Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacteria, Phages, and Culture Media
2.2. Phage Genome Sequencing and Analysis
2.3. Multilocus Sequence Typing (MLST) of E. faecalis Strains
2.4. Examination of Antibacterial Activity of E. faecalis to S. agalactiae
2.5. Analysis of Phage Lytic Activity
2.6. Analysis of Bacterial Densities in S. agalactiae and E. faecalis Coculture with Phage Mixtures
2.7. Detection of Bacteria on Chromogenic Selective Agar after S. agalactiae and E. faecalis Coculture with Phage Mixtures
2.8. Statistical Analysis
3. Results and Discussion
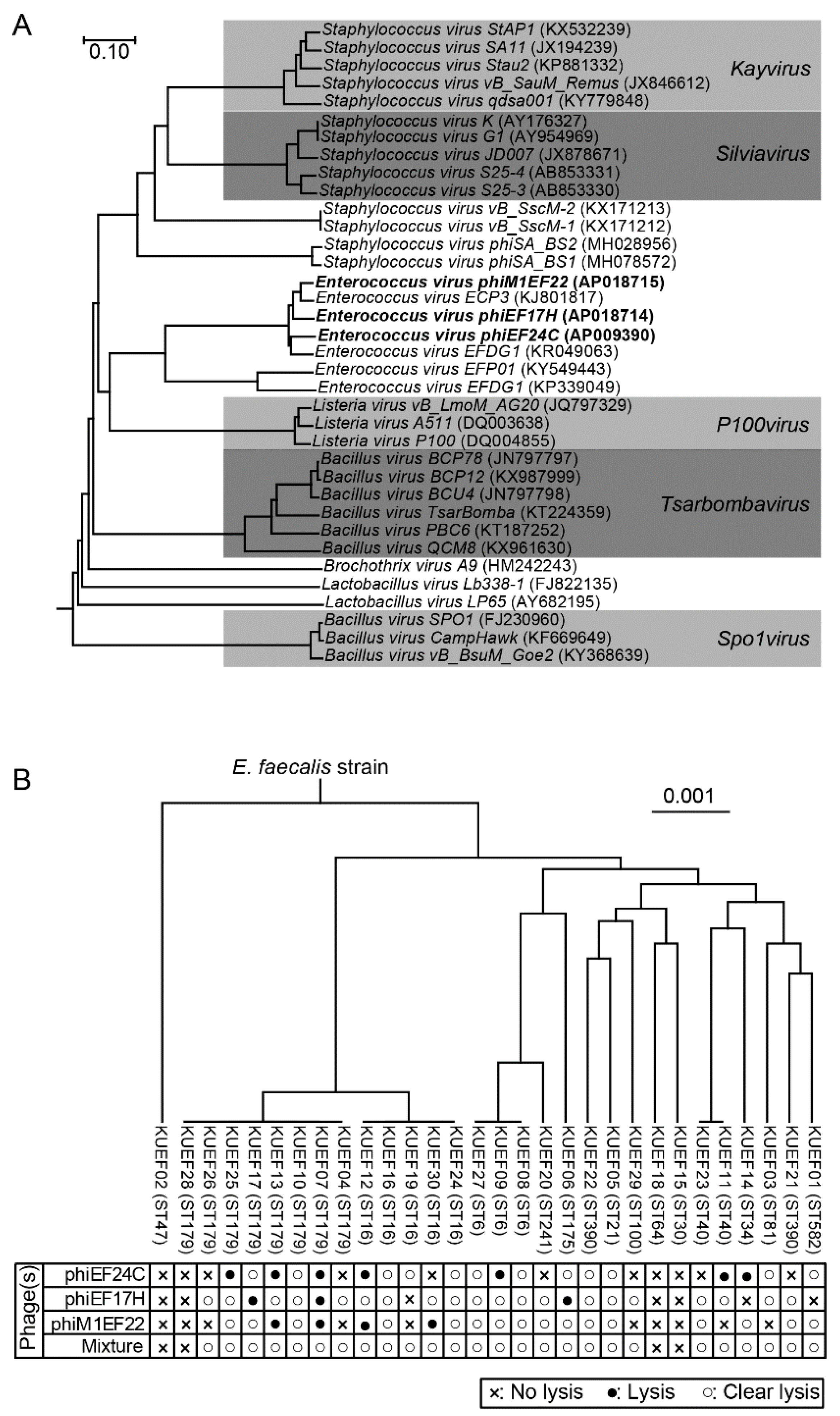
3.1. Phage Characteristics
3.2. Characteristics of E. faecalis Strains Isolated from Vaginal Swabs
3.3. Phage Lytic Spectrum and Phage Mixture
3.4. Effect of Phage Mixture on S. agalactiae and E. faecalis Cell Densities in Experimental Enrichment Cultures
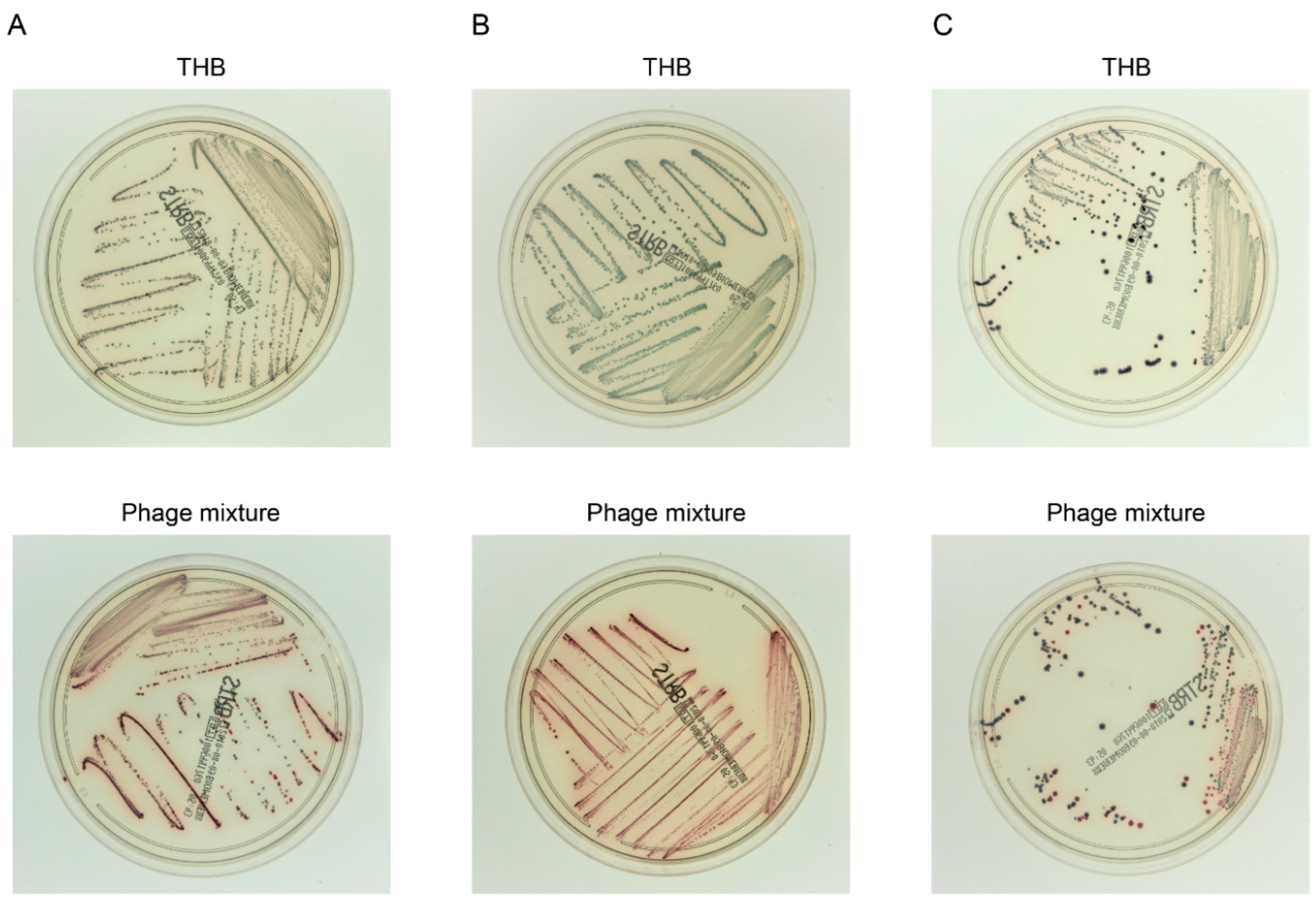
3.5. Efficient Detection of S. agalactiae after Experimental Enrichment Culture in the Presence of the Phage Mixture
3.6. Phage Application Potential in the Clinical Setting
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Morgan, J.A.; Cooper, D.B. Pregnancy, group b streptococcus. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2018; (updated on 11 February 2018). [Google Scholar]
- Edmond, K.M.; Kortsalioudaki, C.; Scott, S.; Schrag, S.J.; Zaidi, A.K.; Cousens, S.; Heath, P.T. Group b streptococcal disease in infants aged younger than 3 months: Systematic review and meta-analysis. Lancet 2012, 379, 547–556. [Google Scholar] [CrossRef]
- Cagno, C.K.; Pettit, J.M.; Weiss, B.D. Prevention of perinatal group b streptococcal disease: Updated cdc guideline. Am. Fam. Physician 2012, 86, 59–65. [Google Scholar] [PubMed]
- Rosa-Fraile, M.; Spellerberg, B. Reliable detection of group b streptococcus in the clinical laboratory. J. Clin. Microbiol. 2017, 55, 2590–2598. [Google Scholar] [CrossRef] [PubMed]
- Dunne, W.M., Jr.; Holland-Staley, C.A. Comparison of nna agar culture and selective broth culture for detection of group b streptococcal colonization in women. J. Clin. Microbiol. 1998, 36, 2298–2300. [Google Scholar] [PubMed]
