Escherichia coli (STEC) strain and enterohemorrhagic E. coli (EHEC) associated with food poisoning outbreaks often accompanied by fatalities in immune-compromised individuals [,,]. As a result, a precise understanding of the developmental regulation of lambdoid phages is important not only for basic science, but also for identifying new leads for drug discovery.
Viral genomes may be considered as compact systems bearing genes that are most necessary and optimized for propagation. This is due to the evolutionary pressure to select minimal genomes that are easy to replicate, and can be effectively packaged into capsids. It is surprising, therefore, that non-essential genes in λ and related phages are still observed. One such region (exo-xis) is found between the well characterized exo and xis genes, and is comprised of several potential open reading frames (ORFs). While the exo-xis region is dispensable for propagation of lambdoid bacteriophages under laboratory conditions [], it is evolutionarily conserved which implicates its possible important role in bacteriophage development [] (Figure 1).
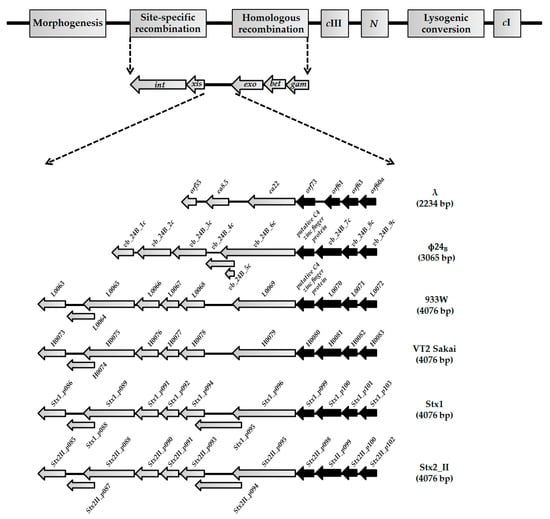
Figure 1.
Genes and open reading frames (ORFs) located between exo and xis genes in genomes of lambdoid bacteriophages: λ (NC_001416), Φ24B (HM208303), 933W (NC_000924), VT2 Sakai (AP000422), Stx1 (NC_004913), and Stx2_II (NC_004914). Black arrows represent highly conserved orf60a–orf73 regions among lambdoid phages (≥70% nucleotide sequence identity). Grey arrows with black borders present genes and ORFs with lower level of identity (<35%) or additional ORFs that occur in the exo-xis regions of Stx bacteriophages.
Since the exo-xis region did not appear to markedly affect lytic development during the first studies, any defined roles for its genetic elements and functional insights into any of gene products were not intensively investigated, and remained almost completely unknown for a considerable period of time. Approximately ten years ago, our team has demonstrated that lysogenization of E. coli cells by phage λ was impaired when the exo-xis region was overexpressed []. Under similar conditions, induction of prophages λ and Φ24B was found to be more effective [,]. The Ea8.5 protein, encoded in this region, appeared to play important role in this regulation [,], and detailed structural studies revealed that it contains a fused homeodomain/zinc-finger fold, suggesting a potential regulatory function []. Intriguingly, when induction of prophages λ and Φ24B was provoked by various agents (mitomycin C or hydrogen peroxide), different expression patterns of genes from the exo-xis region were observed []. This suggested that genes and ORFs from this region may be involved in regulatory processes occurring under, and responding to, various environmental conditions. Oxidative stress appeared to be predominant environmental condition influencing exo-xis mediated phage development []. Therefore, studies on particular genes and ORFs from this region appeared substantiated. Recently, we demonstrated that deletion of orf63 resulted in delayed and less efficient induction of λ and Φ24B prophages. Since this ORF encodes a structured protein, it follows that orf63 is a functional gene [].
In this study of the exo-xis region, we have concentrated on the functions of orf60a and orf61 to obtain the first insights regarding their roles in the regulation of development of lambdoid phages. To make this work compatible with previous reports, λ bacteriophage and the clinically relevant Φ24B bacteriophage were used in this study.
2. Materials and Methods
2.1. Bacterial Strains and Bacteriophages and Plasmids
E. coli MG1655 strain, its derivatives, and bacteriophages used in this work, are listed in Table 1.

Table 1.
Bacterial strains and bacteriophages.
The deletion mutants were constructed as described previously [,] by using E. coli MG1655 (λ) or E. coli MG1655 (Φ24B) strains. The procedure was performed according to the manufacturer’s protocol of the Quick and Easy E. coli Gene Deletion Kit (Gene Bridges, Heidelberg, Germany). In the first step, the nucleotide sequence of orf60a or orf61 has been replaced with FRT-flanked kanamycin resistance cassette. Then, the selection marker was removed in the FLP-recombinase step, leaving 87 nucleotides of the cassette in the place of the original sequence of orf60a or orf61 in the genome of lysogenic E. coli bacteria. All constructs (phage genomes with deletions of either orf60a or orf61) were confirmed by DNA sequencing.
