Rapid Large-Scale Monitoring of Pine Wilt Disease Using Sentinel-1/2 Images in GEE
Abstract
1. Introduction
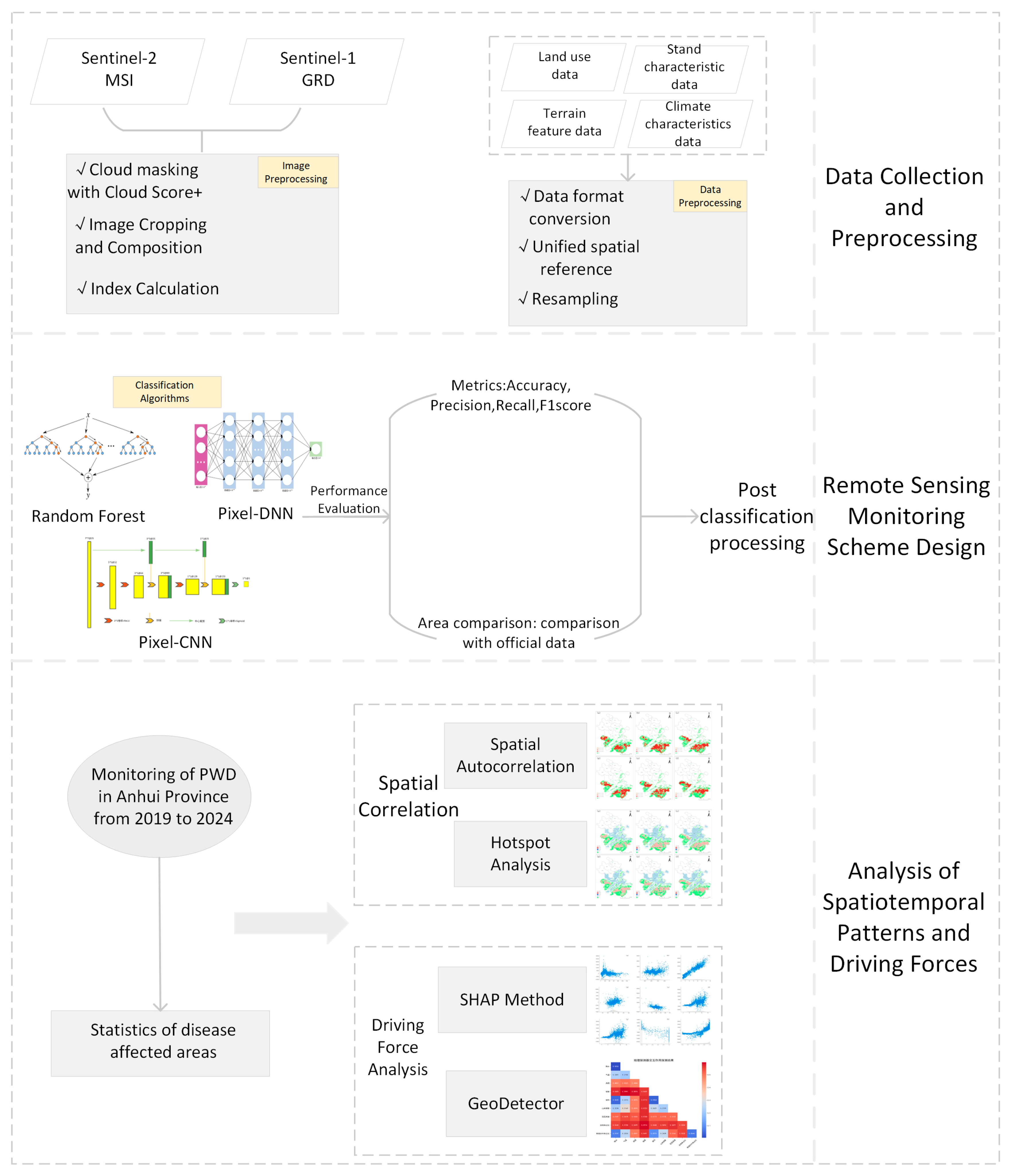
2. Materials and Methods
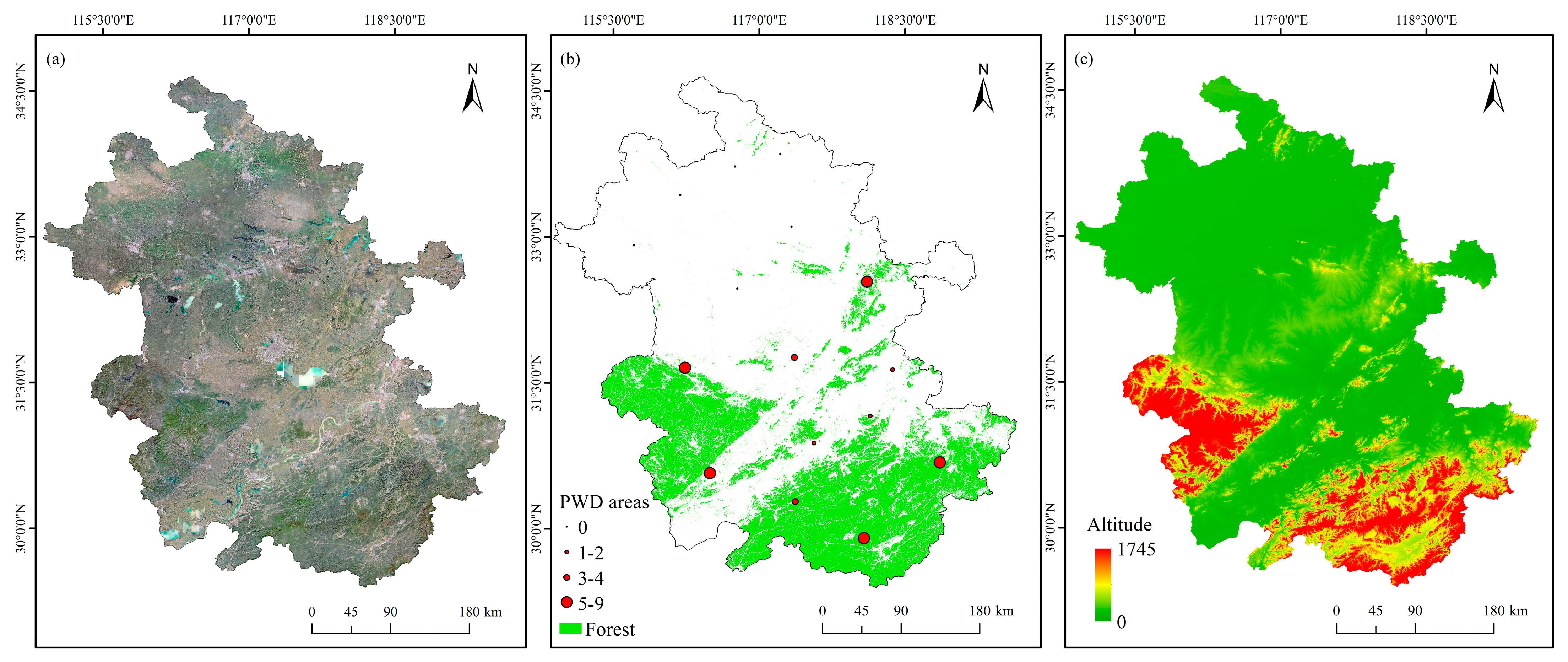
2.1. Study Area
2.2. Data Sources and Preprocessing
