Abstract
In order to protect and utilize the germplasm resource better, it is highly necessary to carry out a study on the genetic diversity of Camellia chekiangoleosa Hu. However, systematic research on population genetics analysis of the species is comparatively rare. Herein, 16 highly variable simple sequence repeat (SSR) markers were used for genetic structure assessment in 12 natural C. chekiangoleosa populations. The genetic diversity of C. chekiangoleosa was low (h = 0.596), within which, central populations (such as Damaoshan (DMS), Sanqingshan (SQS), and Gutianshan (GTS)) at the junction of four main mountain ranges presented high diversity and represented the center of the C. chekiangoleosa diversity distribution; the Hengshan (HS) population in the west showed the lowest diversity, and the diversity of the eastern and coastal populations was intermediate. C. chekiangoleosa exhibited a high level of genetic differentiation, and the variation among populations accounted for approximately 24% of the total variation. The major reasons for this situation are the small population scale and bottleneck effects in some populations (HS and Lingshan (LS)), coupled with inbreeding within the population and low gene flow among populations (Nm = 0.796). To scientifically protect the genetic diversity of C. chekiangoleosa, in situ conservation measures should be implemented for high-diversity populations, while low-diversity populations should be restored by reintroduction.
1. Introduction
The genus Camellia L. includes many valuable shrubs, economic trees, and endangered species belonging to the family Theaceae, among which Camellia japonica L. is an internationally known ornamental flower [1]. In addition, C. oleifera Abel., a species of oil tea camellia, is considered one of the four major woody oil tree species [2]. Oil tea camellia generally refers to a tree species in the genus Camellia with a high oil content and high production and cultivation value [3]. In China, oil tea camellia has a cultivation history of more than 2300 years, and it has been used as an oil tree species for more than 1000 years [4]. Currently, there are more than 20 species of oil tea camellias that are mainly planted in China [5]. As the two major oil tea camellia species, C. oleifera and C. chekiangoleosa Hu. have been usually cultivated in southern China [6]. However, compared with the former, no varieties or superior clones of C. chekiangoleosa have been developed, and it is urgent to utilize the germplasm resource of this species [7].
Camellia chekiangoleosa is a typical diploid oil tea camellia species (2 n = 30). The species is a relative of C. japonica and C. oleifera with excellent ornamental and oil value due to its beautiful flower color and excellent oil quality [8,9]. This species has become an important economic tree in southern China [7]. C. chekiangoleosa is still growing in the wild, without sufficient studies on related breeding and horticultural culture [8,10]. Early plant taxonomists believed that C. chekiangoleosa was naturally distributed in the mountains at an altitude of 600~1400 m (117° 34′~120°14′ E and 28°08′~29°13′ N) at the junction of five provinces (southern Zhejiang, southern Anhui, northeastern Jiangxi, southern Hunan, and northern Fujian), and its distribution once was narrow but continuous [11]. However, because of low natural fruit yield and great ornamental value, C. chekiangoleosa are often felled and transplanted in the natural environment. The large-scale excavation of mountains for farmland has also seriously damaged the natural habitat of C. chekiangoleosa, resulting in a sharp reduction in its natural resources. As early as the 1990s, due to the shrinking wild habitat, the concentrated distribution of C. chekiangoleosa is very rare. Nowadays, it has been listed as a precious and endangered plant in Zhejiang Province, China, and has been listed as a key protected plant in Fujian Province, China [12]. Today, the survival situation of C. chekiangoleosa is even very serious, and it is urgent to protect its natural resources.
The key to protecting biodiversity is to maintain the genetic diversity or evolutionary potential of species. The genetic variation levels and population genetic structures of plant populations result from the comprehensive actions of various factors, such as their evolutionary history, distribution range, life form, breeding mode, and seed dispersion mechanism, which are closely related to adaptability and evolutionary potential [13]. Therefore, the assessment of genetic variation levels and spatial structure of plants is the basis for exploring their adaptability, speciation process, patterns, and evolutionary mechanisms, and is related to the formulation of strategies and measures for species protection and population restoration [14,15]. A large amount of evidence shows that the average level of genetic variation in narrowly distributed species is significantly lower than that in widely distributed species [16,17]. However, even among narrowly distributed species, diversity levels and genetic structures are very different [18]. The population genetic structure of a species is also a product of its long-term evolution, and the population genetic structure observed in the endemic habitats of many species may reflect distinct events in their evolutionary history. For example, species with fragmented habitats are more prone to inbreeding depression and reduced diversity [19]. Therefore, the formulation of species protection strategies, on the basis of a full understanding of species diversity levels and population genetic structures, is a reasonable scientific approach. In addition, understanding the population genetic structures of cultivated species and wild relatives can help us to maintain their genetic integrity and diversity during breeding [20], thus allowing the in-depth discovery and utilization of various genes and genotype resources in the population and the prediction and scientific use of the variation of important economic traits.
The genetic diversity of C. chekiangoleosa, based on inter-simple sequence repeat (ISSR) markers, was carried out [12]. The previous research results are worth referencing; however, because of the limitations of the applied research methods and sample numbers, the results are relatively limited. Due to highly polymorphic, reproducible, abundant, inherited, co-dominant, and distributed genomes, simple sequence repeat (SSR) markers are very effective molecular markers in population genetics [17]. In recent years, there have been many research reports on the population genetic structure of species in the genus Camellia using SSR markers, such as those of C. petelotii (C.W. Chi), C. sinensis (L.) (Kuntze), C. huana (T.L. Ming and W.J. Zhang), and C. oleifera [21,22,23,24]. Therefore, under the premise of clarifying the resource distribution, the purpose of this study was to use SSR markers to analyze population genetic structure of C. chekiangoleosa. This approach can provide a molecular basis for the protection of C. chekiangoleosa natural resources and provide a technical reference for their rational utilization.