Bacteriophage suspensions were routinely stored in Tris-HCl-Magnesium sulfate buffer (TM buffer; 10 mM Tris-HCl, 10 mM MgSO4, pH 7.2) at 4 °C. E. coli MG1655 strain was selected as a host for bacteriophage infection. Bacteria were cultured in the Luria-Bertani (LB) medium at 30 °C.
2.2. Prophage Induction Experiments
Bacteria lysogenic with tested phages were grown in LB medium at 30 °C, with a shaking, until the OD600 reached 0.1, and then treated with 0.2 µg/mL mitomycin C or 1 mM hydrogen peroxide to induce prophages. Following the induction step, at indicated times (every 30 min), 0.5 mL samples were withdrawn, mixed with chloroform, and centrifuged (2000× g for 5 min at room temperature). Afterwards, serial dilutions were prepared in TM buffer and 2.5 μL of each dilution was spotted onto plates with double-layer LB agar (phage λ) or LB agar supplemented with 2.5 μg/mL chloramphenicol (Φ24B phage), that were prepared according to a procedure described previously []. A separate set of analogous experiments with each lysogenic strain was performed without addition of the induction agent. Such control experiments allowed for estimating the levels of spontaneous prophage induction. Following an overnight incubation of plates at 37 °C, the relative phage titer (PFU/mL) was determined by subtracting the phage titer of the non-induced culture from the phage titer of a respective induced variant.
2.3. One-Step Growth Experiment
To examine the intracellular life cycle of analyzed phages, a procedure described previously [,], was employed, with a few minor modifications. Briefly, bacteria were grown in LB medium at 30 °C, with shaking, until the OD600 reached 0.2. At this stage, 10 mL of bacterial culture was centrifuged (2000× g for 10 min at 4 °C) and the obtained pellet was suspended in 1 mL of LB enriched with 3 mM NaN3 (Sigma-Aldrich, St. Louis, MO, USA). Following a 5 min incubation at 30 °C, the phage lysate was added to an multiplicity of infection (m.o.i.) of 0.05, and then incubated again for 10 min. Afterwards, the sample was washed 3 times with LB supplemented with 3 mM NaN3, and centrifuged each time (3000× g for 10 min at 4 °C). After unadsorbed phages were removed, 250 μL of the suspension was added to 25 mL of LB medium prewarmed to 30 °C (time 0), and cultivated at this temperature with shaking. To estimate the number of infection centers, 0.2 mL culture samples were collected at 5, 10, and 15 min post-infection, and mixed with 0.8 mL indicator bacteria and 2 mL 0.7% top agar (prewarmed to 45 °C), supplemented with MgSO4 (phage λ) or MgSO4 and CaCl2 (phage Φ24B), to a final concentration of 10 mM each. The mixtures were then poured onto LB plates (phage λ) or LB plates enriched with 2.5 μg/mL chloramphenicol (phage Φ24B). The phage titer was determined by collecting 0.5 mL samples that were prepared by shaking in a chloroform mixture followed by centrifugation (2000× g for 5 min). Phage lysates from this step were diluted in TM buffer, and titrated under permissive conditions. Following an overnight incubation at 37 °C, the burst size (number of virions released from single infected cell) was estimated as a ratio of a phage titer to the titer of infection centers.
2.4. Measurement of the Efficiency of Phage Adsorption
Bacteria were grown in LB medium at 30 °C, with shaking, until the OD600 reached 0.1, upon which samples of 1 mL were centrifuged (2000× g for 10 min at 4 °C) and the pellets subsequently dissolved in 0.15 mL of 0.85% NaCl. This mixture was centrifuged (2000× g for 10 min at 4 °C) and pellets were suspended in 0.15 mL LB medium supplemented with 10 mM MgSO4 (phage λ) or 10 mM MgSO4 + 10 mM CaCl2 (phage Φ24B). Following a 15 min incubation at 30 °C, tested bacteriophages were added to an m.o.i. of 0.1, and such mixtures were incubated at 30 °C. At specified times after the addition of phage lysate, the samples were withdrawn, centrifuged in a microcentrifuge (12,000× g for 1 min at room temperature) and supernatants were titrated on indicator E. coli bacteria. Plates were incubated at 37 °C overnight. In this method, the efficiency of phage adsorption was estimated by using the ln (Pt/P0) equation described previously [], where Pt and P0 are phage concentrations at indicated times and time zero (immediately after addition of bacteriophages), respectively.
