Abstract
The rhizospheric microbial community plays a crucial role in the growth and ecological adaptation of truffles. Although extensive research has been conducted on bacterial communities in truffle habitats, the spatial variation and functional implications across different regions and soil compartments remain poorly understood in the current literature. In this study, soil bacterial communities were analyzed using 16S rRNA high-throughput sequencing across truffle-producing (Tuber sinense) and non-producing sites in Quercus aliena forests in Panzhihua, China. To capture microhabitat-level variation, soils were classified into three compartments: rhizosphere soil tightly adhering to ectomycorrhizal roots (TRS), rhizoplane soil loosely attached to roots (TRE), and bulk soil from truffle zones without visible roots (TBS), with corresponding controls (RS, RE, BS) collected from truffle-free forests. An alpha diversity analysis revealed that truffle-producing soils harbored significantly higher bacterial richness than control soils, while beta diversity indicated more clustered community composition in truffle-associated soils. A taxonomic analysis showed that T. sinense occurrence was associated with enrichment of specific bacterial taxa, including Chloroflexi, Anaeromyxobacteraceae, and Bradyrhizobium, whereas widespread taxa such as Firmicutes and Bacteroidota were more abundant in control soils. To further identify microbial indicators closely associated with truffle presence, we employed random forest modeling, which highlighted Pseudomonas, Streptomyces, and Bradyrhizobium as key genera distinguishing truffle-associated rhizospheres. These findings suggest that T. sinense may influence the composition of rhizospheric microbial communities, thereby constructing a favorable rhizospheric microenvironment. This work provides new insights into the microbial ecology of T. sinense and lays a foundation for future truffle domestication and cultivation efforts.
1. Introduction
The genus Tuber comprises ectomycorrhizal fungi of significant ecological and economic value, primarily forming symbiotic associations with tree roots in forest ecosystems []. Tuber sinense, commonly referred to as the Chinese black truffle, is predominantly distributed in southwestern China, particularly in Yunnan and Sichuan provinces, where it associates with Pinus spp. and Quercus spp., or their mixed forests []. Historically, T. sinense was considered part of the T. indicum complex due to morphological similarity and limited molecular data [,]. Recent multi-locus molecular analyses of over 1000 truffle specimens from China have clarified that the black truffles previously assigned to the Tuber indicum complex comprise two distinct species: T. sinense, confined to southwestern China, and T. formosanum, distributed widely across East Asia []. This clarification is crucial for truffle ecology and domestication studies, as it allows researchers to target species-specific ecological patterns and microbial associations, particularly for T. sinense, which is the focus of the present study.
In truffle-producing ecosystems, truffle mycelia form a characteristic structure known as “shiro”. This refers to a porous, spongy, and well-aerated soil zone, resulting from the interaction between Tuber mycelia, host plant roots, and the humus layer []. In the truffle shiro, after establishing ectomycorrhizal associations with plant roots, Tuber mycelia extend into the surrounding soil, aiding in nutrient uptake for the host plant []. These hyphae can also release volatile organic compounds (VOCs), which may inhibit the growth of neighboring herbaceous plants and shrubs, resulting in vegetation-sparse “burnt areas” []. Such interactions further alter the microbial community composition within the truffle shiro [,,].
As a hypogeous ectomycorrhizal fungus, T. sinense is surrounded by a diverse array of microorganisms that contribute to and are influenced by the truffle–host–soil system [,]. Previous studies have underscored the pivotal role of microbial interactions in ectomycorrhizal symbioses, demonstrating that soil bacteria and fungi can significantly affect truffle colonization, nutrient acquisition, and the formation of fruiting bodies [,]. Such interactions contribute to the specificity and structural differentiation of rhizosphere microbial communities associated with distinct Tuber species []. Notably, certain mycorrhiza-helper bacteria (MHB) have been shown to play dual roles in promoting mycorrhizal establishment [,] and suppressing phytopathogens []. Despite these insights, the structure and functional roles of microbial communities associated with T. sinense remain poorly characterized, especially in natural truffle habitats in China.
Understanding the diversity, composition, and key microbial taxa in T. sinense-associated soils is crucial for elucidating the ecological dynamics of truffle symbioses. Such knowledge could also inform strategies to improve truffle cultivation and enhance yield in managed plantations. In this study, we aimed to investigate the microbial diversity and identify key bacterial groups in soils associated with T. sinense, thereby providing a foundation for ecological insight and cultivation improvement of this endemic truffle species.