2. Materials and Methods
2.1. Population Sample Information
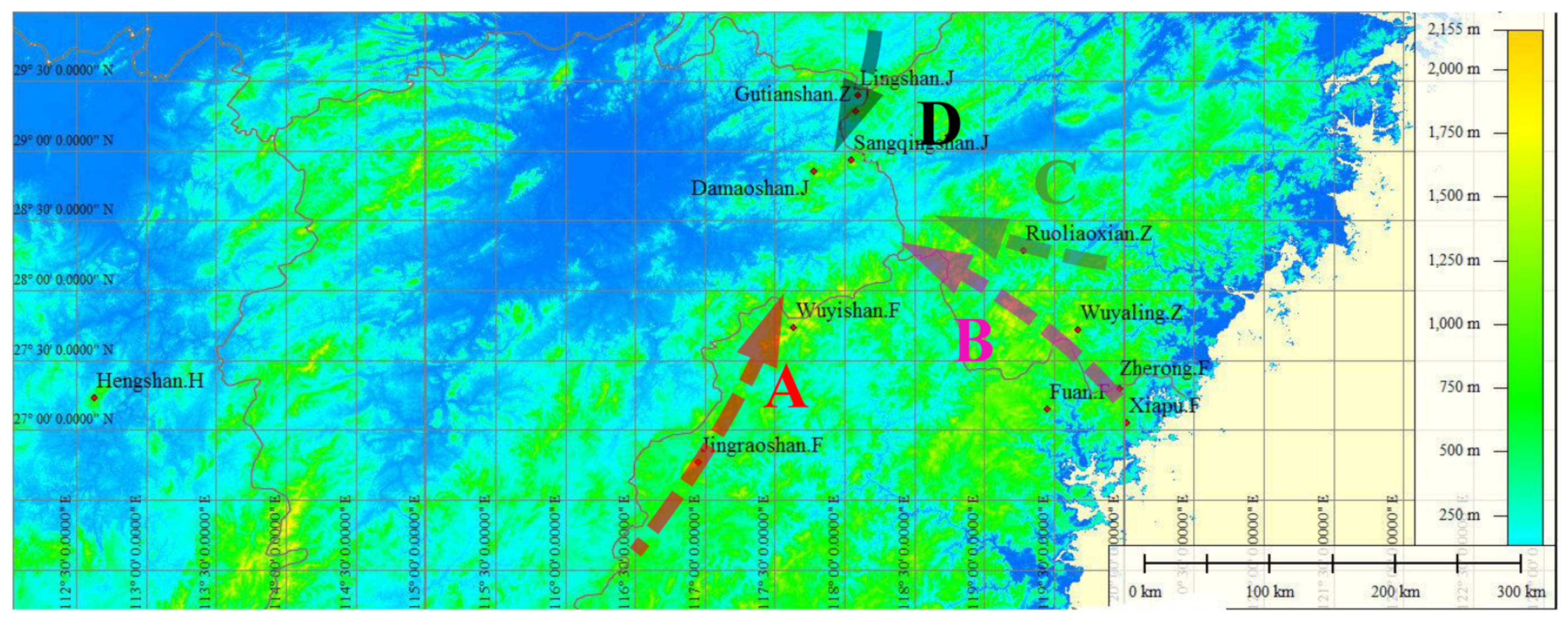
Samples were taken from 12 existing natural populations of C. chekiangoleosa within a certain scale in 4 provinces within the Jiangxi, Zhejiang, Fujian, and Hunan Provinces (Supplementary Materials Table S1). An appropriate sample size was selected according to the population size, but at least 30 single plants were sampled in each population; the distance between individuals was more than 20~30 m; the sampled plants showed normal growth without diseases or pests. Young tissue samples (young leaves, buds, or flower buds) were collected and stored via the silica gel drying method. Based on the longitude and latitude of the sampling points indicated by a GarminGPS60 device (WGS84 coordinate system), and the sampling map (Figure 1) was drawn by using Global Mapper [25]. The distribution of the geographic information and sample quantities at each sampling point (correspondence between population name and code) are also shown in Table S1.
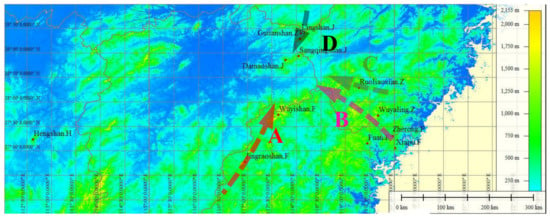
Figure 1.
Geographic distribution of the 12 sampled populations of C. chekiangoleosa. The dots in the figure are the collection points of each test sample. A—Wuyi mountain range population; B—Donggong mountain range population; C—Xianxia mountain range population; D—Huaiyu mountain range population.
2.2. Experimental Methods
Total DNA was extracted via the cetyl trimethylammonium bromide (CTAB) method [26], and a NanoDrop-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to detect the concentration of DNA. After electrophoresis in 0.8% agarose gelatin, the integrity of the purified DNA was determined, and qualified DNA samples were stored at −20 °C.
In this study, 16 pairs of highly polymorphic EST-SSR primers (Supplementary Materials Table S2) for C. chekiangoleosa were selected out from our previous research [11]. All primers were synthesized by Shanghai Generay Biotech Co., Ltd. The polymerase chain reaction (PCR) system had a volume of 10 µL, which included 1 µL of 10 × PCR buffer, 1.5 mM Mg2+, 100 µM dNTPs, upstream and downstream primers at 0.5 µM, 1.25 U of Taq polymerase (5 U/µL), and 50 ng of DNA. The amplification reaction procedure was as follows: pre denaturation at 94 °C for 5 min; 30 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s; a final extension at 72 °C for 1 min; then, storage at 8 °C. During the test, the annealing temperature was increased or decreased according to the melting temperature (Tm) of each primer. The PCR products were uniformly added to 8% polyacrylamide gels, along with a 50 bp labeled fragment size marker (TIANGEN, Beijing, China); standard electrophoresis was performed at a 120 V constant voltage for 2 h; the product was detected by silver staining. Digital photographs were recorded, and Quality One software (Bio–Rad Laboratories Inc., California, USA) was used to quantitatively analyze the fragment size of each detection site according to the standard molecular weight of the 50 bp marker; finally, the data were finally counted.
