Morphology and Molecular Phylogenetic Characterization of Novel Tar Spot Disease-Causing Fungi on Fabaceae Trees in Thailand
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Morphological Study, and Herbarium Deposit
2.2. DNA Extraction, PCR Amplification, and Sequencing
2.3. Sequence Alignment and Phylogenetic Analyses
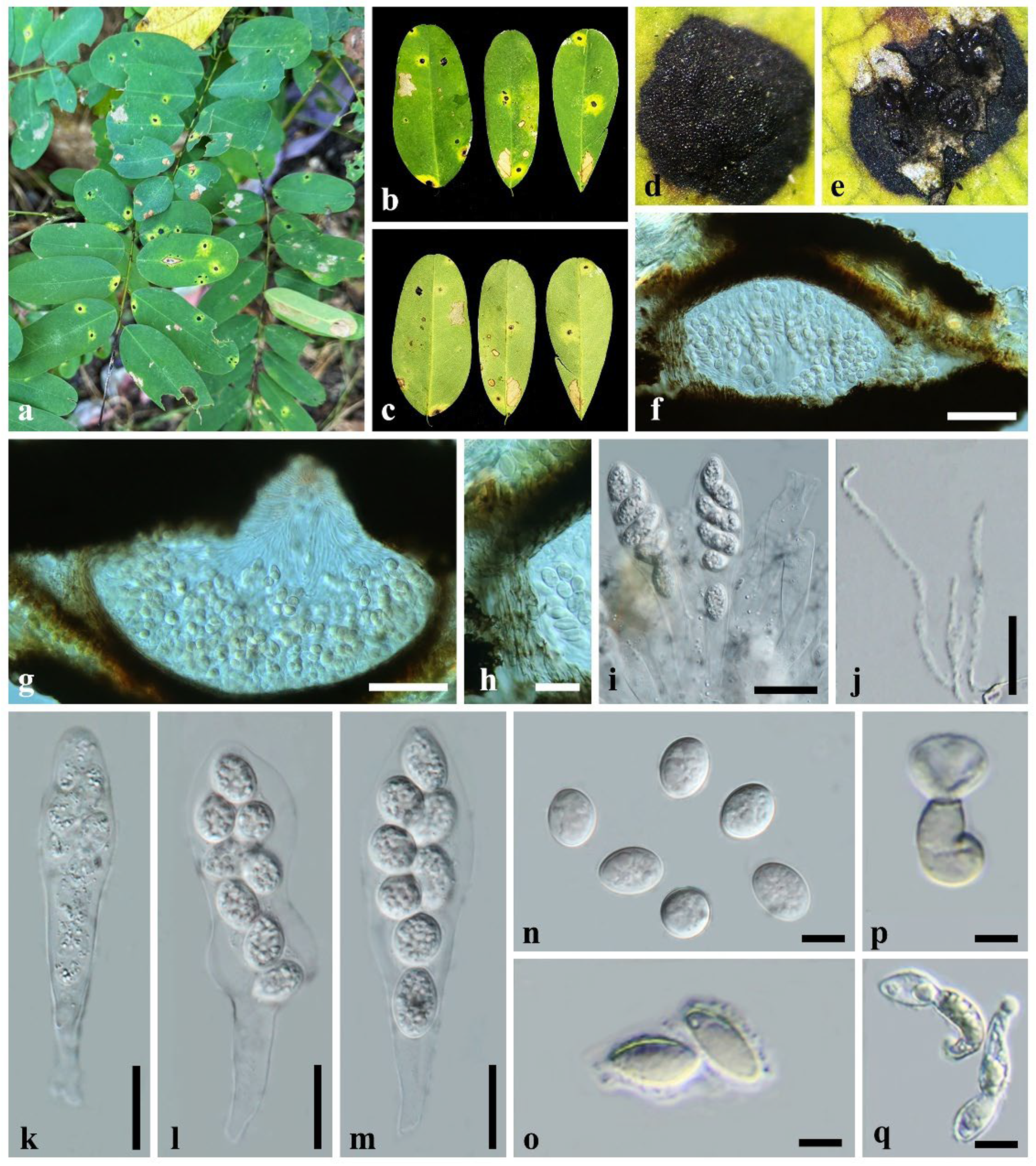
3. Results
3.1. Phylogenetic Analysis
3.2. Taxonomy
4. Discussion
- 1.