2. Materials and Methods
2.1. Study Site and Soil Sample Collection
Soil samples were collected from a natural T. sinense habitat located in a Quercus forest (Quercus aliena) in Panzhihua City, China. The region features a mountainous terrain under a subtropical monsoon climate. The sampling area was characterized by active T. sinense colonization (26°31′27″ N, 101°46′45″ E), and five independent sampling sites were selected. At each site, three types of soil compartments associated with Tuber sinense ectomycorrhizae were collected in triplicate:
Tuber Rhizosphere (TRS): tightly adhering soil particles that remain attached to T. sinense ectomycorrhizal roots even after gentle shaking. This compartment is directly influenced by root and mycorrhizal exudates and represents the most intimate microbe–host interaction zone.
Tuber Rhizoplane (TRE): soil that is loosely attached to the ectomycorrhizal root surface and can be removed by manual shaking. This zone reflects immediate root proximity but with reduced host influence compared to TRS.
Tuber Bulk Soil (TBS): soil collected ~40 cm away from ectomycorrhizal roots within the same truffle-colonized zone but free of visible roots or mycelia. This serves as a site-level baseline to assess the broader effects of T. sinense colonization on surrounding microbial communities.
As controls, soils from a nearby Quercus forest lacking truffle colonization were sampled at corresponding distances from Quercus forest roots (26°17′29″ N, 101°37′47″ E), designated as bulk control (BS), rhizoplane control (RE), and rhizosphere control (RS).
Fresh ectomycorrhizal roots of T. sinense were carefully excavated with sterile tools, and soils from each microhabitat were immediately placed into sterile sampling bags, labeled, and transported on ice. In the laboratory, samples were homogenized under sterile conditions, sieved (2.0 mm), and stored at −80 °C until analysis. For rhizosphere soil, tightly attached particles were separated using PBS buffer and sonication, followed by centrifugation.
2.2. DNA Extraction, PCR Amplification, and High-Throughput Sequencing
Total genomic DNA was extracted from all soil samples using the FastDNA™ SPIN Kit for Soil (MP Biomedicals, Santa Ana, CA, USA) following the manufacturer’s protocol []. DNA concentration and purity were assessed using a Nanodrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), and samples with suboptimal quality were re-extracted. DNA integrity was confirmed by agarose gel electrophoresis. The bacterial 16S rRNA gene V3–V4 hypervariable regions were amplified using universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) []. Amplicons were sequenced using the Illumina MiSeq platform. Raw sequencing data were quality-filtered using Cutadapt (version 1.9.1) [], and chimeric reads were removed using UCHIME (version 4.2) []. High-quality reads were clustered into operational taxonomic units (OTUs) at 97% sequence similarity using the UPARSE algorithm (version 7.1) []. Representative sequences from each OTU were taxonomically annotated using the SILVA database through the Mothur pipeline (Version 1.33.3) [,]. Singletons were removed prior to diversity and statistical analyses to avoid potential sequencing errors. Multiple sequence alignment was performed using MUSCLE (Version 3.8.31) to construct phylogenetic trees for downstream analyses [].
2.3. Microbial Diversity and Community Structure Analysis
Alpha diversity indices, including richness estimators (Chao1 and ACE), diversity indices (Shannon and Simpson), and Faith’s phylogenetic diversity (PD), were calculated using QIIME (version 1.17) []. Rarefaction and rank-abundance curves were generated in R. Statistical comparisons of diversity indices among sample types were conducted to assess richness and evenness. Beta diversity was evaluated using UniFrac distance matrices and visualized via UPGMA clustering and principal coordinates analysis (PCoA) in R, utilizing the packages WGCNA, stats, and ggplot2 (version 3.2.1) []. These analyses provided insights into the microbial community dissimilarities among different soil compartments and between truffle-influenced and control soils.
2.4. Identification of Differential and Indicator Taxa
Differentially abundant taxa across sample types and regions were identified using LEfSe (version 1.0) [], which integrates non-parametric Kruskal–Wallis and Wilcoxon tests with LDA threshold of 3.5 for effect size estimation. Taxa with significant abundance differences were visualized and interpreted as potential microbial indicators of truffle-associated habitats. To identify the most influential taxa differentiating truffle-associated microhabitats, random forest classification was applied using the randomForest package in R []. Biomarkers were identified by comparing TRS, TRE, and TBS against their respective controls and among different collection sites, highlighting microbial taxa potentially shaped by the presence of T. sinense.