2.5. Efficiency of Lysogenization
Host E. coli bacteria were grown in LB medium at 30 °C, with shaking, until the OD600 reached 0.2. Next, 1 mL of culture was centrifuged (2000× g for 10 min at room temperature), and the obtained pellet was dissolved in 1 mL of TM buffer and, again, centrifuged (2000× g for 5 min at room temperature) and suspended. Following a short incubation at 30 °C, bacteriophages were added to an m.o.i. of 1, 5, and 10 and, next, the mixtures were again incubated at 30 °C. Serial dilutions were prepared in TM buffer and 40 μL of each dilution was poured onto LB plates. After overnight incubation at 37 °C, 96 colonies were passaged separately each in a well of a 96-well plate with 200 μL of LB medium. Each plate was shaken at 37 °C until the OD600 reached 0.1 and irradiated with UV light at 50 J/m2 for 20 seconds to induce prophages, followed by an incubation at 37 °C for 1 h. After induction, lysogens were mixed with chloroform, centrifuged (2000× g for 10 min at 4 °C), and 2.5 μL of each top layer was spotted on a freshly prepared plate with double layer LB agar (phage λ) or LB agar supplemented with 2.5 μg/mL chloramphenicol (phage Φ24B). Plates were incubated at 37 °C overnight. The efficiency of lysogenization was determined as a percent of lysogens among all tested 96 bacterial colonies. Each experiment was repeated three times. Lysogens were also verified by testing their resistance to superinfection, as indicated previously [].
2.6. Survival of Cells after Bacteriophage Infection
To determine the survival of the wild-type E. coli strain after phage infection, a previously published method [,] was used with only minor modifications. Briefly, bacteria were grown in LB medium at 30 °C, with shaking, until the OD600 reached 0.2. Following centrifugation (2000× g for 10 min at 4 °C), pellets were washed with 0.85% NaCl, and then suspended in 1.2 mL of LB medium enriched with 10 mM MgSO4 (phage λ) or 10 mM MgSO4 + 10 mM CaCl2 (phage Φ24B). Next, samples were incubated for 15 min at 30 °C, and tested bacteriophages were added to an m.o.i. of 1, 5 or 10. Following another incubation at 30 °C, serial dilutions of the initial samples were prepared in 0.85% NaCl, and 40 μL of each dilution was plated on LB agar plates and incubated at 37 °C overnight. The number of viable bacterial cells was calculated on the basis of counted colonies. The fraction of surviving bacterial cells in a population infected with the tested phages was calculated in relation to the control experiment, in which TM buffer was used instead of phage lysate.
2.7. Measurement of Bacterial Viability during Prophage Induction Experiments
Bacterial viability was measured following a published procedure that is briefly described here []. E. coli lysogenic with tested phages was grown in LB medium at 30 °C, with shaking, until the OD600 reached 0.1, and then treated with 1 mM hydrogen peroxide as an induction agent. At various times post-induction, samples of 2 × 108 cells/mL were withdrawn and centrifuged (8000× g for 10 min at room temperature). Pellets were washed and suspended in 0.85% NaCl. Samples prepared were stained with LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes, Eugene, OR, USA), which provides an estimate of live bacteria under the assumption they have intact cell membranes. Following the manufacturer’s protocol, fluorescence measurements were performed in a microplate reader using an excitation wavelength of 485 nm and emission wavelengths of 530 and 630 nm. Presented values indicate the percent of live bacteria normalized to results of control experiments (cultures without induction agent) which, at each time, were assumed as 100% live bacteria.
3. Results
3.1. Sequences of orf60a and orf61, and Their Putative Products, Are Conservative among Lambdoid Phages
Since the exo-xis region presents a similar organization and sequence conservation among lambdoid phages [,,,], we began by determining if sequences of orf60a and orf61, as well as putative products of these ORFs, were similar among phages from this group. We observed that scores of pairwise nucleotide alignments of orf60a indicate generally high similarities (>90% identity) between this ORF and six lambdoid phages (Table 2).

Table 2.
Scores of pairwise alignments of the nucleotide sequences of orf60a from six analyzed lambdoid phages: λ phage (NC_001416), Φ24B phage (HM208303), 933W phage (NC_000924), VT2 Sakai phage (AP000422), Stx1 converting phage (NC_004913), and Stx2 converting phage II (NC_004914).
A similar analysis of the predicted amino acid sequences of the putative Orf60a protein demonstrated a high degree of similarity among tested phages (>90% identity) (Table 3).

Table 3.