- Parasitic on Dalbergia Ficus, Myrcia, Myrciaria, and Pterocarpus species..................................................................................................................................................2
- 1′.
- Parasitic on Psidium species, ascospores thin-walled, short-ellipsoidal covered with thin-walled gelatinous sheath..............................................................................N. subcircinans
- 2.
- Parasitic on Dalbergia, Ficus, Myrcia, and Pterocarpus species..............................................3
- 2′.
- Parasitic on Myrciaria species, clavate-fusoid asci...........................................N. myrciariae
- 3.
- Parasitic on Dalbergia, Ficus, Myrcia and Pterocarpus, species..............................................4
- 3′.
- Parasitic on Myrcia species, lunate-reniform to half-moon shape ascospores with thick walled ....................................................................................................................N. truncatispora
- 4.
- Parasitic on Myrcia species…………………………………………….……………………..5
- 4′.
- Parasitic on Dalbergia, Ficus and Pterocarpus, species…………….…………..……………6
- 5.
- Elliptic-Oblong microguttulate ascospores...................................................... N. cerradensis
- 5′.
- Thin walledLunate ascospores …… ..................................................................... N. myrciae
- 6.
- Parasitic on Ficus species………………………………………………………….………….7
- 6′.
- Parasitic on Dalbergia and Pterocarpus………………………………………………………8
- 7.
- Parasitic on Ficus species, globose to ellipsoidal ascospores with gelatinous ........N. religiosa
- 7′.
- Parasitic on Ficus species, globose to subglobose ascospores without gelatinous sheath..................................................................................................................................... N. fici
- 8.
- Parasitic on Dalbergia species………………………….………………………. N. dalbergiae
- 8′.
- Parasitic on Pterocarpus species…………………….…………….N. pterocarpi-macrocarpae
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Q.-T.; Guo, M.-J.; Lv, T.; Zhou, H.; Wang, S.; Wang, S.-J.; Lin, Y.-R.; Gronefeld, S.; Kirschner, R.; Piepenbring, M.; et al. Phylogeny and Taxonomy of Rhytisma-like Species Worldwide. Fungal Divers. 2023, 120, 77–119. [Google Scholar] [CrossRef]
- Cannon, P.F. A Revision of Phyllachora and Some Similar Genera on the Host Family Leguminosae; Mycological Papers; CAB International: Wallingford, UK, 1991; pp. 1–302. [Google Scholar]
- Dos Santos, M.D.M.; de Noronha Fonseca, M.E.; Silva Boiteux, L.; Câmara, P.E.A.S.; Dianese, J.C. ITS Phylogeny and Taxonomy of Phyllachora Species on Native Myrtaceae from the Brazilian Cerrado. Mycologia 2016, 108, 1141–1164. [Google Scholar] [CrossRef] [PubMed]
- Cannon, P.F. Diversity of the Phyllachoraceae with Special Reference to the Tropics. Biodivers. Trop. Microfungi 1997, 8, 255–278. [Google Scholar]
- Theissen, F.; Sydow, H. Die Dothideales. Kritisch-Systematische Original Untersuchungen. Ann. Mycol. 1915, 13, 147–746. [Google Scholar]