2.3. Data Analysis
The electrophoretic bands were read according to codominant markers, and the sizes of the allele fragments at each point were recorded. The data were imported into Excel according to the requirements. The polymorphism information content index (PIC) and linkage disequilibrium were calculated by using PowerMarker 3.25 (bioinformatics research center; North Carolina State University, Raleigh, NC, USA) [27], and Bonferroni correction was performed to adjust the P value. Popgene 1.32 software [28] was used to calculate the following: number of observed alleles (Na); number of effective alleles (Ne); Nei’s gene diversity (h); the Shannon diversity index (I); observed heterozygosity (Ho); expected heterozygosity (He); the inbreeding index (F). The genetic differentiation index (GST, based on He; FST, based on an infinite allele model, IAM; RST, based on a stepwise mutation model—SMM), gene flow (Nm = (1−FST)/4FST), and allele richness (AR) were calculated across all populations at each locus and over all loci using FSTAT 2.9.3 [29].
The genotype data format was transformed by using Genelex 6.4 [30]; an analysis of molecular variance (AMOVA) was carried out for the sources of different levels of genetic variation within and among populations using Arlequin 3.5 [31]. The results of the analysis of the genetic relationships between populations were displayed by factorial correspondence analysis (FCA) with the analysis software Genetix 4.05 (CNRSUMR 5000; Universite Montpellier II, Montpellier, France) [32]. The isolation by distance pattern (IBD) was detected by Mantel tests with 1000 permutations based on matrices of pairwise genetic differentiation (Multilocus estimates of genetic differentiation, Fst, expressed as Fst/(1−Fst)), and geographic distance among populations were performed in NTSYS 2.10e [33,34,35]. STRUCTURE 2.3.1 software [36] was used to infer the population genetic structure with the allelic model. Moreover, the number of distinguishable groups was initially set between k = 1 and k = 15, and the false positioning points were independent. The length of the burn-in period at the beginning of the Markov chain Monte Carlo simulation algorithm (MCMC) was set to 50,000 times, and the MCMC after non-counting iteration was then set to 50000 times for the estimation of the α value to determine whether a separate population existed (α ≤ 0.2 was the standard for the classification of the test samples). A suitable value of was selected according to the size of ∆K. ∆K is the difference in LnP(D) based on adjacent K values. Finally, Bottleneck 1.2 software [37] was used to determine whether each group had experienced a bottleneck effect in its recent history. A two-phase mutation model (TPM) and a stepwise mutation model (SMM) were used for Wilcoxon signed rank tests. The parameter settings included a 90% SMM and 10% TPM with 12% variation in TPMs and 1000 repeats.
3. Results
3.1. Genetic Diversity
Using 16 pairs of primers to detect 528 individuals in 12 geographical populations of C. chekiangoleosa (Table 1), a total of 74 alleles were obtained. The amplified alleles of a single primer ranged from 3 to 7. The average and effective allele values were 4.625 and 2.682, respectively. The highest amplified effective allele value obtained was for CC_eSSR03 (3.311), and the lowest obtained was for CC_eSSR16 (1.545). The average PIC value of the 16 loci was 0.616, reflecting a high polymorphism level, among which CC_eSSR16 showed the lowest PIC (0.352), and CC_eSSR03 showed the highest (0.698). These results were consistent with the effective equivalent gene comparison result, and the average h at the species level reached 0.596.

Table 1.
Information on the polymorphism of 16 microsatellite loci amplified in C. chekiangoleosa.
The results of the genetic diversity analysis of each geographical population of C. chekiangoleosa are shown in Table 2. Overall, the genetic diversity levels of different geographical populations of C. chekiangoleosa were quite different. AR ranged from 1.625 (HS) to 3.982 (GTS), with an average of 3.098, and the average He was 0.458. The SQS population presented the highest level of genetic diversity and the highest value of He, at 0.575. The lowest He value was found in the HS group, at 0.160. The 12 populations were ranked from high to low, according to the values He, as follows: SQS > DMS > GTS > LS/RLX > WYS > XP > tr > JRS > WYL > FA > HS. The diversity of LS and RLX was similar. The diversity parameter I ranged from 0.290 to 1.029 in the populations, with an average of 0.787. Both He and I show that the diversity of each population was maintained at an intermediate level. The results for the 12 populations sorted according to He and I were different, but the highest values appeared among DMS, SQS, and GTS, and the lowest values were obtained in the HS population, which also showed the lowest percentage of polymorphic loci among the 12 populations (43.75%). The above results indicate the reliability of genetic diversity evaluation.

Table 2.
Genotypic variation and heterozygosity statistics of 16 loci of C. chekiangoleosa.
The fixed index (F), also known as the inbreeding coefficient, can reflect the gene flow and inbreeding of the population. The F value of the natural C. chekiangoleosa population in this study was greater than 0, and the F value of the HS population was 0.087, which was the closest value to 0. The fixed indexes (F) of the RLX population and WYL population were relatively large, at 0.345 and 0.360, respectively, but those of the other populations were lower; the average F was 0.205, which generally indicated that the population deviated from the Hardy–Weinberg equilibrium, as reflected in the excess of homozygotes (Table 2).
3.2. Genetic Differentiation
The level of population genetic differentiation was analyzed based on Nei’s genetic diversity, IAM, and SMM. The statistics showed that the average GST, FST, and RST were 0.234, 0.239, and 0.253, respectively. It indicated that there was a high level of genetic differentiation among the C. chekiangoleosa populations (Table 1). Additionally, the AMOVA of the components within and among populations performed with Arlequin 3.5 software also showed that there was significant genetic variation (p < 0.001), where 24.15% of the genetic variation existed among populations, while 75.85% of the genetic variation occurred within populations (Table 3). In addition, the gene flow (Nm), estimated based on FST, was consistent with that estimated based on GST (Table 1; Supplementary Materials Table S3), with values of 0.796 and 0.731, respectively, representing the numbers of individuals that entered or exited a population in each generation among all populations studied. The measured values showed that the gene flow among populations was low, indicating relatively little exchange of genetic information among populations. The pairwise FST values between populations are shown in Supplementary Materials Table S4. The genetic differentiation between the SQS and DMS populations was lowest, at only 0.0901. The close geographical distances of the populations and their altitude may be the reasons for this phenomenon.

Table 3.
Analysis of molecular variance (AMOVA) within and among populations.