- Park, C.J.; Vandel, N.M.; Ruprai, D.K.; Martin, E.A.; Gates, K.M.; Coker, D. Detection of group b streptococcal colonization in pregnant women using direct latex agglutination testing of selective broth. J. Clin. Microbiol. 2001, 39, 408–409. [Google Scholar] [CrossRef] [PubMed]
- Binghuai, L.; Yanli, S.; Shuchen, Z.; Fengxia, Z.; Dong, L.; Yanchao, C. Use of maldi-tof mass spectrometry for rapid identification of group b streptococcus on chromid strepto b agar. Int. J. Infect. Dis. 2014, 27, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Baden, M.; Higashiyama, T.; Ikemoto, T.; Okada, Y. Evaluation of direct latex agglutination of selective broth for detection of group b streptococcal carriage in pregnant women. J. Jpn. Soc. Clin. Microbiol. 2016, 26, 7–13. [Google Scholar]
- Kurtböke, D. Actinophages as indicators of actinomycete taxa in marine environments. Antonie Van Leeuwenhoek 2005, 87, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Muldoon, M.T.; Teaney, G.; Li, J.; Onisk, D.V.; Stave, J.W. Bacteriophage-based enrichment coupled to immunochromatographic strip-based detection for the determination of salmonella in meat and poultry. J. Food Prot. 2007, 70, 2235–2242. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Coppenhagen-Glazer, S.; Shlezinger, M.; Kott-Gutkowski, M.; Adini, O.; Beyth, N.; Hazan, R. Complete genome sequence of enterococcus bacteriophage eflk1. Genome Announ. 2015, 3, e01308–e01315. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Rashel, M.; Maeda, Y.; Takemura, I.; Sugihara, S.; Akechi, K.; Muraoka, A.; Wakiguchi, H.; Matsuzaki, S. Isolation and characterization of a novel enterococcus faecalis bacteriophage phief24c as a therapeutic candidate. FEMS Microbiol. Lett. 2008, 278, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Gelman, D.; Shlezinger, M.; Dessal, A.L.; Coppenhagen-Glazer, S.; Beyth, N.; Hazan, R. Defeating antibiotic- and phage-resistant enterococcus faecalis using a phage cocktail in vitro and in a clot model. Front. Microbiol. 2018, 9, 326. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Rashel, M.; Takemura, I.; Wakiguchi, H.; Matsuzaki, S. In silico and in vivo evaluation of bacteriophage phief24c, a candidate for treatment of Enterococcus faecalis infections. Appl. Environ. Microbiol. 