- MacCready, J.S.; Roggenkamp, E.M.; Gdanetz, K.; Chilvers, M.I. Elucidating the Obligate Nature and Biological Capacity of an Invasive Fungal Corn Pathogen. MPMI 2023, 36, 411–424. [Google Scholar] [CrossRef]
- Yang, C.-L.; Xu, X.-L.; Liu, Y.-G.; Hyde, K.D.; Mckenzie, E.H.C. A New Species of Phyllachora (Phyllachoraceae, Phyllachorales) on Phyllostachys heteroclada from Sichuan, China. Phytotaxa 2019, 392, 186. [Google Scholar] [CrossRef]
- Sutton, B.C.; Hodges, C.S. Hawaiian Forest Fungi. III. A New Species, Gloeocoryneum Hawaiiense, on Acacia Koa. Mycologia 1983, 75, 280–284. [Google Scholar] [CrossRef]
- Gabel, A.W. Host-Parasite Relations of Phyllachora Species on Native Grasses. Mycologia 1989, 81, 702–708. [Google Scholar] [CrossRef]
- Dayarathne, M. Neophyllachora Gen Nov. (Phyllachorales), Three New Species of Phyllachora from Poaceae and Resurrection of Polystigmataceae (Xylariales). Mycosphere 2017, 8, 1598–1625. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Phillips, A.J.L.; Xu, J.; Balasuriya, A.; Hyde, K.D.; Stępień, Ł.; Harischandra, D.L.; Karunarathna, A.; Yan, J.; Weerasinghe, J.; et al. Defining a Species in Fungal Plant Pathology: Beyond the Species Level. Fungal Divers. 2021, 109, 267–282. [Google Scholar] [CrossRef]
- Mueller, D.S.; Wise, K.A.; Sisson, A.J.; Allen, T.W.; Bergstrom, G.C.; Bissonnette, K.M.; Bradley, C.A.; Byamukama, E.; Chilvers, M.I.; Collins, A.A.; et al. Corn Yield Loss Estimates Due to Diseases in the United States and Ontario, Canada, from 2016 to 2019. Plant Health Prog. 2020, 21, 238–247. [Google Scholar] [CrossRef]
- Broders, K.; Iriarte-Broders, G.; Bergstrom, G.C.; Byamukama, E.; Chilvers, M.; Cruz, C.; Dalla-Lana, F.; Duray, Z.; Malvick, D.; Mueller, D.; et al. Phyllachora Species Infecting Maize and Other Grass Species in the Americas Represents a Complex of Closely Related Species. Ecol. Evol. 2022, 12, e8832. [Google Scholar] [CrossRef] [PubMed]
- Tennakoon, D.S.; Kuo, C.-H.; Maharachchikumbura, S.S.N.; Thambugala, K.M.; Gentekaki, E.; Phillips, A.J.L.; Bhat, D.J.; Wanasinghe, D.N.; De Silva, N.I.; Promputtha, I.; et al. Taxonomic and Phylogenetic Contributions to Celtis formosana, Ficus ampelas, F. septica, Macaranga tanarius and Morus australis Leaf Litter Inhabiting Microfungi. Fungal Divers. 2021, 108, 1–215. [Google Scholar] [CrossRef]
- Literatus, I.C.E.; Maharachchikumbura, S.S.N.; Withee, P.; Haituk, S.; Tamakaew, N.; Nguanhom, J.; Monkhung, S.; Thiyagaraja, V.; Cheewangkoon, R. Taxonomic and Phylogenetic Appraisal of a New Holomorphic Neophyllachora Species from Chiang Mai, Thailand. Phytotaxa 2023, 600, 259–271. [Google Scholar] [CrossRef]
- Gardner, S.; Sidisunthorn, P.; Anusarnsunthorn, V. (Eds.) A Field Guide to Forest Trees of Northern Thailand; Kobfai Publ. Project: Bankok, Thailand, 2000; ISBN 978-974-7798-29-6. [Google Scholar]
- Index Fungorum. Available online: https://www.indexfungorum.org/names/Names.asp (accessed on 20 August 2024).