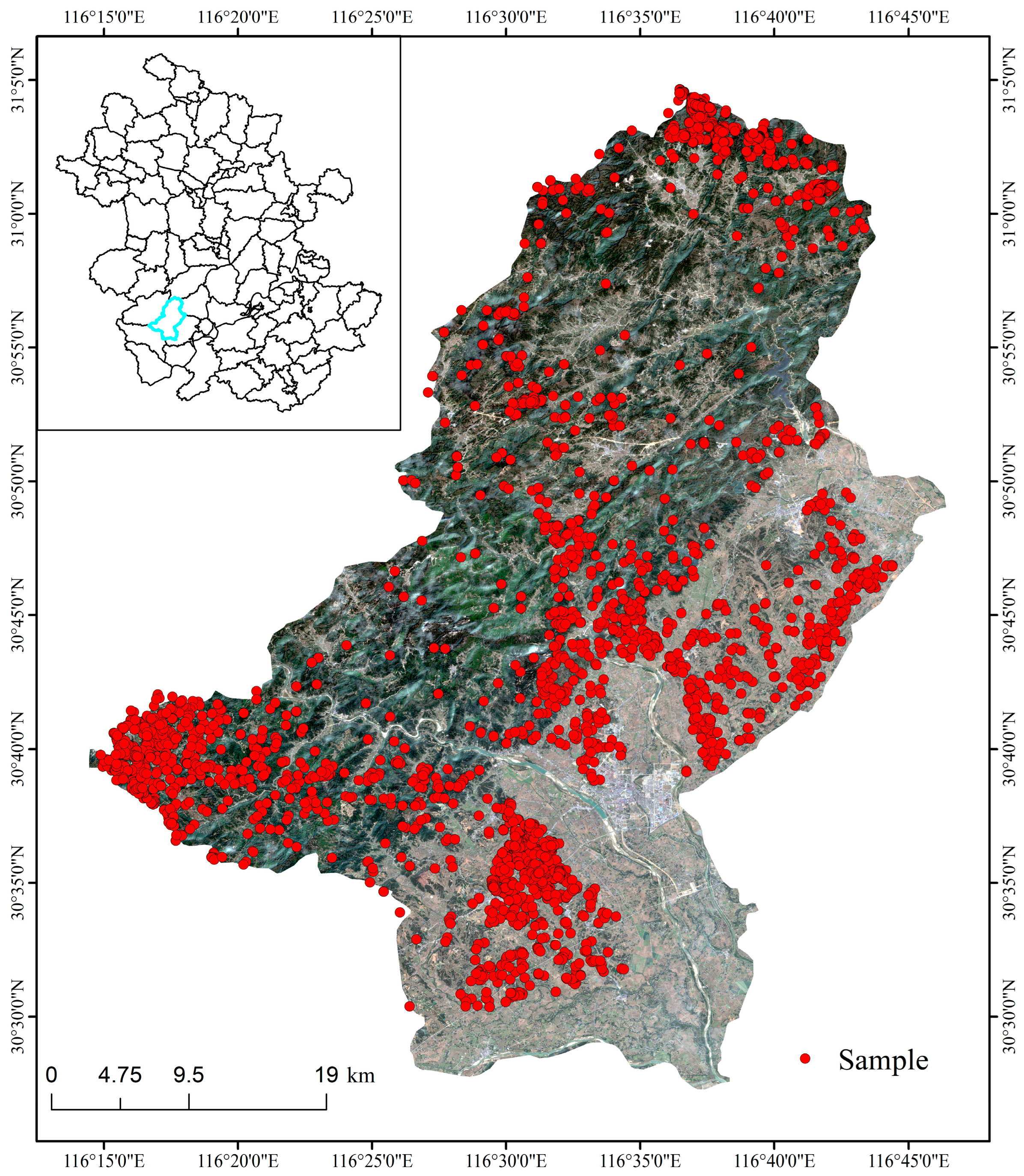
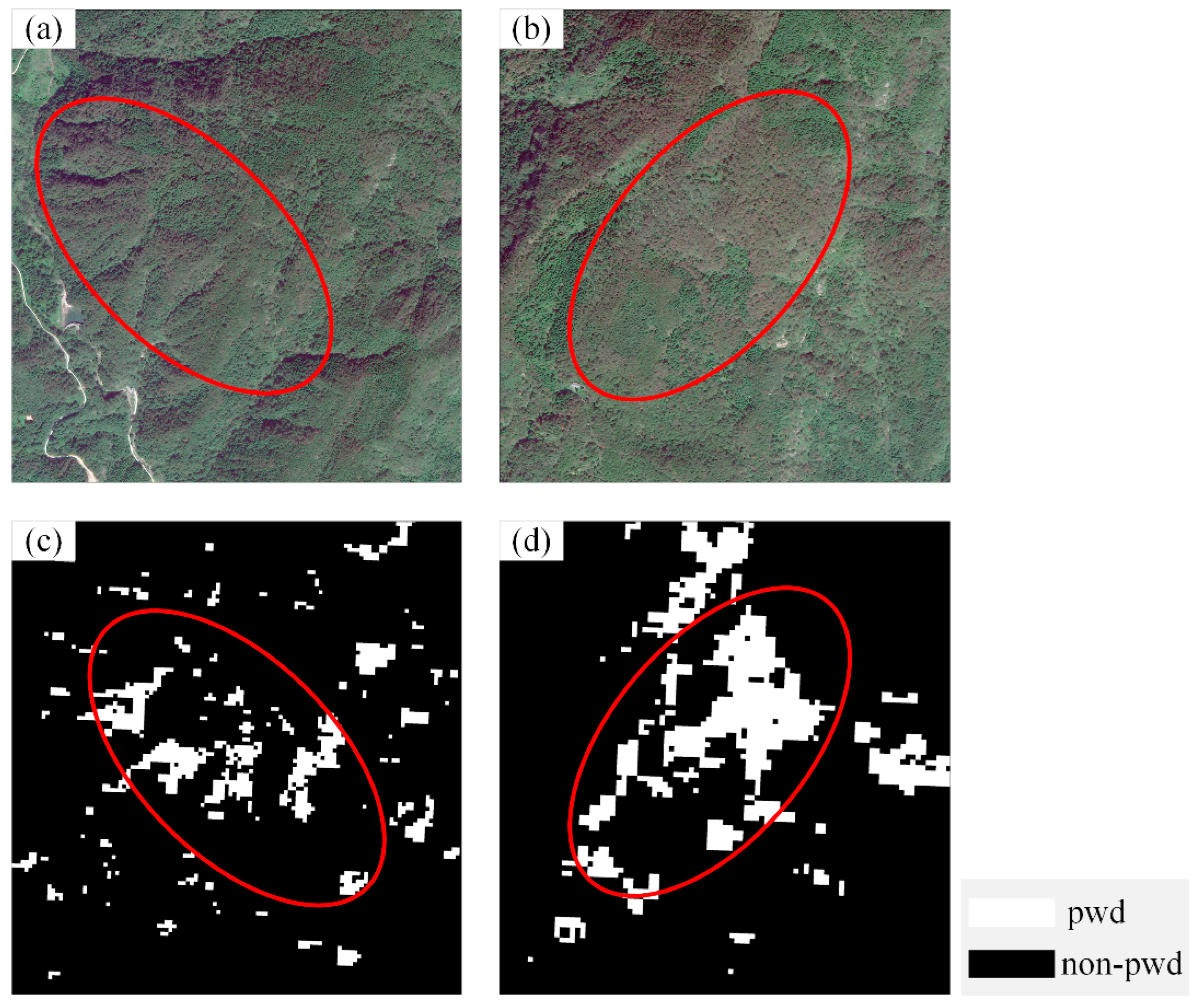
2.2.1. Remote Sensing Imagery Data and Sample Preparation
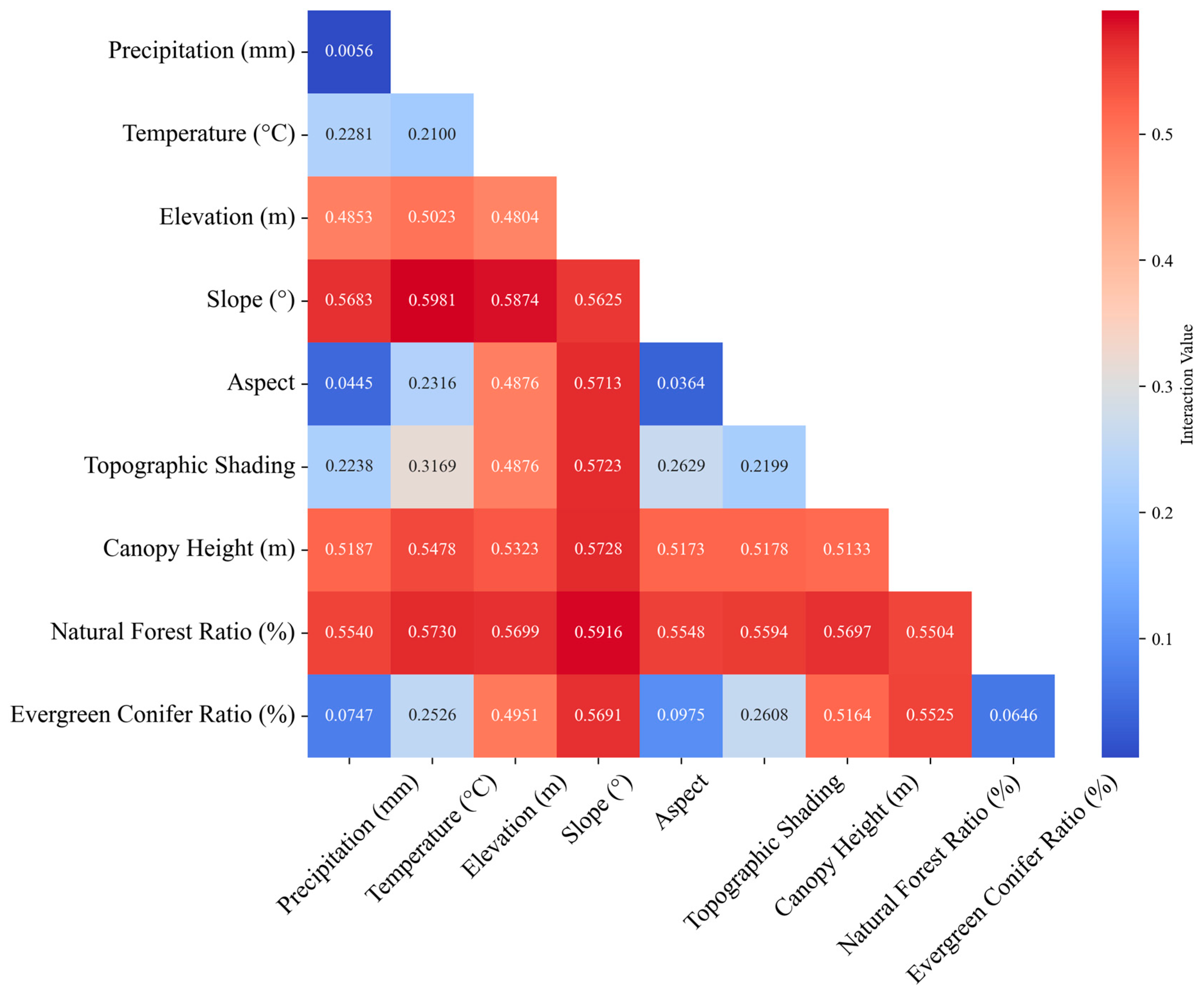
2.2.2. Influencing Factors
2.3. Methods
2.3.1. Feature Settings