In this study, the TPM model and the SMM model of the Wilcoxon signed rank test [37] were used to analyze the bottleneck effect. According to the data (see Table 2), with the exception of the HS and LS groups, which recently experienced bottleneck effects (p < 0.05), no other groups experienced bottleneck effects.
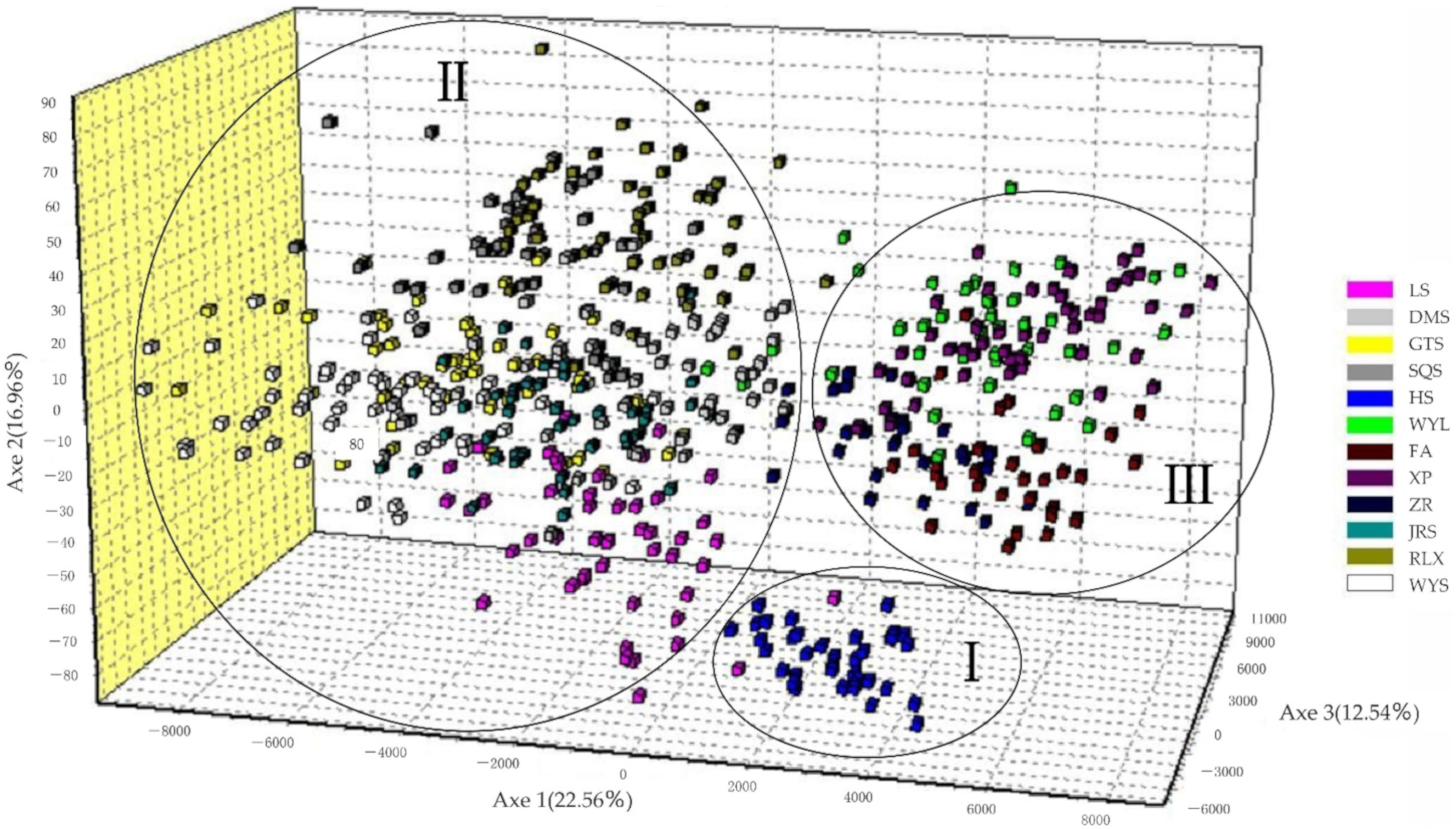
According to 3D graphics of factorial correspondence analysis, the 12 populations were divided into 3 groups: Group I, Group II, and Group III. Group I was the western cluster, and only contained HS groups; Group II was a central cluster with DMS at its core, which was distributed on the ridge connecting the Huaiyu Mountain Range to Wuyi Mountain Range in the south, and included the LS, GTS, SQS, DMS, RLX, WYS, and LRS populations; Group III was an eastern coastal cluster with WYL at its core, including the WYL, ZR, FA, and XP populations (Figure 2). There were certain genetic differences among the groups, especially between the HS population and the other populations.
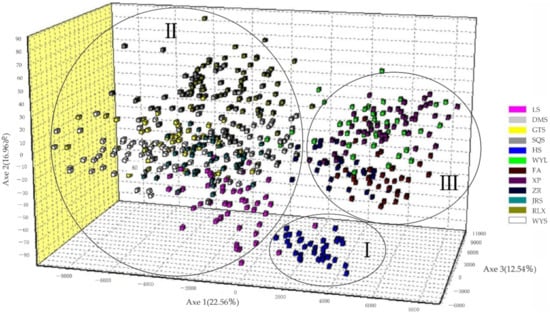
Figure 2.
Factorial correspondence analysis of the C. chekiangoleosa population. Axes 1, 2, and 3 explain 22.56, 16.96, and 12.54% of the genetic variation, respectively.