- Vilgalys, R.; Hester, M. Rapid Genetic Identification and Mapping of Enzymatically Amplified Ribosomal DNA from Several Cryptococcus Species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS Primers with Enhanced Specificity for Basidiomycetes—Application to the Identification of Mycorrhizae and Rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT Online Service: Multiple Sequence Alignment, Interactive Sequence Choice and Visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-Oriented Conversion of DNA and Protein Alignments. Nucleic Acids Res. 2010, 38, W14–W18. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, USA, 14 November 2010; pp. 1–8. [Google Scholar]
- Rambaut, A. FigTree 2012. FigTree v1.3.1. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 5 November 2024).
- Sydow, H.; Sydow, P. Beschreibungen Neuer Südafrikanischer Pilze. Ann. Mycol. 1912, 10, 33–45. [Google Scholar]
- De Souza Neto, J.D.; Dos Santos, E.K.; Lucas, E.; Vetö, N.M.; Barrientos-Diaz, O.; Staggemeier, V.G.; Vasconcelos, T.; Turchetto-Zolet, A.C. Advances and Perspectives on the Evolutionary History and Diversification of Neotropical Myrteae (Myrtaceae). Bot. J. Linn. Soc. 2022, 199, 173–195. [Google Scholar] [CrossRef]
- Zerega, N.J.C.; Clement, W.L.; Datwyler, S.L.; Weiblen, G.D. Biogeography and Divergence Times in the Mulberry Family (Moraceae). Mol. Phylogenetics Evol. 2005, 37, 402–416. [Google Scholar] [CrossRef]
- Doyle, J.J.; Luckow, M.A. The Rest of the Iceberg. Legume Diversity and Evolution in a Phylogenetic Context. Plant Physiol. 2003, 131, 900–910. [Google Scholar] [CrossRef] [PubMed]
- Eric Schranz, M.; Mohammadin, S.; Edger, P.P. Ancient Whole Genome Duplications, Novelty and Diversification: The WGD Radiation Lag-Time Model. Curr. Opin. Plant Biol. 2012, 15, 147–153. [Google Scholar] [CrossRef]
- Braun, U.; Cook, R.T.A. Taxonomic Manual of the Erysiphales (Powdery Mildews); CBS Biodiversity Series; CBS-KNAW Fungal Biodiversity Centre: Utrecht, The Netherlands, 2012; ISBN 978-90-70351-89-2. [Google Scholar]
- Takamatsu, S. Studies on the Evolution and Systematics of Powdery Mildew Fungi. J. Gen. Plant Pathol. 2018, 84, 422–426. [Google Scholar] [CrossRef]
- Niinomi, S.; Takamatsu, S.; Havrylenko, M. Molecular Data Do Not Support a Southern Hemisphere Base of Nothofagus Powdery Mildews. Mycologia 2008, 100, 716–726. [Google Scholar] [CrossRef]


| Species | Sample Code | LSU | SSU | ITS |
|---|---|---|---|---|
| Camarotella costaricensis | MM-21 | KX430490 | KX451851 | KX451900 |
| Coccodiella calatheae | MP5133T | MF460370 | MF460376 | MF460366 |
| Coccodiella melastomatum | CMU78543 | U78543 | ||
| Coccodiella miconiae | ppMP1342 | KX430506 | KX451871 | MF460365 |
| Coccodiella miconiicola | SO-15 | MF460374 | MF460380 | MF460369 |
| Coccodiella toledoi | MM-165 | KX430488 | KX451865 | KX451917 |
| Neophyllachora cerradensis | UB21823 | — | — | KC683470 |
| Neophyllachora cerradensis | UB21908T | — | — | KC683471 |