- Training samples and validation samples production
- 2.
- Original Feature Construction
- 3.
- The Recursive Feature Elimination
2.3.2. Monitoring Models
- 1.
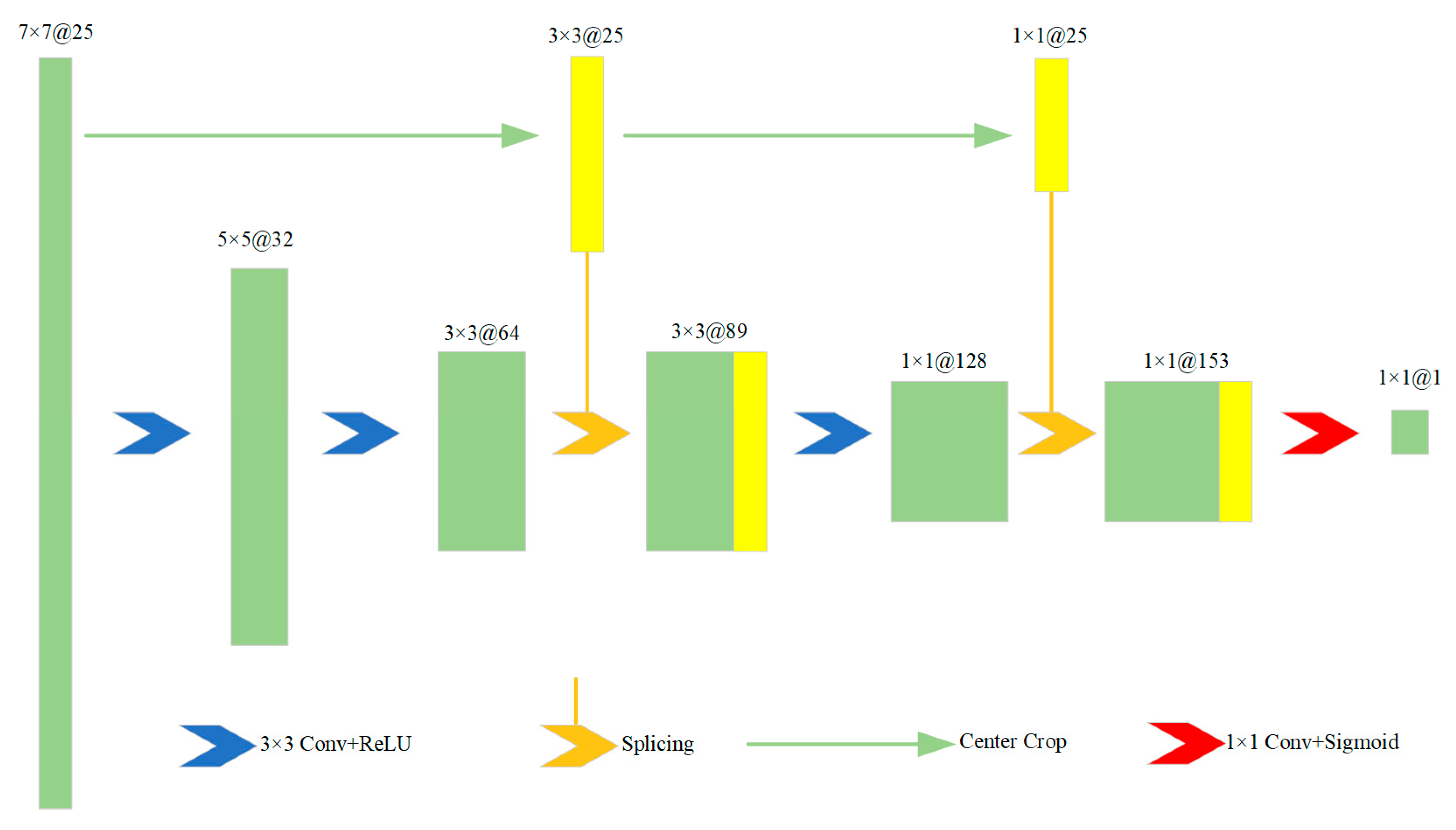
- Convolutional Neural Network Model
- 2.