3. Results
3.1. Comparison of Bacterial Community Composition Between Tuber sinense-Producing and Control Areas
Based on OTU annotation results (Figure 1), Proteobacteria dominates across compartments, but Firmicutes are largely confined to control areas. The bacterial communities across different samples were dominated at the phylum level by Proteobacteria (19.7–33.2%), Acidobacteriota (10.3–22.6%), and Actinobacteriota (5.9–16.1%). However, the relative abundance of these dominant phyla varied markedly among samples. Proteobacteria were more abundant in rhizosphere and root surface soils (TRS, TRE, RS, RE) than their controls (TBS, BS). Additionally, the abundance of Proteobacteria was generally lower in T. sinense-producing soils (TRS, TRE, TBS) than in their corresponding control sites (RS, RE, BS).
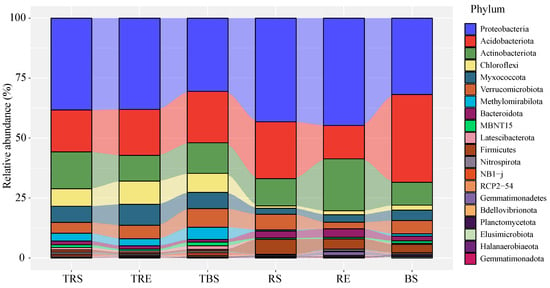
Figure 1.
Relative abundance of bacterial phyla in producing and control areas of Tuber sinense. Samples from the producing areas (rhizosphere soil, TRS; rhizoplane soil, TRE; bulk soil, TBS). Samples from control areas (rhizosphere control, RS; rhizoplane control, RE; bulk control, BS).
Acidobacteriota showed lower abundance in producing site rhizosphere and bulk soils (TRS, TBS) than in the controls (RS, BS), whereas their abundance was higher on the root surface (TRE) of producing areas compared to the control (RE). In contrast, Actinobacteriota exhibited an opposite distribution pattern to Acidobacteriota across the sample types. Other major phyla, such as Chloroflexi, Myxococcota, and Methylomirabilota, were relatively more abundant in producing sites, while Bacteroidota and Firmicutes were more abundant in control areas.
At the genus level (Figure 2), key functional genera, such as Bradyrhizobium, Anaeromyxobacter, Acidibacter, Acidothermus, Arthrobacter, Haliangium, Pedomicrobium, Sphingomonas, and Rhodoplanes, exhibited high relative abundance. Among these, Haliangium and Anaeromyxobacter were more abundant in the producing areas (TRS, TRE, TBS), while Rhodoplanes and Sphingomonas were more enriched in the control areas (RS, RE, BS). Additionally, Mesorhizobium showed a higher relative abundance in producing areas, whereas Mycobacterium was more abundant in controls. Within producing areas, some genera, such as Sphingomonas, Pseudomonas, Mesorhizobium, Burkholderia, and Bacillus, showed a decreasing trend from rhizosphere (TRS) to root surface (TRE) to bulk soil (TBS).
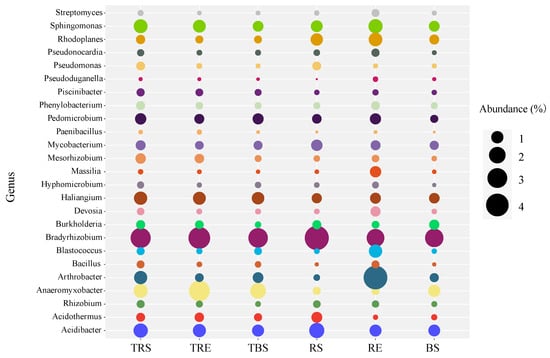
Figure 2.
Relative abundance of bacterial genera in producing and control areas of Tuber sinense. Samples from the producing areas (rhizosphere soil, TRS; rhizoplane soil, TRE; bulk soil, TBS). Samples from control areas (rhizosphere control, RS; rhizoplane control, RE; bulk control, BS).
3.2. Alpha Diversity of Bacterial Communities in Producing and Control Areas
The alpha diversity analysis (Table 1) revealed that all samples had Good’s coverage values > 95%, indicating sufficient sequencing depth. Shannon and Simpson indices varied little across samples, suggesting similar evenness and diversity. However, the total number of observed OTUs was higher in producing areas (TRS, TRE, TBS) compared to controls (RS, RE, BS), with the highest richness observed in the rhizosphere soil (TRS).

Table 1.
Alpha diversity of bacterial in producing areas and non-producing areas of Tuber sinense.
Both Chao1 and ACE indices were significantly higher in producing areas, especially in the TRS compartment, indicating greater estimated community richness. Notably, Chao1 values appeared disproportionately high compared to observed OTUs and sequencing depth. This may be attributed to a large number of low-abundance amplicon sequence variants (ASVs) detected in truffle-producing samples, especially in TRS, which inflated the richness estimators. The Good’s coverage for TRS was relatively lower (0.95), supporting the presence of many rare taxa not fully captured at the sequencing depth used. Therefore, while the Chao1 index suggests higher richness, it may partly reflect undersampling of a long-tail microbial community distribution rather than actual numerical abundance. The PD whole tree index, reflecting phylogenetic diversity, remained relatively stable across producing areas, suggesting closely related bacterial communities. In contrast, PD values varied more among control samples, reflecting greater phylogenetic complexity.