| Neophyllachora dalbergiae | CDEP-50T | PV241510 | — | PV241504 |
| Neophyllachora dalbergiae | CDEP-51 | PV241511 | — | PV241505 |
| Neophyllachora fici | MFLU 19-2702 | — | — | MW114384 |
| Neophyllachora fici | NCYU 19-0061 | — | — | MW114385 |
| Neophyllachora fici | NCYU 19-0326 | — | — | MW114386 |
| Neophyllachora myrciae | UB21292 | — | — | KC683463 |
| Neophyllachora myrciae | UB22192 | — | — | KC683476 |
| Neophyllachora myrciariae | UB21781T | — | — | KC683469 |
| Neophyllachora pterocarpi-macrocarpae | CDEP-52T | PV241512 | PV241165 | PV241506 |
| Neophyllachora pterocarpi-macrocarpae | CDEP-53 | PV241513 | PV241166 | PV241507 |
| Neophyllachora pterocarpi-macrocarpae | CDEP-54 | PV241514 | PV241167 | PV241508 |
| Neophyllachora pterocarpi-macrocarpae | CDEP-55 | PV241515 | PV241168 | PV241509 |
| Neophyllachora subcircinans | UB09748 | — | — | KC683441 |
| Neophyllachora subcircinans | UB21347 | — | — | KC683466 |
| Neophyllachora subcircinans | UB21747 | — | KC902622 | KC683467 |
| Neophyllachora truncatispora | UB14083 | — | KC902614 | KC683448 |
| Neophyllachora religiosa | MFLU 23-0258 | — | — | OQ821004 |
| Phyllachora arthraxonis | MHYAU:072 | MG269803 | — | MG269749 |
| Phyllachora arundinellae | MHYAU:108 | MG269815 | — | MG269761 |
| Phyllachora capillipediicola | MHYAU 20089 | MG356698 | — | KY498084 |
| Phyllachora chloridis | MFLU 15-0173T | MF197499 | MF197505 | KY594026 |
| Phyllachora chloridis-virgatae | MHYAU 20136 | MG356685 | — | KY498122 |
| Phyllachora chongzhouensis | SICAU 24-0044 | PP785312 | PP785323 | PP785301 |
| Phyllachora chrysopogonicola | MFLU 16-2096T | MF372146 | — | MF372145 |
| Phyllachora cynodonticola | MFLU 16-2977T | MF197501 | MF197507 | KY594024 |
| Phyllachora cynodontis | MHYAU:20043 | KY498081 | — | KY471329 |
| Phyllachora dendrocalami-hamiltoniicola | MHYAU 221 | MK614118 | — | — |
| Phyllachora dendrocalami-membranacei | MHYAU 220 | MK614117 | — | MK614102 |
| Phyllachora flaccidudis | IFRD9445T | ON072101 | ON072097 | ON075524 |
| Phyllachora graminis | SICAU 24-0051 | PP785306 | PP785317 | PP785295 |
| Phyllachora heterocladae | MFLU 18-1221T | MK296472 | MK296468 | MK305902 |
| Phyllachora huiliensis | SICAU 24-0048 | PP785308 | PP785319 | PP785297 |
| Phyllachora imperatae | MHYAU:014 | MG269800 | — | MG269746 |
| Phyllachora indosasae | MHYAU 125 | MG195662 | — | MG195637 |
| Phyllachora isachnicola | MHYAU:179T | MH018563 | — | MH018561 |
| Phyllachora jiaensis | IFRD9448T | ON075440 | ON072100 | ON075527 |
| Phyllachora keralensis | MHYAU:20082 | MG269792 | — | KY498106 |
| Phyllachora maydis | BPI 893231 | — | — | KU184459 |
| Phyllachora maydis | BPI 910560 | — | — | MG881846 |
| Phyllachora miscanthi | SICAU 24-0050 | PP785305 | PP785316 | PP785294 |
| Phyllachora neidongensis | SICAU 24-0046 | PP785314 | PP785325 | PP785303 |
| Phyllachora panicicola | MFLU 16-2979T | MF197503 | MF197504 | KY594028 |
| Phyllachora pogonatheri | MHYAU:071 | MG269802 | — | MG269748 |