- Deep Neural Network Model
- 3.
- Random Forest Model
- 4.
- Accuracy Metrics Evaluation and Classification Processing
2.3.3. Spatiotemporal Pattern Analysis
- 1.
- Spatial autocorrelation analysis
- 2.
- High/Low Clustering and Hot Spot Analysis
2.3.4. Driving Factor Analysis
- 1.
- SHAP contribution analysis
- 2.
- Geodetector
3. Results and Discussion
3.1. Model Performance of Different Remote Sensing Monitoring Models
3.2. Accuracy Validation of the Optimal Classification Model
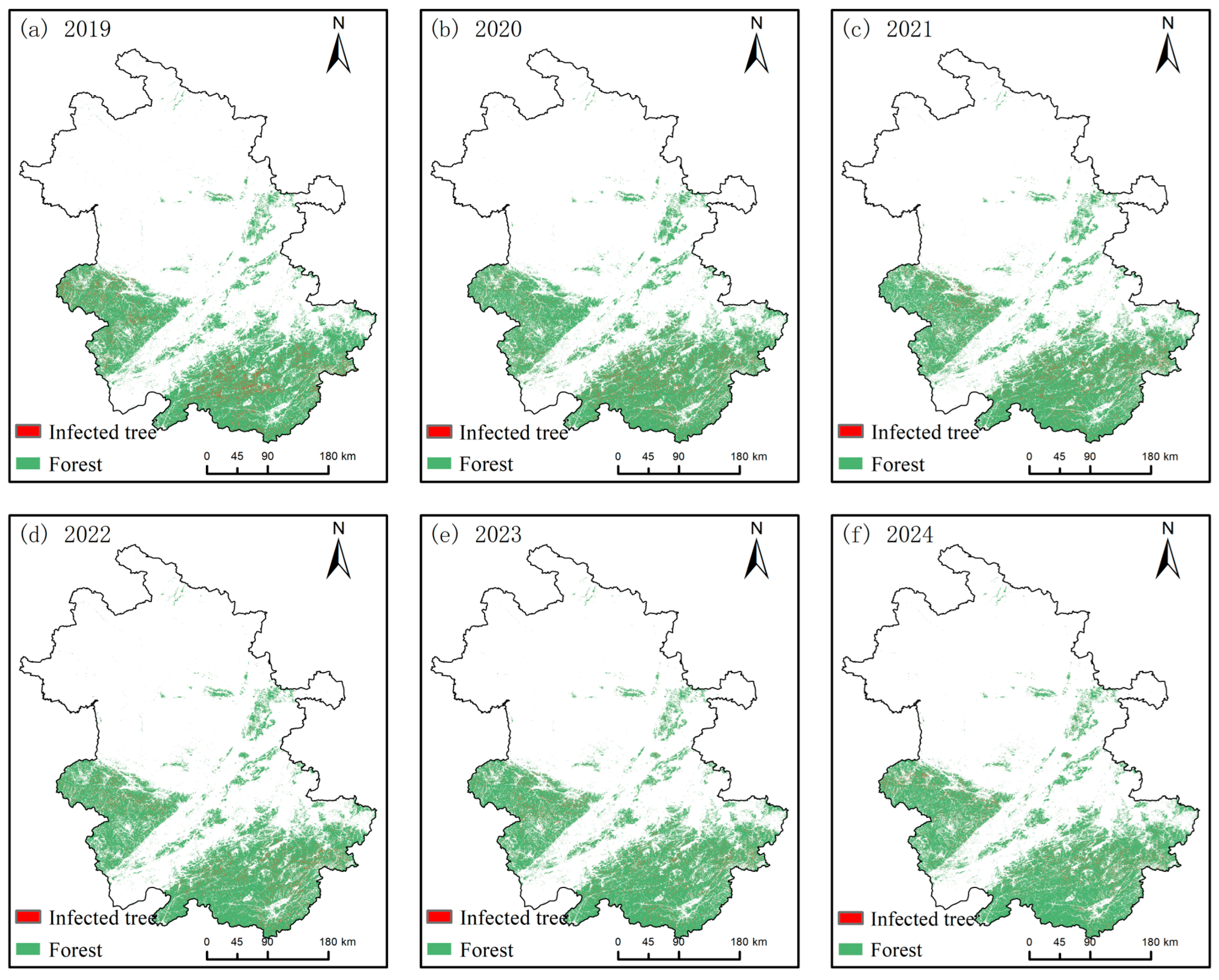
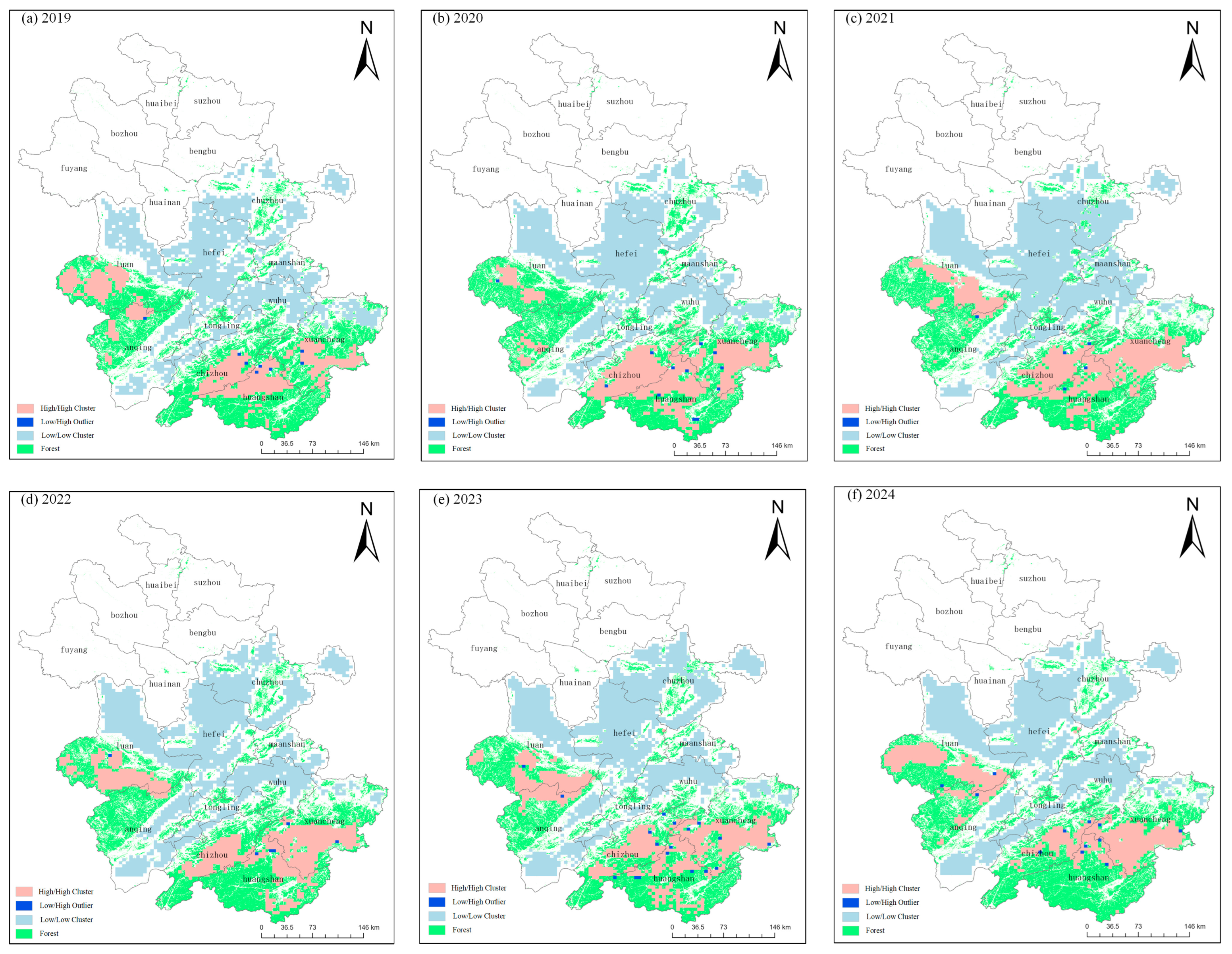
3.3. Spatiotemporal Dynamics of Pine Wilt Disease in Anhui Province
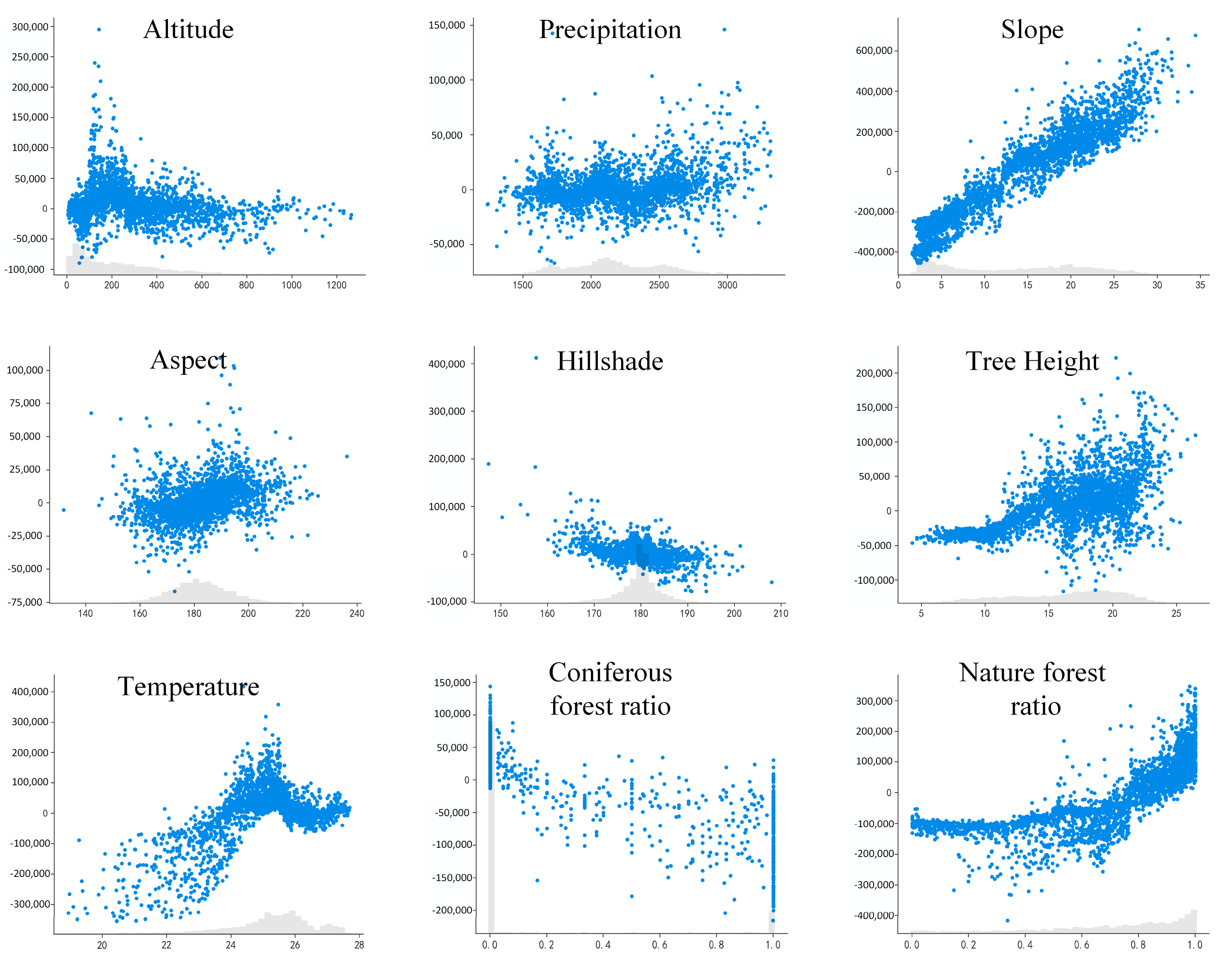
3.4. Driving Factors Analysis of Pine Wilt Disease in Anhui Province
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PWD | Pine Wilt Disease |
| GEE | Google Earth Engine |
| TM | Template Matching |
| CNN | Convolutional neural network |
| 3D-CNN | 3D convolutional neural network |
| 3D-RsCNN | 3D convolutional neural network with enhanced residual structures |
| RPN | Region Proposal Network |
| RFE | The Recursive Feature Elimination |
| IW | Interferometric Wide Swath |
| VV | Vertical–Vertical |
| VH | Vertical–Horizontal |
| GNPFD | The Global Natural/Planted Forest Dataset |
| IGBP | The Annual International Geosphere–Biosphere Programme |
| SRTM | The Shuttle Radar Topography Mission |
| GNDVI | Green Normalized Difference Vegetation Index |
| SAVI | Soil-Adjusted Vegetation Index |
| RGI | Red–Green Index |
| MSR2 | Modified Simple Ratio 2 |
| NDVIgreen | Normalized Difference Vegetation Index (Green) |
| NDVInir | Normalized Difference Vegetation Index (NIR-based) |
| NDVIswir | Normalized Difference Vegetation Index (SWIR-based) |
| REIP | Red Edge Inflection Point |
| TCG | Tasseled Cap Greenness |
| TCW | Tasseled Cap Wetness |
| BWDRVI | Blue-Wide Dynamic Range Vegetation Index |
| GLI | Green Leaf Index |
| NDVIre | Normalized Difference Vegetation Index (Red Edge) |
| SLAVI | Specific Leaf Area Vegetation Index |
| NDMI | Normalized Difference Moisture Index |
| NBR | Normalized Burn Ratio |
| DSWI | Disease Water Stress Index |
| RDVI | Renormalized Difference Vegetation Index |
| NDre1 | Normalized Difference Red Edge (1) |
| NDre2 | Normalized Difference Red Edge (2) |
| NDre3 | Normalized Difference Red Edge (3) |
| RVSI | Red-Edge Vegetation Stress Index |
| GARI | Green Atmospherically Resistant Index |
| ARI | Anthocyanin Reflectance Index |
| PBI | Plant Biochemical Index |
| MNDWI | Modified Normalized Difference Water Index |
| SHAP | SHapley Additive exPlanations |
References
- Hou, Y.; Ding, Y. Dynamic analysis of pine wilt disease model with memory diffusion and nonlocal effect. Chaos Solitons Fractals 2024, 179, 114480. [Google Scholar] [CrossRef]
- Wang, G.; Aierken, N.; Chai, G.; Yan, X.; Chen, L.; Jia, X.; Wang, J.; Huang, W.; Zhang, X. A novel BH3DNet method for identifying pine wilt disease in Masson pine fusing UAS hyperspectral imagery and LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2024, 134, 104177. [Google Scholar] [CrossRef]
- Li, N.; Huo, L.; Zhang, X. Using only the red-edge bands is sufficient to detect tree stress: A case study on the early detection of PWD using hyperspectral drone images. Comput. Electron. Agric. 2024, 217, 108665. [Google Scholar] [CrossRef]
- Zang, Z.; Wang, G.; Lin, H.; Luo, P. Developing a spectral angle based vegetation index for detecting the early dying process of Chinese fir trees. ISPRS J. Photogramm. Remote Sens. 2021, 171, 253–265. [Google Scholar] [CrossRef]