2008, 74, 4149–4163. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, J.; Takemura, I.; Satoh, M.; Kato, S.; Ujihara, T.; Akechi, K.; Matsuzaki, S.; Daibata, M. Improved adsorption of an Enterococcus faecalis bacteriophage phief24c with a spontaneous point mutation. PLoS ONE 2011, 6, e26648. [Google Scholar] [CrossRef] [PubMed]
- de la Rosa, M.; Perez, M.; Carazo, C.; Pareja, L.; Peis, J.I.; Hernandez, F. New granada medium for detection and identification of group b streptococci. J. Clin. Microbiol. 1992, 30, 1019–1021. [Google Scholar] [PubMed]
- Heelan, J.S.; Struminsky, J.; Lauro, P.; Sung, C.J. Evaluation of a new selective enrichment broth for detection of group b streptococci in pregnant women. J. Clin. Microbiol. 2005, 43, 896–897. [Google Scholar] [CrossRef] [PubMed]
- Nasukawa, T.; Uchiyama, J.; Taharaguchi, S.; Ota, S.; Ujihara, T.; Matsuzaki, S.; Murakami, H.; Mizukami, K.; Sakaguchi, M. Virus purification by cscl density gradient using general centrifugation. Arch. Virol. 2017, 162, 3523–3528. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. Dfast: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. Dfast and daga: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. Viptree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Garbajosa, P.; Bonten, M.J.; Robinson, D.A.; Top, J.; Nallapareddy, S.R.; Torres, C.; Coque, T.M.; Cantón, R.; Baquero, F.; Murray, B.E.; et al. Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J. Clin. Microbiol. 2006, 44, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Maiden, M.C. Bigsdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 2010, 11, 595. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Vijayakumar, P.P.; Muriana, P.M. A microplate growth inhibition assay for screening bacteriocins against Listeria monocytogenes to differentiate their mode-of-action. Biomolecules 2015, 5, 1178–1194. [Google Scholar] [CrossRef] [PubMed]
- Kanda, Y. Investigation of the freely available easy-to-use software “ezr” for medical statistics. Bone Marrow Transplant. 2013, 48, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the international committee on taxonomy of viruses (ictv). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [PubMed]