Scores of pairwise alignments of the predicted amino acid sequences of Orf60a from six analyzed lambdoid phages: λ phage (NC_001416), Φ24B phage (HM208303), 933W phage (NC_000924), VT2 Sakai phage (AP000422), Stx1 converting phage (NC_004913), and Stx2 converting phage II (NC_004914).
Analogous analyses of orf61 indicated even higher similarities of both nucleotide and amino acid sequences. For this ORF, and its putative product, nucleotide, and amino acids similarities were high for all six tested phages (Table 4 and Table 5, respectively). From these comparisons, it is possible that orf60a and orf61 are true genes that encode functional proteins (Table 3 and Table 5, respectively).

Table 4.
Scores of pairwise alignments of the nucleotide sequences of orf61 from six analyzed lambdoid phages: λ phage (NC_001416), Φ24B phage (HM208303), 933W phage (NC_000924), VT2 Sakai phage (AP000422), Stx1 converting phage (NC_004913), and Stx2 converting phage II (NC_004914).

Table 5.
Scores of pairwise alignments of the predicted amino acid sequences of Orf61 from six analyzed lambdoid phages: λ phage (NC_001416), Φ24B phage (HM208303), 933W phage (NC_000924), VT2 Sakai phage (AP000422), Stx1 converting phage (NC_004913), and Stx2 converting phage II (NC_004914).
3.2. Influence of orf60a and orf61 on Prophage Induction with Various Inductors
Either deletion or overexpression of the exo-xis region affects the induction of lambdoid prophages [,,,]. Here, we have tested if specific deletions of orf60a or orf61, alone, can alter induction of λ and Φ24B prophages by mitomycin C or hydrogen peroxide (Figure 2).
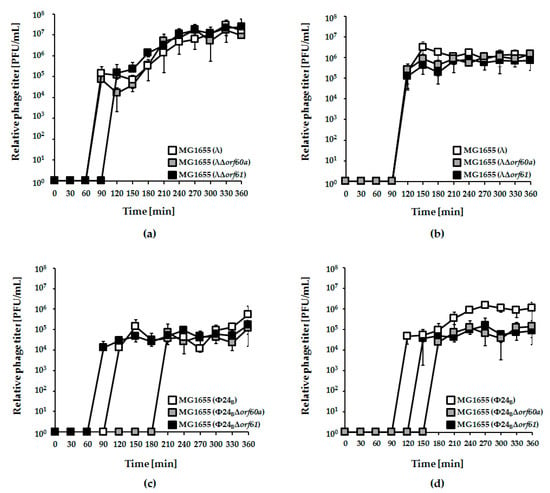
Figure 2.
Development of bacteriophages λ (panels (a) and (b)) and Φ24B (panels (c) and (d)), either wild-type (white squares), Δorf60a (grey squares), or Δorf61 (black squares) after induction of lysogenic Escherichia coli MG1655 strain with 0.2 µg/mL mitomycin C (panels (a) and (c)) or 1 mM hydrogen peroxide (panels (b) and (d)). The presented results are mean values from three independent experiments (biological samples), with error bars indicating the standard deviation (S.D.).
Irrespective of the kind of the inductor used in experiments, deletion of either orf60a or orf61 had only minor effects on induction of λ prophage and phage development (Figure 2, panels a and b). For Φ24B, however, the effects were more dramatic. Induction of the Φ24BΔorf60a prophage by both mitomycin C and hydrogen peroxide was significantly delayed, and the efficiency of lytic development was lower, relative to wild-type phage (Figure 2, panels c and d). Therefore, orf60a appears to play a significant role in the control of prophage induction. In accordance with the induction experiments, we have also observed higher survival of E. coli host cells after induction of mutant Φ24B prophages, relative to their wild-type counterparts (Figure 3). Although an opposite trend was observed at time 1 h in experiments with λ and at time 3 h with Φ24B, these differences were not statistically significant (Figure 3), thus, we conclude as described above.
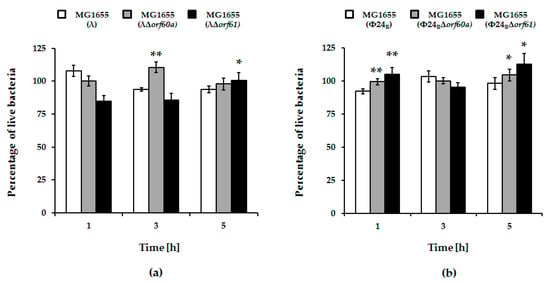
Figure 3.