- Pan, J.; Ye, X.; Shao, F.; Liu, G.; Liu, J.; Wang, Y. Impacts of pine species, infection response, and data type on the detection of Bursaphelenchus xylophilus using close-range hyperspectral remote sensing. Remote Sens. Environ. 2024, 315, 114468. [Google Scholar] [CrossRef]
- Jung, J.-M.; Yoon, S.; Hwang, J.; Park, Y.; Lee, W.-H. Analysis of the spread distance of pine wilt disease based on a high volume of spatiotemporal data recording of infected trees. For. Ecol. Manag. 2024, 553, 121612. [Google Scholar] [CrossRef]
- Zhang, R.; Xia, L.; Chen, L.; Xie, C.; Chen, M.; Wang, W. Recognition of wilt wood caused by pine wilt nematode based on U-Net network and unmanned aerial vehicle images. Trans. Chin. Soc. Agric. Eng 2020, 36, 61–68. [Google Scholar]
- Li, X.; Liu, Y.; Huang, P.; Tong, T.; Li, L.; Chen, Y.; Hou, T.; Su, Y.; Lv, X.; Fu, W. Integrating Multi-Scale Remote-Sensing Data to Monitor Severe Forest Infestation in Response to Pine Wilt Disease. Remote Sens. 2022, 14, 5164. [Google Scholar] [CrossRef]
- Zhao, X.; Qi, J.; Xu, H.; Yu, Z.; Yuan, L.; Chen, Y.; Huang, H. Evaluating the potential of airborne hyperspectral LiDAR for assessing forest insects and diseases with 3D Radiative Transfer Modeling. Remote Sens. Environ. 2023, 297, 113759. [Google Scholar] [CrossRef]
- Yu, R.; Luo, Y.; Zhou, Q.; Zhang, X.; Wu, D.; Ren, L. A machine learning algorithm to detect pine wilt disease using UAV-based hyperspectral imagery and LiDAR data at the tree level. Int. J. Appl. Earth Obs. Geoinf. 2021, 101, 102363. [Google Scholar] [CrossRef]
- Oide, A.H.; Nagasaka, Y.; Tanaka, K. Performance of machine learning algorithms for detecting pine wilt disease infection using visible color imagery by UAV remote sensing. Remote Sens. Appl. Soc. Environ. 2022, 28, 100869. [Google Scholar] [CrossRef]
- Iordache, M.-D.; Mantas, V.; Baltazar, E.; Pauly, K.; Lewyckyj, N. A Machine Learning Approach to Detecting Pine Wilt Disease Using Airborne Spectral Imagery. Remote Sens. 2020, 12, 2280. [Google Scholar] [CrossRef]
- Abdel-Rahman, E.M.; Mutanga, O.; Adam, E.; Ismail, R. Detecting Sirex noctilio grey-attacked and lightning-struck pine trees using airborne hyperspectral data, random forest and support vector machines classifiers. ISPRS J. Photogramm. Remote Sens. 2014, 88, 48–59. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Tao, H.; Cunjun, L.; Dan, Z.; Shiqing, D.; Haitang, H.; Xinluo, X.; Jing, W. Deep learning-based dead pine tree detection from unmanned aerial vehicle images. Int. J. Remote Sens. 2020, 41, 8238–8255. [Google Scholar] [CrossRef]
- Deng, X.; Tong, Z.; Lan, Y.; Huang, Z. Detection and Location of Dead Trees with Pine Wilt Disease Based on Deep Learning and UAV Remote Sensing. AgriEngineering 2020, 2, 294–307. [Google Scholar] [CrossRef]
- Li, H.; Chen, L.; Yao, Z.; Li, N.; Long, L.; Zhang, X. Intelligent Identification of Pine Wilt Disease Infected Individual Trees Using UAV-Based Hyperspectral Imagery. Remote Sens. 2023, 15, 3295. [Google Scholar] [CrossRef]
- Li, F.; Liu, Z.; Shen, W.; Wang, Y.; Wang, Y.; Ge, C.; Sun, F.; Lan, P. A Remote Sensing and Airborne Edge-Computing Based Detection System for Pine Wilt Disease. IEEE Access 2021, 9, 66346–66360. [Google Scholar] [CrossRef]
- Park, H.G.; Yun, J.P.; Kim, M.Y.; Jeong, S.H. Multichannel Object Detection for Detecting Suspected Trees With Pine Wilt Disease Using Multispectral Drone Imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 8350–8358. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, J.; Sun, H.; Lu, X.; Huang, J.; Wang, S.; Fang, G. Satellite Remote Sensing Identification of Discolored Standing Trees for Pine Wilt Disease Based on Semi-Supervised Deep Learning. Remote Sens. 2022, 14, 5936. [Google Scholar] [CrossRef]