| Phyllachora pomigena | CBS 193.33 | MH866860 | — | MH855409 |
| Phyllachora qualeae | UB 21159 | — | — | KU682781 |
| Phyllachora sandiensis | IFRD9446T | ON075528 | ON072098 | ON075525 |
| Phyllachora sinobambusae | MHYAU 085 | MG195655 | — | MG195630 |
| Phyllachora sphaerocaryi | MHYAU 178T | MK614114 | — | MK614100 |
| Phyllachora thysanolaenae | MFLU 16-2071 | — | MF372147 | — |
| Phyllachora virgataes | IFRD9447T | ON075439 | ON072099 | ON075526 |
| Phyllachora yushaniae-falcatiauritae | MHYAU 123 | MG195656 | — | MG195631 |
| Phyllachora yushaniae-polytrichae | MHYAU 122 | MG195657 | MH992455 | MG195632 |
| Polystigma pusillum | MM-19 | KX430489 | KX451850 | KX451899 |
| Telimena bicincta | MM-108 | KX430473 | KX451857 | KX451906 |
| Telimena bicincta | MM 133 | KX430478 | KX451861 | KX451910 |
| Neophyllachora Species | Phyllachora s.l. | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Morphological Characteristics | N. cerradensis | N. fici | N. myrciae | N. myrciariae | N. subcircinans | N. truncatispora | N. religiosa | N. dalbergiae | N. pterocarpi-macrocarpae | P. pterocarpi |
| Pseudostromata (mm in diam) | 2–4 | 2–3 | 3–6 | 0.5–1.5 | 1–4 | 2–5 | 2–3 | 1–3 | 2–3 | 0.75–2 |
| Shape of Ascomata | Ampulliform to Globose | Globose to Subglobose | Globose to Ampulliform | Ampulliform | Ampulliform | Globose to Ovoid | Globose to Subglobose | Globose to sub globose | Globose to Ampulliform | Rotundate to illegular |
| Position of Ascomata | Occasionally coalescing | Epiphyllous | Occasionally coalescent | Immersed in the pseudostromata | Immersed in the pseudostromatic tissue | Immersed in pseudostroma | Epiphyllous | Epiphyllous | Epiphyllous | Epiphyllous |
| Peridium thickness (µm) | 17–21 | 20–25 | - | 19–24 | 15–21 | - | 15–40 | 25–35 | 22–40 | - |
| Size of Ascomata (µm) | 260–362 × 168–264 | 150–300 × 200–400 | 205–485 × 148–207 | 161–432 × 126–267 | 329–417 × 193–275 | 160–200 × 100–350 | 150–300 × 200–400 | 272–329 × 509–620 | 213–337 × 456–497 | 150–200 |
| Size of Asci (µm) | 69–92 × 17–28 | 90–100 × 15–19 | 89–117 × 13–19 | 63–90 × 11–15 | 73–100 × 12–19 | 69–117 × 14–25 | 55–185 × 11–26 | 90–100 × 19–25 | 81–106 × 19–29 | 45–80 × 15–25 |
| Shape of Asci | Fusoid | Cylindrical to Fusiform | Fusoid | Clavate–Fusoid | Cylindrical | - | Cylindrical to Fusiform | Cylindrical to clavate | Cylindrical to clavate | Cylindrical clavate to clavate |
| Width of Paraphyses (µm) | 1.6–3 | 1.5–2.5 | 2.5–4.5 | 2–3 | 2–3.5 µm | 2.5–5 | 1.5–2.5 | 1.1–2.3 | 2.8–5.0 | - |
| Septate of Paraphyses | Septate | Aseptate | Septate | Septate | Septate | Septate | Septate | Aseptate | Septate | - |
| Size of Ascospores (µm) | 15–22 × 6–9 | 12–13 × 10–11 | 14–18 × 5–7 | 14–19 × 5–8 | 11–16 × 7–9 | 18–26 × 7–8 | 8–15 × 5–12 | 13–17 × 11–13 | 18–21 × 9–12 | 14–18 × 8–11 |
| Shape of Ascospores | Elliptic–Oblong | Globose to Subglobose | Lunate | Elliptical | Oblong to Ellipsoid | Sublunate to Fusoid | Globose to elliptical | Globose to elliptical with a central concave depression | Globose, ovoid, ellipsoid, pyriform, fusiform, amygdaliform | Ellipsoid |