- Klumpp, J.; Lavigne, R.; Loessner, M.J.; Ackermann, H.W. The spo1-related bacteriophages. Arch. Virol. 2010, 155, 1547–1561. [Google Scholar] [CrossRef] [PubMed]
- Nes, I.F.; Diep, D.B.; Holo, H. Bacteriocin diversity in streptococcus and enterococcus. J. Bacteriol. 2007, 189, 1189–1198. [Google Scholar] [CrossRef] [PubMed]
- van Houte, S.; Buckling, A.; Westra, E.R. Evolutionary ecology of prokaryotic immune mechanisms. Microbiol. Mol. Biol. Rev. 2016, 80, 745–763. [Google Scholar] [CrossRef] [PubMed]
- Rosen, G.H.; Randis, T.M.; Desai, P.V.; Sapra, K.J.; Ma, B.; Gajer, P.; Humphrys, M.S.; Ravel, J.; Gelber, S.E.; Ratner, A.J. Group b streptococcus and the vaginal microbiota. J. Infect. Dis. 2017, 216, 744–751. [Google Scholar] [CrossRef] [PubMed]
- Rick, A.M.; Aguilar, A.; Cortes, R.; Gordillo, R.; Melgar, M.; Samayoa-Reyes, G.; Frank, D.N.; Asturias, E.J. Group b streptococci colonization in pregnant guatemalan women: Prevalence, risk factors, and vaginal microbiome. Open Forum Infect. Dis. 2017, 4, ofx020. [Google Scholar] [CrossRef] [PubMed]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial changes during pregnancy, birth, and infancy. Front Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef] [PubMed]
- Ando, H.; Lemire, S.; Pires, D.P.; Lu, T.K. Engineering modular viral scaffolds for targeted bacterial population editing. Cell Syst. 2015, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.; Tremblay, D.; Moineau, S. Long-term bacteriophage preservation. WFCC Newsl. 2004, 38, 35–40. [Google Scholar]
- Lobocka, M.B.; Glowacka, A.; Golec, P. Methods for bacteriophage preservation. Methods Mol. Biol. 2018, 1693, 219–230. [Google Scholar] [PubMed]
- Ulisse, S.; Peccio, A.; Orsini, G.; Di Emidio, B. A study of the shelf-life of critical culture media. Vet. Ital. 2006, 42, 237–247. [Google Scholar] [PubMed]
- Carvalho Mda, G.; Facklam, R.; Jackson, D.; Beall, B.; McGee, L. Evaluation of three commercial broth media for pigment detection and identification of a group b Streptococcus (Streptococcus agalactiae). J. Clin. Microbiol. 2009, 47, 4161–4163. [Google Scholar] [CrossRef] [PubMed]