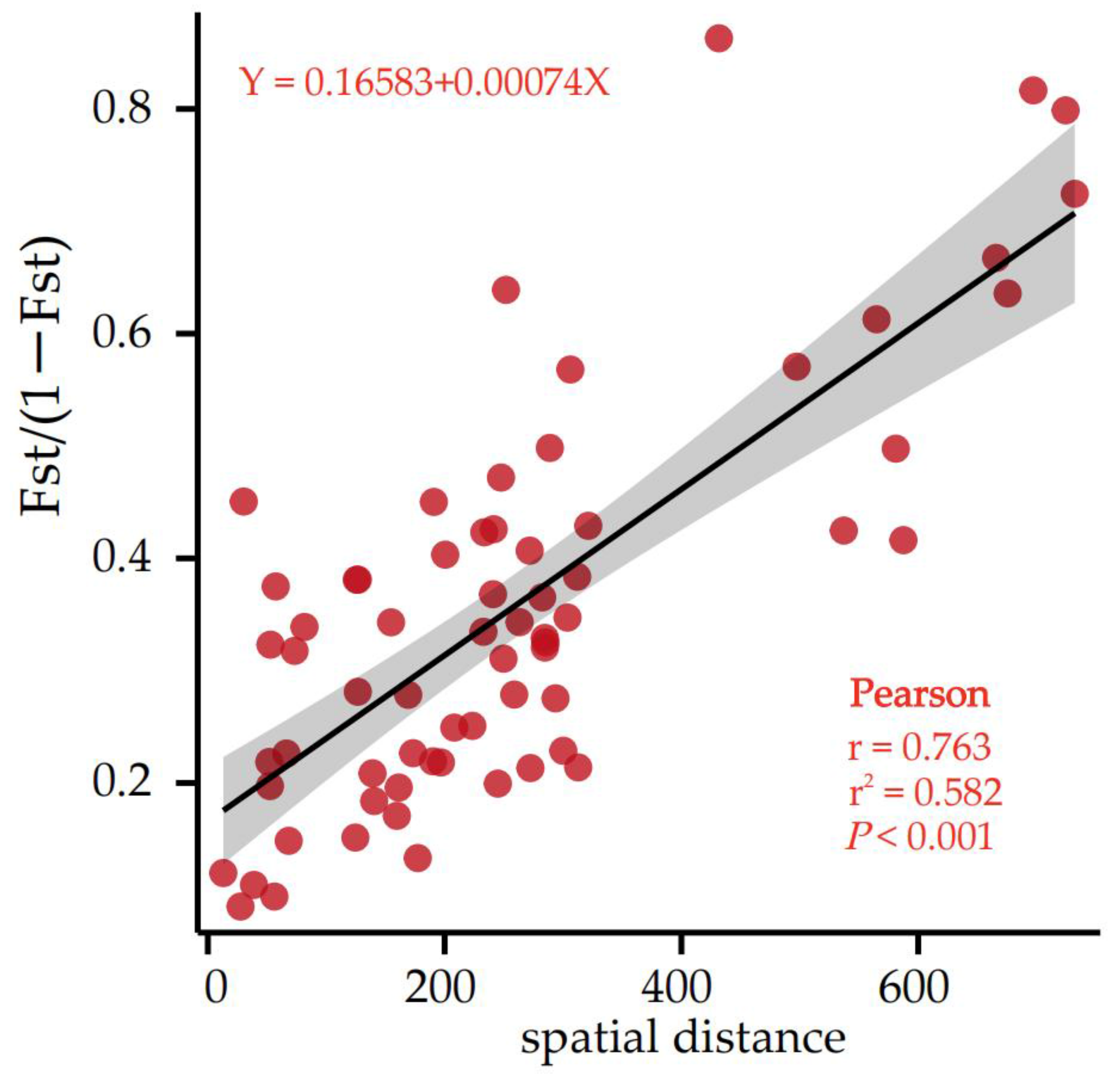
In addition, the correlation analysis between genetic differentiation (FST/1 − FST) and spatial geographical distance was conducted (Supplementary Materials Table S3) to check whether isolation by distance (IBD) existed. The correlation analysis showed that the correlation between the 2 groups was very significant (r = 0.763, p < 0.001), indicating that geographical isolation is the main cause of genetic differentiation (Figure 3).
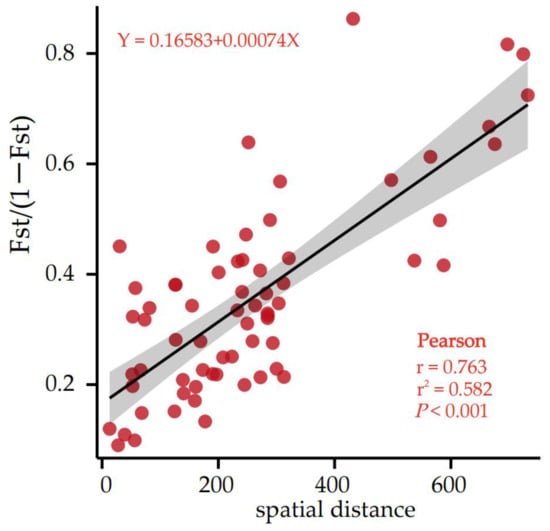
Figure 3.
Matrix plot of the Mantel test of genetic differentiation and geographic distance.
3.3. Genetic Structure
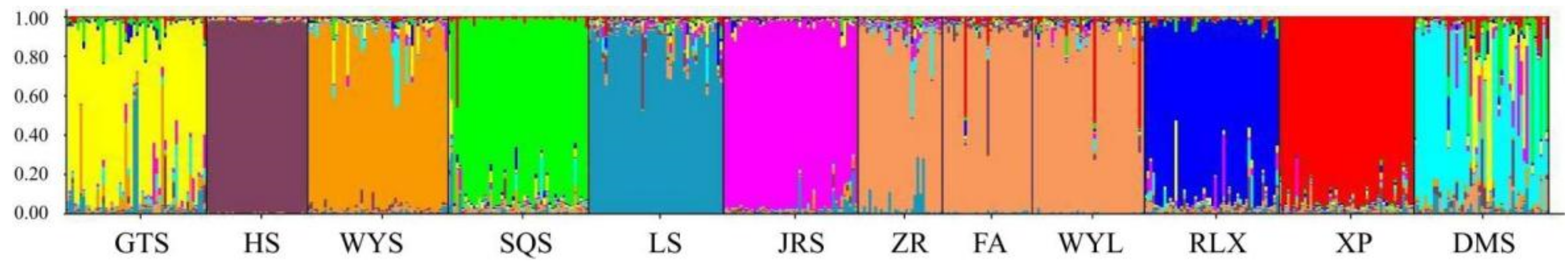
The genetic structure of the 12 studied populations was inferred using the STRUCTURE software. The most probable division with the highest ΔK value was detected at K = 10 (Supplementary Materials, Figure S1a,b). Figure 4 showed the genetic structure of the 12 C. chekiangoleosa populations, simulated based on the mathematical model. Each color represents a genetic cluster, and the colored segments show the individual’s estimated ancestry proportion to each of the genetic clusters. The 12 populations were clustered into 10 clusters. The DMS population presented the most gene introgression from the GTS, WYS, SQS, LS, and XP populations. The GTS population presented the most gene introgression from the WYS population and LS population. However, the ZR, FA, and WYL populations were mixed, and the degree of differentiation between these populations was extremely weak. Genes from the XP and LS populations mainly infiltrated the mixed group composed of the last three populations. In the SQS, LS, and XP populations, there were also some individuals with similar genetic backgrounds to the GTS population, and the HS population was the most independent.