Percentage of live E. coli MG1655 cells lysogenic with λ (panel (a)) and Φ24B (panel (b)), either wild-type (white columns), Δorf60a (grey columns), or Δorf61 (black columns) during prophages induction with 1 mM hydrogen peroxide. The presented results are mean values from three biological experiments with error bars indicating S.D. Statistical analysis was performed by using Student’s t-test. Significant differences between fractions of bacterial cells lysogenic with wild-type phages and their deletion mutants are marked by asterisks, p < 0.05 (*) or p < 0.01 (**).
3.3. Effects of orf60a and orf61 Deletions on Phage Infection
Next, we tested the effects of orf60a or orf61 deletions on infection of host cells by λ and Φ24B phages. In one-step growth experiments, no significant effects were observed (Figure 4).
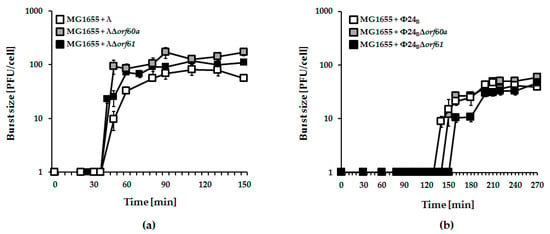
Figure 4.
One-step growth experiments with λ (panel (a); white squares), Φ24B (panel (b); white squares), and recombinant phage mutants bearing deletions of orf60a (panels (a) and (b); grey squares) or orf61 (panels (a,b); black squares), infecting E. coli MG1655 host. Mean values from three independent experiments with error bars indicating S.D. are shown.
However, when post-infection host survival was assessed, it was significantly higher for mutant Φ24B phages (either devoid of orf60a or orf61) relative to wild-type phage (Figure 5; panel b). Again, similar deletions tested with phage λ were less pronounced (Figure 5; panel a).
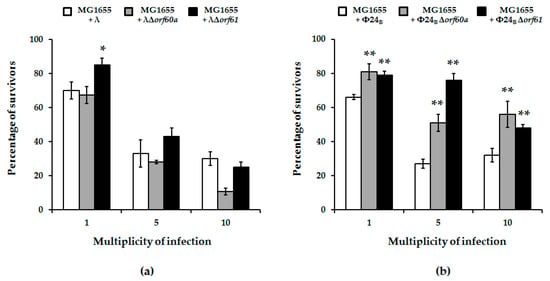
Figure 5.
Percentage of survivors of E. coli MG1655 bacteria after infection of wild-type phages: λ (panel (a); white columns) and Φ24B (panel (b); white columns) or their deletion mutants: λΔorf60a (panel (a); grey columns), λΔorf61 (panel (a); black columns), Φ24BΔorf60a (panel (b); grey columns), and Φ24BΔorf61 (panel (b); black columns). Results are presented as mean values ± S.D. from three biological experiments. A t-test was performed for results from each multiplicity of infection (m.o.i.). The significance of differences between fractions of bacterial cells infected with wild-type phages and their deletion mutants are marked by asterisks, p < 0.05 (*) or p < 0.01 (**).
To explore possible causes for enhanced bacterial survival upon mutant phage infection, we assessed the efficiency of lysogenization on host cells. Formation of lysogenes was more effective in the absence of orf60a or orf61. This was true for both λ and Φ24B phages, but the effects were more pronounced in the case of Φ24B (Figure 6). We therefore conclude that these ORFs have regulatory roles that determine the decision to enter the lytic or lysogenic stage of bacteriophage development.
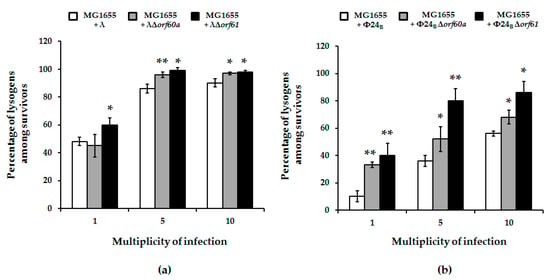
Figure 6.
Fraction of lysogens among survivors of E. coli MG1655 bacteria after infection with wild-type bacteriophages: λ (panel (a); white columns) and Φ24B (panel (b); white columns) or their deletion mutants: λΔorf60a (panel (a); grey columns), λΔorf61 (panel (a); black columns), Φ24BΔorf60a (panel (b); grey columns), and Φ24BΔorf61 (panel (b); black columns). Results are presented as mean values ± S.D. from three independent experiments. A t-test was performed for results from each m.o.i. Statistically significant differences between wild-type phage and its deletion mutants are marked by asterisks, p < 0.05 (*) or p < 0.01 (**).