- He, X.; Zhou, Y.; Zhao, J.; Zhang, D.; Yao, R.; Xue, Y. Swin Transformer Embedding UNet for Remote Sensing Image Semantic Segmentation. IEEE Trans. Geosci. Remote Sens. 2022, 60, 4408715. [Google Scholar] [CrossRef]
- Lee, M.-G.; Cho, H.-B.; Youm, S.-K.; Kim, S.-W. Detection of Pine Wilt Disease Using Time Series UAV Imagery and Deep Learning Semantic Segmentation. Forests 2023, 14, 1576. [Google Scholar] [CrossRef]
- Yu, R.; Luo, Y.; Li, H.; Yang, L.; Huang, H.; Yu, L.; Ren, L. Three-Dimensional Convolutional Neural Network Model for Early Detection of Pine Wilt Disease Using UAV-Based Hyperspectral Images. Remote Sens. 2021, 13, 4065. [Google Scholar] [CrossRef]
- Long, L.; Chen, Y.; Song, S.; Zhang, X.; Jia, X.; Lu, Y.; Liu, G. Remote Sensing Monitoring of Pine Wilt Disease Based on Time-Series Remote Sensing Index. Remote Sens. 2023, 15, 360. [Google Scholar] [CrossRef]
- Tamiminia, H.; Salehi, B.; Mahdianpari, M.; Quackenbush, L.; Adeli, S.; Brisco, B. Google Earth Engine for geo-big data applications: A meta-analysis and systematic review. ISPRS J. Photogramm. Remote Sens. 2020, 164, 152–170. [Google Scholar] [CrossRef]
- Peng, S.; Ding, Y.; Liu, W.; Li, Z. 1 km monthly temperature and precipitation dataset for China from 1901 to 2017. Earth Syst. Sci. Data 2019, 11, 1931–1946. [Google Scholar] [CrossRef]
- Xu, Q.; Zhang, X.; Li, J.; Ren, J.; Ren, L.; Luo, Y. Pine Wilt Disease in Northeast and Northwest China: A Comprehensive Risk Review. Forests 2023, 14, 174. [Google Scholar] [CrossRef]
- Konapala, G.; Kumar, S.V.; Khalique Ahmad, S. Exploring Sentinel-1 and Sentinel-2 diversity for flood inundation mapping using deep learning. ISPRS J. Photogramm. Remote Sens. 2021, 180, 163–173. [Google Scholar] [CrossRef]
- Pasquarella, V.J.; Brown, C.F.; Czerwinski, W.; Rucklidge, W.J. Comprehensive quality assessment of optical satellite imagery using weakly supervised video learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Vancouver, BC, Canada, 17–24 June 2023; pp. 2125–2135. [Google Scholar]
- Zhao, F.; Sun, R.; Zhong, L.; Meng, R.; Huang, C.; Zeng, X.; Wang, M.; Li, Y.; Wang, Z. Monthly mapping of forest harvesting using dense time series Sentinel-1 SAR imagery and deep learning. Remote Sens. Environ. 2022, 269, 112822. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, C.; Gu, X.; Duporge, I.; Hughey, L.F.; Stabach, J.A.; Skidmore, A.K.; Hopcraft, J.G.C.; Lee, S.J.; Atkinson, P.M.; et al. Deep learning enables satellite-based monitoring of large populations of terrestrial mammals across heterogeneous landscape. Nat. Commun. 2023, 14, 3072. [Google Scholar] [CrossRef]
- Huang, J.; Lu, X.; Chen, L.; Sun, H.; Wang, S.; Fang, G. Accurate Identification of Pine Wood Nematode Disease with a Deep Convolution Neural Network. Remote Sens. 2022, 14, 913. [Google Scholar] [CrossRef]
- Wang, S.; Cao, X.; Wu, M.; Yi, C.; Zhang, Z.; Fei, H.; Zheng, H.; Jiang, H.; Jiang, Y.; Zhao, X.; et al. Detection of Pine Wilt Disease Using Drone Remote Sensing Imagery and Improved YOLOv8 Algorithm: A Case Study in Weihai, China. Forests 2023, 14, 2052. [Google Scholar] [CrossRef]
- Solórzano, J.V.; Mas, J.F.; Gao, Y.; Gallardo-Cruz, J.A. Land Use Land Cover Classification with U-Net: Advantages of Combining Sentinel-1 and Sentinel-2 Imagery. Remote Sens. 2021, 13, 3600. [Google Scholar] [CrossRef]
- Yang, F.; Zeng, Z. Refined fine-scale mapping of tree cover using time series of Planet-NICFI and Sentinel-1 imagery for Southeast Asia (2016–2021). Earth Syst. Sci. Data 2023, 15, 4011–4021. [Google Scholar] [CrossRef]
- Mantas, V.; Fonseca, L.; Baltazar, E.; Canhoto, J.; Abrantes, I. Detection of Tree Decline (Pinus pinaster Aiton) in European Forests Using Sentinel-2 Data. Remote Sens. 2022, 14, 2028. [Google Scholar] [CrossRef]