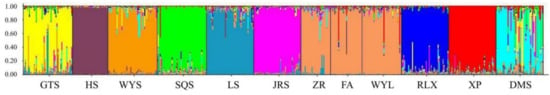
Figure 4.
Population genetic structure based on the Bayesian clustering model among 528 samples of C. chekaingoleosa at K = 10. Each color represents a genetic cluster.
4. Discussion
4.1. Evaluation of Genetic Diversity in Camellia chekiangoleosa
Previous research has shown that the geographical distribution range of a species often presents a strong correlation with its genetic diversity [23]. Especially, compared with widely distributed plant species, species with narrow geographical ranges or endangered species theoretically show lower genetic diversity [38]. In this study, C. chekiangoleosa showed low genetic diversity (h = 0.596) at the species level, that was higher than the average He (when the calculation was based only on polymorphism and double allele loci, this parameter was equivalent to Nei’s genetic diversity h) of endangered species (0.420) but lower than that of widely distributed species (0.620) [17]. The results are consistent with previous studies. The diversity level according to similar markers was even lower than that in other widely distributed related species. A total of 96 pairs of EST-SSR primers were used to evaluate the genetic diversity of C. sinensis in the main production areas of China, and the value of h reached 0.64 [39]. When 4 pairs of SSR molecular markers were used to evaluate the genetic diversity of the ancient C. japonica population, the He value reached 0.84 [40], which was higher than the genetic diversity of the wild populations of C. chekiangoleosa. Meanwhile, the assessment of population genetic diversity can reveal the species adaptability to the different environment [41]. Among the C. chekiangoleosa populations, the SQS, DMS, LS, and GTS populations, with high genetic diversity, are concentrated in the area linking the Wuyi Mountain Range population and the Huaiyu Mountain Range population in the north, which we believe is suitable establish areas for C. chekiangoleosa. In addition, because of the barrier imposed by the Heng Mountains, the diversity of the HS population in the west was lowest under the pressure accumulation of environmental selection.
It is helpful to analyze the reasons for the low genetic diversity of C. chekiangoleosa especially when compared with related species. In the genus Camellia, some endangered tree species with narrow distributions also show low genetic diversity [42]. When researchers evaluated the diversity of 12 wild populations of the narrowly distributed tree species C. huana with chloroplast fragments and 12 pairs of SSR primers, the obtained He, based on SSRs, was 0.466 [22]. Chen analyzed the genetic diversity of 7 populations of 3 endangered Camellia species (C. chrysanthoides (H.T. Chang), C. micrantha (S. Yun Liang and Y.C. Zhong), C. parvipetala (J.Y. Liang and Z.M. Su)) based on SSRs, and the calculated He values were 0.379~0.543 [43]. Li and Lu carried out genetic diversity studies on the endangered tree species C. petelotii and its variant C. petelotii var. microcarpa (S.L. Mo) (T.L. Ming and W.J. Zhang), and the obtained He values based on SSR markers were 0.546 and 0.533, respectively [20,44]. The genetic diversity of the above related species was thus shown to be similar or slightly higher than that recorded in our study. This is related to the fact that most populations of C. chekiangoleosa are not only affected by land loss but are also seriously disturbed by human beings. In addition, certain inbreeding phenomena were identified in each population, and the F values of the WYL and RLX populations reached 0.36 and 0.345, indicating that excess homozygosity is one of the internal reasons for the reduction in genetic diversity [45]. Some individual populations, such as the HS population, have experienced bottleneck effects and genetic drift, resulting in the loss of genetic diversity.
4.2. Population Genetic Structure Analysis Reveals Significant Differentiation of Populations
Compared with the genetic differentiation coefficients of 106 plant species based on SSR molecular markers [17], the genetic differentiation level of C. chekiangoleosa populations (FST = 0.239) was close to the average level of narrowly distributed species (FST = 0.23), but lower than widely distributed species (FST = 0.25). Therefore, C. chekiangoleosa presents a high differentiation level as a narrowly distributed species. AMOVA showed that 24.15% of the observed variation occurred among the populations of C. chekiangoleosa, which was similar to findings in the related species C. huana with a narrow distribution [23]. The result also reflected the fact that the population variation of C. chekiangoleosa was mainly attributed to the variation between individuals within the population. Moreover, our results (GST = 0.234) are lower than previous studies on the genetic differentiation of C. chekiangoleosa (GST = 0.3758) [12], which may be caused by different markers, and the population chosen in our study had higher coverage density.
Natural selection is the most important evolutionary driving force leading to population differentiation, while gene flow is an important factor hindering population differentiation [46]. In addition to the distribution characteristics mentioned above, the diffusion mechanism of plant pollen and seeds also impacts the average gene flow between populations [47]. The seeds of C. chekiangoleosa exhibit similar large and heavy traits to C. japonica and C. flavida (H.T. Chang), which are mainly transmitted by gravity [40,48]. C. chekiangoleosa is mainly distributed in high mountains at altitudes of 600~1400 m and is pollinated by insects. Therefore, mountains hinder the diffusion of seeds and pollen to a certain extent, thus limiting the spread of seeds and pollen [49]. The FST-based gene flow value (Nm) of C. chekiangoleosa was only 0.796, which is in line with the above statement. Slatkin concluded that, if the Nm of migrating individuals per generation is less than 1, genetic drift will become the dominant factor dividing the genetic structure of a population [50,51]. In this study, there was a significant positive correlation between spatial distance and genetic differentiation that reflected a significant geographical isolation effect among the populations of C. chekiangoleosa, which was consistent with the division of population genetic structure caused by lower gene flow [51].