| Color of Ascospores | Hyaline | Hyaline | Hyaline | Hyaline | Hyaline to Light olivaceous | Hyaline | Hyaline to Light olivaceous | Hyaline | Hyaline | Hyaline |
| Ascospores arrangement | Biseriate | 1–2 seriate | Biseriate to multi-seriate | Obliquely biseriate | Mostly uniseriate, sometimes with biseriate | - | Mostly uniseriate, sometimes with biseriate | Obliquely uni- or biseriate | Irregularly biseriate | 1–2 seriate |
| Ascospores wall | Covered by a thin gelatinous sheath | Absent | Thin-walled | Covered by a thin gelatinous sheath | Thin wall surrounded by a gelatinous sheath | Wall thickenings at both acute ends | Covered by a thick gelatinous sheath | Covered by a gelatinous sheath, smooth to rough | Covered by a gelatinous sheath, smooth to rough | - |
| Guttules | Microguttulate cytoplasm | - | - | Irregularly guttulate | Centrally guttulate | - | Irregularly guttulate | Guttulate | Guttulate | - |
| Asexual morph | Coelomycete | Unknown | Coelomycete | Unknown | Unknown | Coelomycete | Unknown | Unknown | Unknown | - |
| Host | Leaves of Myrcia torta | Leaves of Ficus septica | Myrcia sp. | Leaves of Myrciaria delicatula | Psidium sp. | Leaves of Myrcia camapuanensis | Leaves of Ficus religiosa | Dalbergia sp. | Pterocarpus macrocarpus | Pterocarpus angolensis |
| Distribution | Brazil | Taiwan | Brazil | Brazil | Brazil | Brazil | Thailand | Thailand | Thailand | South Africa |
| References | [3] | [14] | [3] | [3] | [3] | [3] | [15] | Present study | Present study | [26] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haituk, S.; Karunarathna, A.; Harishchandra, D.L.; Arayapichart, S.; Nakashima, C.; Kodsueb, R.; Monkhung, S.; Cheewangkoon, R. Morphology and Molecular Phylogenetic Characterization of Novel Tar Spot Disease-Causing Fungi on Fabaceae Trees in Thailand. Forests 2025, 16, 556. https://doi.org/10.3390/f16040556
Haituk S, Karunarathna A, Harishchandra DL, Arayapichart S, Nakashima C, Kodsueb R, Monkhung S, Cheewangkoon R. Morphology and Molecular Phylogenetic Characterization of Novel Tar Spot Disease-Causing Fungi on Fabaceae Trees in Thailand. Forests. 2025; 16(4):556. https://doi.org/10.3390/f16040556
Chicago/Turabian StyleHaituk, Sukanya, Anuruddha Karunarathna, Dulanjalee Lakmali Harishchandra, Saruta Arayapichart, Chiharu Nakashima, Rampai Kodsueb, Sararat Monkhung, and Ratchadawan Cheewangkoon. 2025. "Morphology and Molecular Phylogenetic Characterization of Novel Tar Spot Disease-Causing Fungi on Fabaceae Trees in Thailand" Forests 16, no. 4: 556. https://doi.org/10.3390/f16040556
APA StyleHaituk, S., Karunarathna, A., Harishchandra, D. L., Arayapichart, S., Nakashima, C., Kodsueb, R., Monkhung, S., & Cheewangkoon, R. (2025). Morphology and Molecular Phylogenetic Characterization of Novel Tar Spot Disease-Causing Fungi on Fabaceae Trees in Thailand. Forests, 16(4), 556. https://doi.org/10.3390/f16040556