- Bárta, V.; Lukeš, P.; Homolová, L. Early detection of bark beetle infestation in Norway spruce forests of Central Europe using Sentinel-2. Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102335. [Google Scholar] [CrossRef]
- Kuang, J.; Yu, L.; Zhou, Q.; Wu, D.; Ren, L.; Luo, Y. Identification of Pine Wilt Disease-Infested Stands Based on Single- and Multi-Temporal Medium-Resolution Satellite Data. Forests 2024, 15, 596. [Google Scholar] [CrossRef]
- Meng, R.; Gao, R.; Zhao, F.; Huang, C.; Sun, R.; Lv, Z.; Huang, Z. Landsat-based monitoring of southern pine beetle infestation severity and severity change in a temperate mixed forest. Remote Sens. Environ. 2022, 269, 112847. [Google Scholar] [CrossRef]
- Tong, T.; Simei, L.; Linyuan, L.; Tao, L.; Huaguo, H. Remote sensing recognition of pine wilt disease in Pinus massoniana forest combined with microwave and optical time-series images. J. Beijing For. Univ. 2024, 46, 40–52. [Google Scholar]
- Yan, K.; Zhang, D. Feature selection and analysis on correlated gas sensor data with recursive feature elimination. Sens. Actuators B Chem. 2015, 212, 353–363. [Google Scholar] [CrossRef]
- Caraballo-Vega, J.A.; Carroll, M.L.; Neigh, C.S.R.; Wooten, M.; Lee, B.; Weis, A.; Aronne, M.; Alemu, W.G.; Williams, Z. Optimizing WorldView-2, -3 cloud masking using machine learning approaches. Remote Sens. Environ. 2023, 284, 113332. [Google Scholar] [CrossRef]
- Li, K.; Wang, J.; Yao, J. Effectiveness of machine learning methods for water segmentation with ROI as the label: A case study of the Tuul River in Mongolia. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102497. [Google Scholar] [CrossRef]
- Aravena Pelizari, P.; Geiß, C.; Groth, S.; Taubenböck, H. Deep multitask learning with label interdependency distillation for multicriteria street-level image classification. ISPRS J. Photogramm. Remote Sens. 2023, 204, 275–290. [Google Scholar] [CrossRef]
- Ebrahimy, H.; Mirbagheri, B.; Matkan, A.A.; Azadbakht, M. Per-pixel land cover accuracy prediction: A random forest-based method with limited reference sample data. ISPRS J. Photogramm. Remote Sens. 2021, 172, 17–27. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Li, X.; Tong, T.; Luo, T.; Wang, J.; Rao, Y.; Li, L.; Jin, D.; Wu, D.; Huang, H. Retrieving the Infected Area of Pine Wilt Disease-Disturbed Pine Forests from Medium-Resolution Satellite Images Using the Stochastic Radiative Transfer Theory. Remote Sens. 2022, 14, 1526. [Google Scholar] [CrossRef]
- Wang, M.; Mao, D.; Wang, Y.; Xiao, X.; Xiang, H.; Feng, K.; Luo, L.; Jia, M.; Song, K.; Wang, Z. Wetland mapping in East Asia by two-stage object-based Random Forest and hierarchical decision tree algorithms on Sentinel-1/2 images. Remote Sens. Environ. 2023, 297, 113793. [Google Scholar] [CrossRef]
- Wieland, M.; Martinis, S.; Kiefl, R.; Gstaiger, V. Semantic segmentation of water bodies in very high-resolution satellite and aerial images. Remote Sens. Environ. 2023, 287, 113452. [Google Scholar] [CrossRef]
- Ren, D.; Li, M.; Hong, Z.; Liu, L.; Huang, J.; Sun, H.; Ren, S.; Sao, P.; Wang, W.; Zhang, J. MASFNet: Multi-level attention and spatial sampling fusion network for pine wilt disease trees detection. Ecol. Indic. 2025, 170, 113073. [Google Scholar] [CrossRef]
- Lu, X.; Huang, J.; Li, X.; Fang, G.; Liu, D. The interaction of environmental factors increases the risk of spatiotemporal transmission of pine wilt disease. Ecol. Indic. 2021, 133, 108394. [Google Scholar] [CrossRef]