3.4. Adsorption of Phage Φ24B is Impaired in the Absence of orf60a or orf61
The earliest stages of infection depend on efficient adsorption of phage on the host cells. No significant effects of deletion of orf60a or orf61 could be found in phage λ (Figure 7; panel a). However, adsorption of Φ24B phages devoid of either orf60a or orf61 was significantly impaired (Figure 7; panel b). Together, the results suggest that Orf60a and Orf61 proteins may be involved in the process of the virion assembly, with impairment of this process leading to the formation of partially defective virions that are less effective at adsorbing onto host cells.
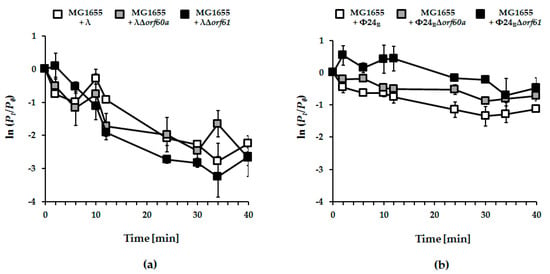
Figure 7.
Kinetics of adsorption of lambdoid bacteriophages: λ (panel (a); white squares), Φ24B (panel (b); white squares) and their mutants bearing deletions of orf60a (panels (a) and (b); grey squares) or orf61 (panels (a) and (b); black squares) on E. coli MG1655 host. Tested bacteriophages were added to bacterial cell suspension to m.o.i. of 0.1. Ratios of unadsorbed bacteriophages were logarithmically transformed according to the ln (Pt/P0) equation, where Pt and P0 are phage concentrations at tested times and time zero, respectively. The presented results are mean values from three biological experiments with S.D. indicated by error bars.
4. Discussion
While lambdoid bacteriophages have been studied for decades, it is remarkable that there are still many genes, particularly in the exo-xis region, whose roles remain unknown. Given that many genes are evolutionarily conserved, it stands to reason that many, if not all, serve important functions in phage development. Likewise, the exo-xis genes also then present several paths towards the development of new therapeutics that target pathogenic/toxicogenic bacteria that are lysogenic for phages like Φ24B.
In this report, we have continued an investigation of genes in the exo-xis region. Previous studies demonstrated that this region, although not essential for phage lytic propagation under laboratory conditions, was still important for precise control of some steps of phage development [,,]. The Ea8.5 protein, for example, participates in the control of gene expression, and its structure suggests regulatory functions [,,]. The orf63 gene also encodes a protein, and appears to control prophage induction and regulation of expression of genes from the exo-xis region []. In this work, we investigated effects of deletions of orf60a and orf61 on development of phage and Shiga toxin-converting phage Φ24B. Our results indicate that these ORFs significantly influence efficiency and timing of prophage induction, as well as the efficiency of lysogenization. Interestingly, differences between mutant and wild-type phages were more pronounced in Φ24B than in λ. When analyzing results of experiments performed to assess efficiency of lysogenization, one might argue that higher fractions of lysogens detected among bacterial cells which survived infection with orf60a and orf61 mutants in comparison to wild-type phages (Figure 6) might result from higher effectivity of adsorption on host cells or less efficient lytic development, rather than actual differences in efficiency of forming prophages. However, contrary to such predictions, we have demonstrated that the mutant phages adsorb on E. coli cells less efficiently than wild-type viruses (Figure 7), and their lytic development is not impaired (Figure 4). Therefore, we propose that lysogenization is more effective in the absence of functional orf60a and orf61 regions.
Although detailed molecular mechanisms of functions of orf60a and orf61 products remain to be elucidated, we speculate that they are involved in the control of gene expression. Our attempts to investigate structures of products of orf60a and orf61 were, as yet, unsuccessful, due to problems with obtaining milligram quantities of soluble folded proteins that were suitable for biochemical and structural studies. However, the observed effects of deletions of these ORFs suggest that they might potentially participate in regulatory networks controlling prophage induction and the lysis-versus-lysogenization decision. Although we could not demonstrate directly that effects of dysfunctions of orf60a and orf61 are due to expression of diffusible gene products (overexpression of these ORFs from a plasmid resulted in production of insoluble protein aggregates), there are some suggestions that these genetic elements code for real protein products. The presence of orf60a- and orf61-derived transcripts could be detected by RT-qPCR []. Since amounts of such transcripts were different under various growth conditions and at various times after initiation of phage lytic development [], one might suggest that the expression levels of orf60a and orf61 can be of importance in modulation of some regulatory processes. Moreover, when λ ORFs were cloned into Gateway vectors, and protein–protein interactions were tested in the yeast two-hybrid system, interactions between the product of orf61 and those of int and orf-314 were detected []. Then, experiments with ribosome profiling, aimed to study the network of proteins encoded by bacteriophage λ, indicated the presence of orf60a and orf61 protein products in E. coli cells after prophage induction in the lysogenic host []. Therefore, one may conclude that these ORFs code for specific proteins. We are aware that still this is not a proof that effects observed in our experiments with mutants in orf60a and orf61 were due to the lack of appropriate diffusible molecules. Nevertheless, although we cannot exclude that deletion of either orf60a or orf61, or both, could cause changes in the phage genome structure or alter expression of other genes, which would result in changes of bacteriophage developed we observed in our experiments, we suggest that, in the light of results presented in this paper, as well as in previously published articles [,,] demonstrating actual expression of both tested ORFs at levels of RNAs and proteins, the hypothesis that orf60a and orf61 code for functional proteins is more likely.