3.3. Beta Diversity of Microbial Communities in Producing and Control Areas
A principal coordinates analysis (PCoA) based on unweighted UniFrac distances was used to explore microbial beta diversity (Figure 3). The first two principal components explained 46.23% and 34.65% of the total variance, respectively. Bacterial communities from producing and control sites were clearly separated, indicating significant compositional differences. Furthermore, bacterial communities in producing areas were more tightly clustered, suggesting higher community similarity among these samples. In contrast, microbial communities in control areas were more dispersed, indicating greater heterogeneity.
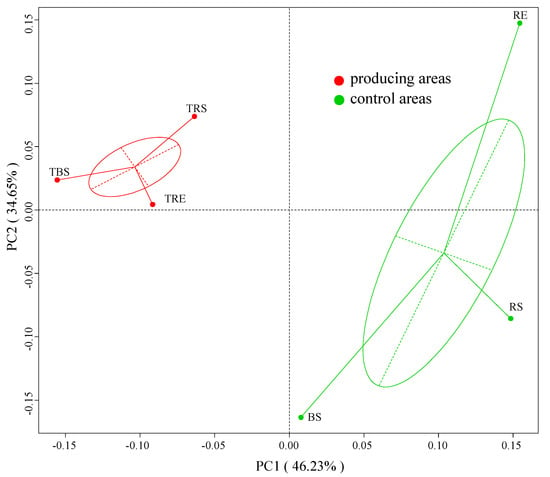
Figure 3.
Beta diversity of bacterial community in producing and control areas of Tuber sinense. Samples from the producing areas (rhizosphere soil, TRS; rhizoplane soil, TRE; bulk soil, TBS). Samples from control areas (rhizosphere control, RS; rhizoplane control, RE; bulk control, BS).
3.4. Spatial Variation in Bacterial Taxa Within Tuber sinense-Producing Regions
To identify region-specific biomarkers within producing sites, LEfSe analysis was performed with an LDA threshold of 3.5 and a p-value < 0.05 based on the Kruskal–Wallis test (Figure 4). A total of 12 bacterial clades showed significant differences across the three regions (TRS, TRE, TBS), including two phyla, three classes, two orders, two families, two genera, and one species. In TRS, six distinctive taxa were identified: Proteobacteria, Alphaproteobacteria, Rhizobiales, Xanthobacteraceae, Bradyrhizobium, and Bradyrhizobium elkanii. TRE was enriched with Alphaproteobacteria and one unidentified phylum. TBS was characterized by the presence of Blastocatellia, Pyrinomonadales, Pyrinomonadaceae, and an unidentified genus g-RB41.
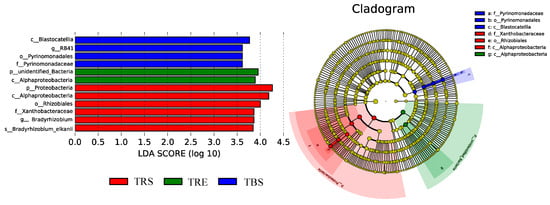
Figure 4.
LEfSe analysis of differential bacterial taxa in different producing regions of Tuber sinense. Samples from the producing areas (rhizosphere soil, TRS; rhizoplane soil, TRE; bulk soil, TBS).
3.5. Identification of Potential Keystone Taxa in Different Producing Regions
A random forest model was constructed using genus-level bacterial data to identify key taxa distinguishing different producing regions (Figure 5). The model demonstrated high predictive accuracy, with AUC values of 96.9%, 100%, and 97.7% for each comparison, indicating strong model reliability. The top 10 genera in each comparison were selected as potential keystone taxa. For TRS vs. TRE, key genera included Catenulispora, Aquisphaera, an unclassified genus of Ilumatobacteraceae, Phenylobacterium, Luteolibacter, Piscinibacter, Kribbella, Streptomyces, Pseudomonas, and Rhizobacter. For TRS vs. TBS, they included Inquilinus, Dokdonella, Bradyrhizobium, Pedosphaera, Pseudomonas, Parapusillimonas, Phenylobacterium, Dyella, Mucilaginibacter, and an unclassified genus of Comamonadaceae. For TRE vs. TBS, important taxa included an unclassified genus of Pyrinomonadaceae, Mucilaginibacter, Dokdonella, Inquilinus, Acidocella, Bradyrhizobium, Granulicella, Rhodanobacter, an unclassified genus of Comamonadaceae, and Pedosphaera.