- Agboluaje, M.; Sauvageau, D. Bacteriophage production in bioreactors. Methods Mol. Biol. 2018, 1693, 173–193. [Google Scholar] [PubMed]
- Krysiak-Baltyn, K.; Martin, G.J.O.; Gras, S.L. Computational modeling of bacteriophage production for process optimization. Methods Mol. Biol. 2018, 1693, 195–218. [Google Scholar] [PubMed]
- Krysiak-Baltyn, K.; Martin, G.J.O.; Gras, S.L. Computational modelling of large scale phage production using a two-stage batch process. Pharmaceuticals (Basel) 2018, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Forde, A.; Hill, C. Phages of life-the path to pharma. Br. J. Pharmacol. 2018, 175, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Los, M.; Czyz, A.; Sell, E.; Wegrzyn, A.; Neubauer, P.; Wegrzyn, G. Bacteriophage contamination: Is there a simple method to reduce its deleterious effects in laboratory cultures and biotechnological factories? J. Appl. Genet. 2004, 45, 111–120. [Google Scholar] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uchiyama, J.; Matsui, H.; Murakami, H.; Kato, S.-i.; Watanabe, N.; Nasukawa, T.; Mizukami, K.; Ogata, M.; Sakaguchi, M.; Matsuzaki, S.; et al. Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening. Viruses 2018, 10, 552. https://doi.org/10.3390/v10100552
Uchiyama J, Matsui H, Murakami H, Kato S-i, Watanabe N, Nasukawa T, Mizukami K, Ogata M, Sakaguchi M, Matsuzaki S, et al. Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening. Viruses. 2018; 10(10):552. https://doi.org/10.3390/v10100552
Chicago/Turabian StyleUchiyama, Jumpei, Hidehito Matsui, Hironobu Murakami, Shin-ichiro Kato, Naoki Watanabe, Tadahiro Nasukawa, Keijiro Mizukami, Masaya Ogata, Masahiro Sakaguchi, Shigenobu Matsuzaki, and et al. 2018. "Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening" Viruses 10, no. 10: 552. https://doi.org/10.3390/v10100552
APA StyleUchiyama, J., Matsui, H., Murakami, H., Kato, S.-i., Watanabe, N., Nasukawa, T., Mizukami, K., Ogata, M., Sakaguchi, M., Matsuzaki, S., & Hanaki, H. (2018). Potential Application of Bacteriophages in Enrichment Culture for Improved Prenatal Streptococcus agalactiae Screening. Viruses, 10(10), 552. https://doi.org/10.3390/v10100552