- Zhang, B.; Ye, H.; Lu, W.; Huang, W.; Wu, B.; Hao, Z.; Sun, H. A Spatiotemporal Change Detection Method for Monitoring Pine Wilt Disease in a Complex Landscape Using High-Resolution Remote Sensing Imagery. Remote Sens. 2021, 13, 2083. [Google Scholar] [CrossRef]
- Lee, T.; Kim, J. Rapid spread and high prevalence of the pine wilt disease around wildfire areas. Trees For. People 2025, 20, 100805. [Google Scholar] [CrossRef]
- Ikegami, M.; Jenkins, T.A.R. Estimate global risks of a forest disease under current and future climates using species distribution model and simple thermal model—Pine Wilt disease as a model case. For. Ecol. Manag. 2018, 409, 343–352. [Google Scholar] [CrossRef]
- Li, Z. Extracting spatial effects from machine learning model using local interpretation method: An example of SHAP and XGBoost. Comput. Environ. Urban Syst. 2022, 96, 101845. [Google Scholar] [CrossRef]
- Wang, J.F.; Li, X.H.; Christakos, G.; Liao, Y.L.; Zhang, T.; Gu, X.; Zheng, X.Y. Geographical Detectors-Based Health Risk Assessment and its Application in the Neural Tube Defects Study of the Heshun Region, China. Int. J. Geogr. Inf. Sci. 2010, 24, 107–127. [Google Scholar] [CrossRef]


| Index Name | Calculation Formula | Reference |
|---|---|---|
| GNDVI | [36] | |
| SAVI | [36] | |
| RGI | [36] | |
| MSR2 | [36] | |
| NDVIgreen | [37] | |
| NDVInir | [37] | |
| NDVIswir | [37] | |
| REIP | [37] | |
| TCG | [37] | |
| TCW | [37] | |
| BWDRVI | [38] | |
| GLI | [38] | |
| NDVIre | [38] | |
| SLAVI | [38] | |
| NDMI | [39] | |
| NBR | [39] | |
| DSWI | [40] | |
| RDVI | [40] | |
| Ndre1 | [3] | |
| Ndre2 | [3] | |
| Ndre3 | [3] | |
| RVSI | [3] | |
| GARI | [38] | |
| ARI | [3] | |
| PBI | [3] | |
| MNDWI | [40] |
| Model | Accuracy | Precision | Recall | F1-Score |
|---|---|---|---|---|
| Pixel-DNN | 0.691 | 0.656 | 0.720 | 0.686 |
| Pixel-CNN | 0.743 | 0.733 | 0.759 | 0.746 |
| RF | 0.789 | 0.779 | 0.805 | 0.791 |
| Year | Statistical Area | Monitored Area | Monitoring Error |
|---|---|---|---|
| 2019 | 110,000 | 137,765 | 25.24% |
| 2020 | 101,333 | 124,358 | 22.72% |
| 2021 | 92,700 | 115,273 | 24.35% |
| Year | Global Moran’s I | Z-Score | p-Value |
|---|---|---|---|
| 2019 | 0.798 | 75.279 | 0 |
| 2020 | 0.850 | 80.113 | 0 |
| 2021 | 0.820 | 77.198 | 0 |
| 2022 | 0.840 | 79.212 | 0 |
| 2023 | 0.807 | 76.077 | 0 |
| Year | Observed Value | Expected Value | Z-Score | p-Value | Clustering Pattern |
|---|---|---|---|---|---|
| 2019 | 0.000863 | 0.0002 | 75.091 | 0 | High Clustering |
| 2020 | 0.000659 | 0.0002 | 79.752 | 0 | High Clustering |
| 2021 | 0.000682 | 0.0002 | 76.902 | 0 | High Clustering |
| 2022 | 0.000711 | 0.0002 | 78.896 | 0 | High Clustering |
| 2023 | 0.000589 | 0.0002 | 75.755 | 0 | High Clustering |
| 2024 | 0.000667 | 0.0002 | 76.951 | 0 | High Clustering |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhi, J.; Li, L.; Fang, Y.; Zhi, D.; Guang, Y.; Liu, W.; Qu, L.; Fu, X.; Zhao, H. Rapid Large-Scale Monitoring of Pine Wilt Disease Using Sentinel-1/2 Images in GEE. Forests 2025, 16, 981. https://doi.org/10.3390/f16060981
Zhi J, Li L, Fang Y, Zhi D, Guang Y, Liu W, Qu L, Fu X, Zhao H. Rapid Large-Scale Monitoring of Pine Wilt Disease Using Sentinel-1/2 Images in GEE. Forests. 2025; 16(6):981. https://doi.org/10.3390/f16060981
Chicago/Turabian StyleZhi, Junjun, Lin Li, Yifan Fang, Dandan Zhi, Yi Guang, Wangbin Liu, Lean Qu, Xinwu Fu, and Haoshan Zhao. 2025. "Rapid Large-Scale Monitoring of Pine Wilt Disease Using Sentinel-1/2 Images in GEE" Forests 16, no. 6: 981. https://doi.org/10.3390/f16060981
APA StyleZhi, J., Li, L., Fang, Y., Zhi, D., Guang, Y., Liu, W., Qu, L., Fu, X., & Zhao, H. (2025). Rapid Large-Scale Monitoring of Pine Wilt Disease Using Sentinel-1/2 Images in GEE. Forests, 16(6), 981. https://doi.org/10.3390/f16060981