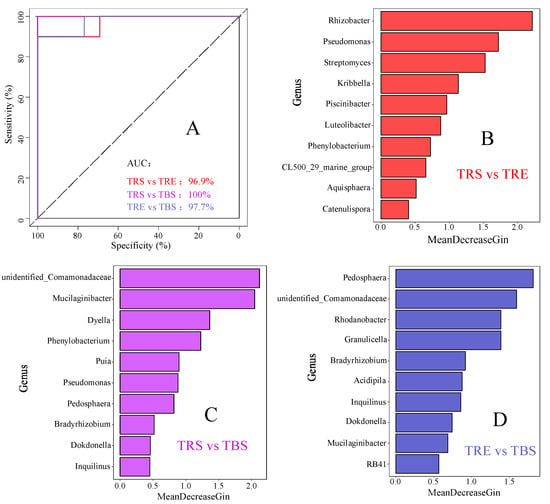
Figure 5.
Bacterial keystone taxa in different regions of Tuber sinense-producing areas predicted by random forest models: (A) model confidence prediction; (B) potential key bacterial taxa distinguishing rhizosphere soil (TRS) and rhizoplane soil (TRE); (C) potential key bacterial taxa distinguishing rhizosphere soil (TRS) and bulk soil (TBS); (D) potential key bacterial taxa distinguishing rhizoplane soil (TRE) and bulk soil (TBS).
4. Discussion
Studies have shown that bacterial communities in truffle-producing and control soils, as well as among different regions, exhibit pronounced heterogeneity [,,]. This may result from the ecological filtering effects of truffle mycelia, which can alter microbial niches and resource availability. Similarly, in our study, bacterial communities in Tuber sinense-producing and control soils displayed differences at the genus level, despite sharing similar dominant taxa at higher taxonomic ranks. Notably, the relative abundances of these dominant taxa varied substantially. Comparable patterns have also been observed in soils with artificially inoculated truffles [,], suggesting that truffle presence consistently influences soil bacterial community composition.
The diversity of bacterial communities between truffle-producing and control soils has yielded inconsistent results in previous studies. Some have reported lower diversity in truffle-associated soils, while others found no significant reduction or even increased diversity [,,]. Such discrepancies may stem from environmental variation among sampling sites, inherent differences in soil microbial communities, or interactions between microbes [].
In our study, alpha diversity analyses indicated that truffle-producing soils had higher bacterial richness compared to control soils, while phylogenetic diversity remained similar, suggesting that the communities were composed of more taxa with close evolutionary relationships. Beta diversity analyses further demonstrated that bacterial communities in producing areas were more clustered and compositionally similar, indicating that truffle presence may exert a selective effect on associated microbial assemblages. These findings are consistent with the results reported in earlier studies [].
Analysis of taxa with significant differences revealed that certain phototrophic (Chloroflexi) and anaerobic (Anaeromyxobacteraceae) groups were enriched in truffle-producing soils, while widespread environmental taxa such as Firmicutes and Bacteroidota were more prevalent in control soils. Although not directly tested here, truffle exudates and volatile organic compounds (VOCs) may influence microbial recruitment. This pattern suggests that T. sinense may selectively enrich or suppress specific bacterial taxa, potentially creating a more favorable microhabitat for its growth and symbiosis.
Within different zones of the truffle-producing area, several beneficial microbial taxa commonly associated with ectomycorrhizal formation were significantly enriched in the rhizosphere. These included members of the order Rhizobiales, genus Bradyrhizobium, and species Bradyrhizobium elkanii. These taxa are known for nitrogen fixation and promotion of ectomycorrhizal formation. Given their consistent presence in productive areas, such bacteria could serve as microbial indicators of habitat suitability or be further explored as bioinoculants to enhance T. sinense domestication efforts. Similar patterns have been reported in earlier studies [,,], underscoring the relevance of root-associated beneficial microbes in truffle symbiosis.
Furthermore, random forest analysis revealed that several genera—such as Pseudomonas, Streptomyces, and Bradyrhizobium—served as key discriminative taxa between rhizosphere, root surface, and bulk soils. These genera have also been reported as dominant taxa in the rhizosphere of other truffle species [,,], supporting the notion that truffle colonization can exert a strong selective pressure on surrounding microbial communities. Such selection may reduce antagonistic interactions and enrich beneficial microbes that support symbiosis, nutrient exchange, or pathogen suppression. Importantly, the consistent enrichment of these beneficial taxa in truffle-associated soils suggests they may play facilitative roles in the formation or maintenance of T. sinense ectomycorrhizae. This raises the possibility that inoculation of these microbial taxa into truffle plantation soils could help engineer a favorable microbiome, potentially enhancing truffle colonization success and yield. Therefore, the bacterial taxa identified in this study may serve as valuable microbial candidates for the development of bio-inoculants or microbiome management strategies aimed at supporting T. sinense cultivation and domestication.