The small-scale distribution of genetic variation within populations, and the large-scale spatial distribution among populations, are two of the important characteristics of population genetic structure [41]. This study showed that 12 geographical populations of C. chekiangoleosa were divided into 10 groups, and there was gene infiltration among the 10 groups, which also reflected the continuity of the population historical distribution of C. chekiangoleosa. The HS population was independent of several other populations, and the genotypes within individuals of this population were relatively distinct due to genetic drift. In addition, HS was located far from other populations, so it can be expected that further genetic drift will be revealed in this population. From the perspective of gene diversity and the Shannon index, it is found that the DMS population showed a relatively high level of diversity. The reason may be that the frequent activities of insects lead to the intensification of gene interaction in the low altitude environment with high average temperature. In addition, the combination of the FA, ZR, and WYL populations may have been related to the fact that they were all located in the Donggong mountain range, and these individuals may have come from the same or similar ancestors. In summary, we infer that C. chekiangoleosa populations can be basically divided into 3 distribution clusters according to the division of mountain ranges: a central cluster, with DMS as the center, which was distributed on the ridge connected with 4 mountain ranges (Figure 1A–D); an eastern coastal cluster with WYL as the center; a western cluster with HS as the center.
4.3. Conservation of Camellia chekiangoleosa Genetic Resources
In summary, this study indicates that the fundamental reason for the endangered status of C. chekiangoleosa is the restriction of gene flow among its populations, resulting in a reduction in genetic diversity and an inability to maintain the ability to adapt to the changing environment [52]. First, similar to other Camellia species, the pollen transmission mode of C. chekiangoleosa depends mainly on insects, while its seeds are mainly transmitted by gravity and animals, which are affected by mountain isolation [48]. Additionally, human activities have exacerbated the fragmentation of the natural habitat of C. chekiangoleosa, which is bound to further reduce the pollen flow between groups. In addition, according to the research of Yang [53], the pollen of C. chekiangoleosa shows a short survival time and a long pollen tube germination time, which is likely to result in abortion if flowers are not pollinated in a timely manner. According to our field investigation, due to the low average temperature in high-altitude mountainous areas, the flowering period of C. chekiangoleosa is long, from November to March of the next year. However, the flowering branches growing on the sunny side of the outer layer of the canopy bloom earlier and often bloom in the inner layer of the canopy, while the flowers in the outer layer are overmature. Therefore, the asynchronous flowering periods within a given stand affect the fruit setting of C. chekiangoleosa, imposing high environmental selection pressure on the existing population. If the protection of the species is not strengthened, its genetic differentiation will further increase, and the selfing rate of the remaining population and the probability of genetic drift will also increase, which will continue to reduce genetic diversity.
According to the results of the genetic diversity assessment and the genetic structure analysis, priority should be given to the local protection of populations with high diversity, such as the DMS, SQS, LS, and GTS populations. Protected areas or stations can be established to strengthen management and eliminate logging and damage. Second, the habitats suitable for the growth and population reproduction of C. chekiangoleosa should be restored with the aim of increasing the heterogeneity and stability of the habitat, which is particularly important for small populations. For populations with low genetic diversity (HS, FA), seeds should be collected within the population (or the most genetically similar population) whenever possible, and artificial seedling cultivation should be carried out according to the biological characteristics of the plants. These plants can then be reintroduced to the original habitat so that the population can slowly recover and grow—on the basis of preserving the original genotype.
5. Conclusions
In our research, natural populations of C. chekiangoleosa, covering the main distributed range, were used to study genetic diversity and differentiation. C. chekiangoleosa, as an endemic species with narrow distribution, has low genetic diversity, but its central population still has relatively high genetic diversity. In addition, the level of genetic differentiation among populations is high, and the genetic structure analysis also found that Hengshan population, central populations, and eastern populations can be divided into independent groups, indicating that the isolation of mountains may be the main reason for the formation of genetic differentiation. Therefore, considering the genetic diversity difference and significant differentiation of C. chekiangoleosa population, appropriate diversity protection strategies should be adopted according to the genetic diversity level and geographical distribution of different populations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/f13020234/s1, Table S1: Geographical distribution of 12 C. chekiangoleosa geographic populations, Table S2: Repeat motif, primer sequence, fragment size, Tm and data sources information for 16 EST-SSR loci, Table S3: Genetic differentiation in C. chekiangoleosa populations by Ney’s, Table S4: Pair-wise FST value and spatial distance matrix of C. chekiangoleosa, Figure S1: (a) The mean log-likelihood value of the data was based on ten repetitions for each K value, (b) The delta K value was changed with each K value.
Author Contributions
This study was carried out with collaboration among all authors. L.-a.X. conceived and designed the experiments; Q.W. and Z.W. performed the experiments; J.H. and Q.W. checked the experimental results; X.L. and H.Z. provided the experimental materials; B.H. and Q.W. wrote the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31860179 and 31260184), Key Research and Development Program of Jiangxi Province, China (20201BBF61003 and 20161BBF60122), Science and Technology Innovation Bases Program of Jiangxi province, China (20212BCD46002), Doctor Initial Project of JiangXi Academy of Forestry (2021521001), Oil-tea special research project of Jiangxi Provincial Department of Forestry (YCYJZX20220103).