One of the most intriguing aspects of our study on the effects orf60a or orf61 deletion was observed at the adsorption phase of Φ24B phage on host cells. One possible hypothesis leading from this observation is that putative products of these ORFs are involved in the control of either expression of particular genes coding for capsid proteins, or participate directly in macromolecular interactions at critical stages during the formation of functional virions. This hypothesis is supported by observations that virions of Φ24B (as well as some other Shiga toxin-converting phages) are significantly more sensitive to UV irradiation than bacteriophage λ, most probably due to the less stable structure of the virion of the former phage which is, thus, more susceptible for damage []. If so, precise control of the virion assembly might be particularly important for Φ24B, and any disturbance in this process might result in significant loss of some functions, like adsorption on host cells.
5. Conclusions
In this study of exo-xis region of lambdoid phages, we have observed that deletions of orf60a and orf61 dysregulate prophage induction and alter the lysis-versus-lysogenization decision of phages λ and Φ24B. Impaired adsorption of virions on host cells was also observed, and specific to phage Φ24B. An explanation for these effects, possibly related to altered gene expression or macromolecular interactions, will require further experiments with purified and active Orf60a and Orf61 proteins.
Author Contributions
Conceived and designed the experiments, A.D., S.B., B.N.-F., A.W. and G.W.; Methodology, S.B. and B.N.-F.; Performed the experiments, A.D.; Analyzed sequences of phage genomes, S.B. and B.N.-F.; Performed the protein analyses, L.W.D.; Prepared the preliminary prophage induction experiments, G.T. and A.N.; Analyzed the data and performed the statistical analysis, A.D., S.B., B.N.-F.; Contributed reagents/materials/analysis tools, A.D., S.B. and B.N.-F; Funding Acquisition, A.W., L.W.D.; Writing—Original Draft Preparation, G.W.
Funding
This research was funded by the National Science Center (Poland), grants no. UMO-2013/09/B/NZ2/02366 and UMO-2015/17/B/NZ9/01724 to A.W, and Natural Sciences and Engineering Research Council (Canada) Discovery Grant 388053 to L.W.D.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ptashne, M. A Genetic Switch: Phage Lambda Revisited, 3rd ed.; Cold Spring Harbor Laboratory Press: Laurel Hollow, NY, USA, 2004; ISBN 0 87969 716 4. [Google Scholar]
- Wegrzyn, G.; Wegrzyn, A. Genetic switches during bacteriophage lambda development. Prog. Nucleic Acid Res. Mol. Biol. 2005, 79, 1–48. [Google Scholar] [PubMed]
- Wegrzyn, G.; Licznerska, K.; Wegrzyn, A. Phage λ—New insights into regulatory circuits. Adv. Virus Res. 2012, 82, 155–178. [Google Scholar] [PubMed]
- Mauro, S.A.; Koudelka, G.B. Shiga toxin: Expression, distribution, and its role in the environment. Toxins 2011, 3, 608–625. [Google Scholar] [CrossRef] [PubMed]
- Hunt, J.M. Shiga toxin-producing Escherichia coli (STEC). Clin. Lab. Med. 2010, 30, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Gyles, C.L. Shiga toxin-producing Escherichia coli: An overview. J. Anim. Sci. 2007, 85, E45–E62. [Google Scholar] [CrossRef] [PubMed]
- Sergueev, K.; Court, D.; Reaves, L.; Austin, S. E. coli cell-cycle regulation by bacteriophage lambda. J. Mol Biol. 2002, 324, 297–307. [Google Scholar] [CrossRef]
- Bloch, S.; Nejman-Falenczyk, B.; Los, J.M.; Baranska, S.; Lepek, K.; Felczykowsk, A.; Los, M.; Wegrzyn, G.; Wegrzyn, A. Genes from the exo-xis region of λ and Shiga toxin-converting bacteriophages influence lysogenization and prophage induction. Arch. Microbiol. 2013, 195, 693–703. [Google Scholar] [CrossRef] [PubMed]