This study is limited by its geographic scope (a single site in southwestern China), which may affect generalizability across different T. sinense habitats. Moreover, functional validation (e.g., metagenomics or metabolite profiling) was not conducted, making it difficult to establish causality between microbial shifts and truffle presence. Future studies should incorporate multi-omics approaches and experimental inoculation trials to better elucidate functional interactions.
5. Conclusions
This study systematically examined the spatial distribution of bacterial communities associated with T. sinense across multiple soil compartments and applied machine learning (random forest) to identify key microbial indicators. The composition and diversity of bacterial communities differed significantly between T. sinense-producing and control soils, and across distinct compartments within producing areas. These differences suggest that T. sinense is associated with the enrichment of symbiosis-promoting and stress-tolerant bacterial taxa—such as Chloroflexi, Anaeromyxobacteraceae, and Bradyrhizobium—which may enhance host–microbe interactions and support truffle development. Our results improve the ecological understanding of truffle–microbe associations and highlight specific bacterial taxa—such as Pseudomonas and Streptomyces—as potential microbial indicators of productive sites. In the context of truffle domestication and artificial cultivation, these taxa may be explored as bioinoculants or site-selection tools. Future research should validate their functional roles in promoting truffle colonization, nutrient cycling, and fruiting body formation in managed plantations.
Author Contributions
Conceptualization, T.M. and H.L.; methodology, T.M. and R.X.; software, Y.C. and H.X.; validation, J.L.; formal analysis, C.P.; resources, T.M.; writing—original draft preparation, T.M.; writing—review and editing H.L., Y.L. and R.L.; visualization, T.M. and R.L.; project administration, Y.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China 52460026; Henan Province Science and Technology Research Project 252102111078; National Modern Agricultural Industry Technology System Project of the Ministry of Finance and the Ministry of Agriculture and Rural Affairs, grant number CARS-13; Major Emergency Response Project for Agricultural Production in Henan Province, grant number 2024ZDYJ001; Independent Innovation Project of Henan Academy of Agricultural Sciences 2025ZC17.
Data Availability Statement
The original contributions presented in the study are included in the article; further inquiries can be directed to the corresponding authors.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Kües, U.; Martin, F. On the road to understanding truffles in the underground. Fungal Genet. Biol. 2011, 48, 555–560. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhang, J.L.; Li, T.; Sun, H.J.; Xiong, W.P.; Li, Y. Chinese black truffles: Tuber yigongense sp. nov., taxonomic reassessment of T. indicum s.l., and re-examination of the T. sinense isotype. Mycotaxon 2018, 133, 183–196. [Google Scholar] [CrossRef]
- García-Montero, L.G.; Díaz, P.; Di Massimo, G.; García-Abril, A. A review of research on Chinese Tuber species. Mycol. Prog. 2010, 9, 315–335. [Google Scholar] [CrossRef]
- Wang, Y.; Tan, Z.M.; Zhang, D.C.; Murat, C.; Jeandroz, S.; Le Tacon, F. Phylogenetic and populational study of the Tuber indicum complex. Mycol. Res. 2006, 110, 1034–1045. [Google Scholar] [CrossRef]
- Fan, L.; Li, T.; Xu, Y.; Yan, X. Species diversity, phylogeny, endemism and geography of the truffle genus Tuber in China based on morphological and molecular data. Pers.-Mol. Phylogeny Evol. Fungi 2022, 48, 175–202. [Google Scholar] [CrossRef]
- Ogawa, M. Microbial ecology of ‘Shiro’ in Tricholoma matsutake (S. Ito & Imai) Sing. and its allied species. V. Tricholoma matsutake in Tsuga sieboldii forests. Trans. Mycol. Soc. Jpn. 1977, 18, 34–46. [Google Scholar]
- Stamets, P. Mycelium Running: How Mushrooms Can Help Save the World; Penguin Random House: New York, NY, USA, 2005; p. 339. [Google Scholar]
- Pacioni, G. Effects of Tuber metabolites on the rhizospheric environment. Mycol. Res. 1991, 95, 1355–1358. [Google Scholar] [CrossRef]
- Streiblová, E.; Gryndlerová, H.; Gryndler, M. Truffle brûlé: An efficient fungal life strategy. FEMS Microbiol. Ecol. 2012, 80, 1–8. [Google Scholar] [CrossRef]
- Taschen, E.; Sauve, M.; Vincent, B.; Parladé, J.; van Tuinen, D.; Aumeeruddy-Thomas, Y.; Assenat, B.; Selosse, M.A.; Richard, F. Insight into the truffle brûlé: Tripartite interactions between the black truffle (Tuber melanosporum), holm oak (Quercus ilex) and arbuscular mycorrhizal plants. Plant Soil 2020, 446, 577–594. [Google Scholar] [CrossRef]
- Chen, J.; Li, J.M.; Tang, Y.J.; Xing, Y.M.; Qiao, P.; Li, Y.; Liu, P.G.; Guo, S.X. Chinese black truffle-associated bacterial communities of Tuber indicum from different geographical regions with nitrogen fixing bioactivity. Front. Microbiol. 2019, 10, 2515. [Google Scholar] [CrossRef]
- Siebyła, M.; Szyp-Borowska, I.; Młodzińska, A. Bacterial communities inhabiting the ascomata of the ectomycorrhizal summer truffle (Tuber aestivum). Appl. Soil Ecol. 2024, 199, 105428. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, J.; Xiong, C.; Li, X.; Chen, Z.; Li, P.; Huang, W. Tuber indicum shapes the microbial communities of ectomycorhizosphere soil and ectomycorrhizae of an indigenous tree (Pinus armandii). PLoS ONE 2017, 12, e0175720. [Google Scholar] [CrossRef] [PubMed]
- Rondolini, M.; Zotti, M.; Bragato, G.; Baciarelli Falini, L.; Reale, L.; Donnini, D. The expanding truffle environment: A study of the microbial dynamics in the old productive site and the new Tuber magnatum picco habitat. J. Fungi 2024, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Barbieri, E.; Ceccaroli, P.; Agostini, D.; Zeppa, S.D.; Gioacchini, A.M.; Stocchi, V. Truffle-associated bacteria: Extrapolation from diversity to function. In True Truffle (Tuber spp.) in the World; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Volume 47, p. 436. [Google Scholar]
- Piñuela, Y.; Alday, J.G.; Oliach, D.; Bolaño, F.; Colinas, C.; Bonet, J.A. Use of inoculator bacteria to promote Tuber melanosporum root colonization and growth on Quercus faginea saplings. Forests 2020, 11, 792. [Google Scholar] [CrossRef]
- Giorgi, V.; Amicucci, A.; Landi, L.; Castelli, I.; Romanazzi, G.; Peroni, C.; Ranocchi, B.; Zambonelli, A.; Neri, D. Effect of bacteria inoculation on colonization of roots by Tuber melanosporum and growth of Quercus ilex seedlings. Plants 2024, 13, 224. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Garbaye, J.; Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 2007, 176, 22–36. [Google Scholar] [CrossRef]
- Seethalakshmi, P.; Kumaresan, T.; Vishnu Prasad Nair, R.; Prathiviraj, R.; Seghal Kiran, G.; Selvin, J. Comparative analysis of commercially available kits for optimal DNA extraction from bovine fecal samples. Arch. Microbiol. 2024, 206, 314. [Google Scholar] [CrossRef]
- Soliman, H.; Ismaeil, M.; Soussa, H.; El-Sayed, W.S. Unveiling organohalide respiration potential in River Nile sediments via 16S rRNA gene amplicon sequencing of endogenous bacterial communities. BMC Microbiol. 2025, 25, 186. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Wu, F.; Ma, S.; Zhou, J.; Han, C.; Hu, R.; Yang, X.; Nie, G.; Zhang, X. Genetic diversity and population structure analysis in a large collection of white clover (Trifolium repens L.) germplasm worldwide. PeerJ 2021, 9, e11325. [Google Scholar] [CrossRef] [PubMed]
- Khleborodova, A.; Gamboa-Tuz, S.D.; Ramos, M.; Segata, N.; Waldron, L.; Oh, S. lefser: Implementation of metagenomic biomarker discovery tool, LEfSe, in R. Bioinformatics 2024, 40, btae707. [Google Scholar] [CrossRef]