- Los, J.M.; Los, M.; Wegrzyn, A.; Wegrzyn, G. Role of the bacteriophage lambda exo-xis region in the virus development. Folia Microbiol. 2008, 53, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.; Nejman-Falenczyk, B.; Dydecka, A.; Łoś, J.M.; Felczykowska, A.; Wegrzyn, A.; Wegrzyn, G. Different expression patterns of genes from the exo-xis region of bacteriophage λ and Shiga toxin-converting bacteriophage Φ24B following infection or prophage induction in Escherichia coli. PLoS ONE 2014, 9, e108233. [Google Scholar] [CrossRef] [PubMed]
- Kwan, J.J.; Smirnova, E.; Khazai, S.; Evanics, F.; Maxwell, K.L.; Donaldson, L.W. The solution structures of two prophage homologues of the bacteriophage λ Ea8.5 protein reveal a newly discovered hybrid homeodomain/zinc-finger fold. Biochemistry 2013, 52, 3612–3614. [Google Scholar] [CrossRef] [PubMed]
- Licznerska, K.; Dydecka, A.; Bloch, S.; Topka, G.; Nejman-Falenczyk, B.; Wegrzyn, A.; Wegrzyn, G. The role of the exo-xis region in oxidative stress-mediated induction of Shiga toxin-converting prophages. Oxid. Med. Cell. Longev. 2016, 2016, 8453135. [Google Scholar] [CrossRef] [PubMed]
- Dydecka, A.; Bloch, S.; Rizvi, A.; Perez, S.; Nejman-Falenczyk, B.; Topka, G.; Gasior, T.; Necel, A.; Wegrzyn, G.; Donaldson, L.W.; et al. Bad phages in good bacteria: Role of the mysterious orf63 of λ and Shiga toxin-converting Φ24B bacteriophages. Front. Microbiol. 2017, 8, 1618. [Google Scholar] [CrossRef] [PubMed]
- Nejman-Falenczyk, B.; Bloch, S.; Licznerska, K.; Dydecka, A.; Felczykowska, A.; Topka, G.; Wegrzyn, A.; Wegrzyn, G. A small, microRNA-size, ribonucleic acid regulating gene expression and development of Shiga toxin-converting bacteriophage Φ24B. Sci. Rep. 2015, 5, 10080. [Google Scholar] [CrossRef] [PubMed]
- Jensen, K.F. The Escherichia coli K-12 wild types W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993, 175, 3401–3407. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Duda, R.L. Bacteriophage lambda PaPa: Not a mother of all lambda phages. Science 1993, 258, 1145–1148. [Google Scholar] [CrossRef]
- Allison, H.E.; Sergeant, M.J.; James, C.E.; Saunders, J.R.; Smith, D.L.; Sharp, R.J.; Marks, T.S.; McCarthy, A.J. Immunity profiles of wild-type and recombinant shiga-like toxin-encoding bacteriophages and characterization of novel double lysogens. Infect. Immun. 2003, 71, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Loś, J.M.; Golec, P.; Wegrzyn, G.; Wegrzyn, A.; Loś, M. Simple method for plating Escherichia coli bacteriophages forming very small plaques or no plaques under standard conditions. Appl. Environ. Microbiol. 2008, 74, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, I.N. Bacteriophage adsorption rate and optimal lysis time. Genetics. 2008, 180, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Wegrzyn, G.; Glass, R.E.; Thomas, M.S. Involvement of the Escherichia coli RNA polymerase α subunit in transcriptional activation by the bacteriophage λ CI and CII proteins. Gene 1992, 122, 1–7. [Google Scholar] [CrossRef]
- Schmidt, H. Shiga-toxin-converting bacteriophages. Res. Microbiol. 2001, 152, 687–695. [Google Scholar] [CrossRef]
- Rajagopala, S.V.; Casjens, S.; Uetz, P. The protein interaction map of bacteriophage lambda. BMC Microbiol. 2011, 11, 213. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jiang, H.; Gu, Z.; Roberts, J.W. High-resolution view of bacteriophage lambda gene expression by ribosome profiling. Proc. Natl. Acad. Sci. USA 2013, 110, 11928–11933. [Google Scholar] [CrossRef] [PubMed]
- Bloch, S.; Nejman-Falenczyk, B.; Topka, G.; Dydecka, A.; Licznerska, K.; Narajczyk, M.; Necel, A.; Wegrzyn, A.; Wegrzyn, G. UV-Sensitivity of Shiga Toxin-Converting Bacteriophage Virions Φ24B, 933W, P22, P27 and P32. Toxins 2015, 7, 3727–3739. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).