- Diaz-Uriarte, R. GeneSrF and varSelRF: A web-based tool and R package for gene selection and classification using random forest. BMC Bioinform. 2007, 8, 328. [Google Scholar] [CrossRef]
- Giorgio, M.; Niccolò, B.G.M.; Benedetta, T.; Luisa, M.; Leonardo, B.F.; Gregory, B.; Pietro, B.; Alberto, A.; Domizia, D.; Emidio, A. Fungal and bacterial diversity in the Tuber magnatum ecosystem and microbiome. Microb. Ecol. 2023, 85, 508–521. [Google Scholar] [CrossRef]
- Sillo, F.; Vergine, M.; Luvisi, A.; Calvo, A.; Petruzzelli, G.; Balestrini, R.; Mancuso, S.; De Bellis, L.; Vita, F. Bacterial communities in the fruiting bodies and background soils of the white truffle Tuber magnatum. Front. Microbiol. 2022, 13, 864434. [Google Scholar] [CrossRef]
- Antony-Babu, S.; Deveau, A.; Van Nostrand, J.D.; Zhou, J.; Le Tacon, F.; Robin, C.; Frey-Klett, P.; Uroz, S. Black truffle-associated bacterial communities during the development and maturation of Tuber melanosporum ascocarps and putative functional roles. Environ. Microbiol. 2014, 16, 2831–2847. [Google Scholar] [CrossRef] [PubMed]
- Alhuthali, S.; Bello, S.K.; Bageel, A.M.; Shori, A.B.; Bataweel, N.M.; Al-Hejin, A.M.; Al-Qarawi, A.A.; Thomas, P.W. Soil physicochemical and metagenomic analyses of bacteria and fungi: Toward desert truffle cultivation in Saudi Arabia. Agronomy 2024, 14, 3021. [Google Scholar] [CrossRef]
- Splivallo, R.; Vahdatzadeh, M.; Maciá-Vicente, J.G.; Molinier, V.; Peter, M.; Egli, S.; Uroz, S.; Paolocci, F.; Deveau, A. Orchard conditions and fruiting body characteristics drive the microbiome of the black truffle Tuber aestivum. Front. Microbiol. 2019, 10, 1437. [Google Scholar] [CrossRef]
- Mello, A.; Ding, G.C.; Piceno, Y.M.; Napoli, C.; Tom, L.M.; DeSantis, T.Z.; Andersen, G.L.; Smalla, K.; Bonfante, P. Truffle brûlés have an impact on the diversity of soil bacterial communities. PLoS ONE 2013, 8, e61945. [Google Scholar] [CrossRef]
- Liu, D.; Pérez-Moreno, J.; He, X.; Garibay-Orijel, R.; Yu, F. Truffle microbiome is driven by fruit body compartmentalization rather than soils conditioned by different host trees. Msphere 2021, 6, e0003921. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.Y.; Peng, M.H.; Wang, J.J.; Ye, W.Y.; Li, Y.L.; Zhang, T.; Wang, A.R.; Zhang, D.M.; Wang, Z.H.; Lu, G.D.; et al. Microbial community associated with ectomycorrhizal Russula symbiosis and dominated nature areas in southern China. FEMS Microbiol. Lett. 2021, 368, fnab028. [Google Scholar] [CrossRef]
- Monaco, P.; Naclerio, G.; Mello, A.; Bucci, A. Role and potentialities of bacteria associated with Tuber magnatum: A mini-review. Front. Microbiol. 2022, 13, 1017089. [Google Scholar] [CrossRef]
- Graziosi, S.; Puliga, F.; Iotti, M.; Amicucci, A.; Zambonelli, A. In vitro interactions between Bradyrhizobium spp. and Tuber magnatum mycelium. Environ. Microbiol. Rep. 2024, 16, e13271. [Google Scholar] [CrossRef]
- Satish, L.; Barak, H.; Keren, G.; Yehezkel, G.; Kushmaro, A.; Ben-Dov, E.; Kagan-Zur, V.; Barak, Z.; Sitrit, Y. The microbiome structure of the symbiosis between the desert truffle Terfezia boudieri and its host plant Helianthemum sessiliflorum. J. Fungi 2022, 8, 1062. [Google Scholar] [CrossRef]
- Liu, D.; Chater, C.C.; Yu, F.; Perez-Moreno, J. Tuber pseudohimalayense ascomata-compartments strongly select their associated bacterial microbiome from nearby pine forest soils independently of their maturation stage. Pedobiologia 2021, 87–88, 150743. [Google Scholar] [CrossRef]
- Deveau, A.; Antony-Babu, S.; Le Tacon, F.; Robin, C.; Frey-Klett, P.; Uroz, S. Temporal changes of bacterial communities in the Tuber melanosporum ectomycorrhizosphere during ascocarp development. Mycorrhiza 2016, 26, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, Q.; Guo, W.; Wu, F.; Chen, J.; Liu, P.; Tian, W.; Qiao, P. Microbial communities of ascocarps and soils in a natural habitat of Tuber indicum. Arch. Microbiol. 2022, 204, 189. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).