Acknowledgments
We are very grateful to the Li-an Xu for initiating research ideas.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fan, M.L.; Kai, Y.; Rui, Z.; Liu, H.Q.; Xiao, G.; Sun, K.Y. Temporal transcriptome profiling reveals candidate genes involved in cold acclimation of Camellia japonica (Naidong). Plant Physiol. Biochem. 2021, 167, 795–805. [Google Scholar] [CrossRef]
- Zhang, J.; Ying, Y.; Yao, X. Effects of turning frequency on the nutrients of Camellia oleifera shell co-compost with goat dung and evaluation of co-compost maturity. PLoS ONE 2019, 14, e0222841. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.Y.; Wang, X.N.; Wang, L.P.; Chen, Y.Z.; Jiang, X.C. Genetic diversity of oil-tea camellia germplasms revealed by ISSR analysis. Int. J. Biomath. 2015, 5, 331–354. [Google Scholar] [CrossRef]
- Zhou, J.; Ai, Z.; Wang, H.; Niu, G.; Yuan, J. Phosphorus Alleviates Aluminum Toxicity in Camellia oleifera Seedlings. Int. J. Agric. Biol. 2019, 21, 237–243. [Google Scholar] [CrossRef]
- Fan, L.L.; Huang, J.S. Investigation on Species and Verieties pattern of Camellia sp. in Suichuan County. South China For. Sci. 2001, 4, 16–18. [Google Scholar] [CrossRef]
- Wang, X.; Zeng, Q.; María, D.; Wang, L.J. Profiling and quantification of phenolic compounds in Camellia seed oils: Natural tea polyphenols in vegetable oil. Food Res. Int. 2017, 102, 184–194. [Google Scholar] [CrossRef] [PubMed]
- He, Y.C.; Wu, M.J.; Dong, L.; Wen, Q.; Li, T.; Li, X.H. Analysis of kernel oil content and variation of fatty acid composition of Camellia chekiangoleosa in the main producing areas. Non-Wood For. Res. 2020, 3, 37–45. [Google Scholar] [CrossRef]
- Guo, H.; Tan, H.Y.; Zhou, J.P. Proximate composition of Camellia chekiangoleosa Hu fruit and fatty acid constituents of its seed oil. J. Zhejiang Univ. 2010, 36, 662–669. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, X. Comparative transcriptomic analysis identifies genes responsible for fruit count and oil yield in the oil tea plant Camellia chekiangoleosa. Sci. Rep. 2018, 8, 6637. [Google Scholar] [CrossRef]
- Zhou, W.C.; Wang, Z.W.; Dong, L.; Wen, Q.; Huang, W.Y.; Li, T.; Ye, J.S.; Xu, L.A. Analysis on the character diversity of fruit and seed of Camellia chekiangoleosa. J. Nanjing For. Univ. Nat. Sci. Ed. 2021, 45, 51–59. [Google Scholar] [CrossRef]
- Wen, Q.; Xu, L.; Gu, Y.; Huang, M.; Xu, L.A. Development of polymorphic microsatellite markers in Camellia chekiangoleosa (Theaceae) using 454-ESTs. Am. J. Bot. 2012, 99, e203–e205. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Li, J.Y.; Zhang, D.L. Assessment of Genetic Diversity and Population Structure of Endangered Camellia chekiangoleosa Hu Using Issr Markers. Pak. J. Bot. 2018, 50, 1965–1970. [Google Scholar]
- Hughes, A.R.; Inouye, B.D.; Johnson, M.T.J.; Underwood, N.; Vellend, M. Ecological consequences of genetic diversity. Ecol. Lett. 2008, 11, 609–623. [Google Scholar] [CrossRef] [PubMed]
- Kobori, H. Conservation Education Based on the Principles of Conservation Biology. Jpn. J. Environ. Educ. 2009, 19, 77–78. [Google Scholar] [CrossRef]
- Gentili, R.; Fenu, G.; Mattana, E.; Citterio, S.; Mattia, F.D.; Bacchetta, G.; Peeters, T. Conservation genetics of two island endemic Ribes spp. (Grossulariaceae) of Sardinia: Survival or extinction? Plant Biol. 2015, 17, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.B. Understanding crop genetic diversity under modern plant breeding. Theor. Appl. Genet. 2015, 128, 2131–2142. [Google Scholar] [CrossRef]
- Nybom, H. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol. Ecol. 2004, 13, 1143–1155. [Google Scholar] [CrossRef]
- Erfanian, M.B.; Sagharyan, M.; Memariani, F.; Ejtehadi, H. Predicting range shifts of three endangered endemic plants of the Khorassan-Kopet Dagh floristic province under global change. Sci. Rep. 2021, 11, 9159. [Google Scholar] [CrossRef]
- Oakley, C.G.; Winn, A.A. Effects of population size and isolation on heterosis, mean fitness, and inbreeding depression in a perennial plant. New Phytol. 2012, 196, 261–270. [Google Scholar] [CrossRef]
- Gentili, R.; Solari, A.; Diekmann, M.; Duprè, C.; Citterio, S. Genetic differentiation, local adaptation and phenotypic plasticity in fragmented populations of a rare forest herb. Peer J. 2018, 6, e4929. [Google Scholar] [CrossRef]
- Lu, X.L.; Chen, H.L.; Liang, X.Y.; Tang, S.Q. Genetic Diversity Analysis of C. nitidissima var. microcarpa and Peripheral Population of Camellia nitidissima. Mol. Plant Breed. 2019, 17, 301–306. [Google Scholar] [CrossRef]
- Liu, S.; Liu, H.; Wu, A.; Hou, Y.; An, Y.; Wei, C.L. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breed. 2017, 37, 93. [Google Scholar] [CrossRef]
- Li, S.; Liu, S.L.; Pei, S.Y.; Ning, M.M.; Tang, S.Q. Genetic diversity and population structure of Camellia huana (Theaceae), a limestone species with narrow geographic range, based on chloroplast DNA sequence and microsatellite markers. Plant Divers. 2020, 42, 343–350. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Liu, C.; Wang, X.; Wang, R.; Tian, Y. Assessment of genetic diversity in Camellia oleifera Abel. accessions using morphological traits and simple sequence repeat (SSR) markers. Breed. Sci. 2020, 70, 586–593. [Google Scholar] [CrossRef]
- Liu, F.L.; Xiao, B. Global Mapper System and Its Application in Marine Geological Survey. Ocean Technol. 2011, 30, 24–26. [Google Scholar]
- Cota-Sánchez, J.H.; Remarchuk, K.; Ubayasena, K. Ready-to-use DNA extracted with a CTAB method adapted for herbarium specimens and mucilaginous plant tissue. Plant Mol. Biol. Rep. 2006, 24, 161–167. [Google Scholar] [CrossRef]
- Liu, K.J.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE Version 1.32 Microsoft Windows-Based Freeware for Populations Genetic Analysis; University of Alberta: Edmonton, AB, Canada, 1999. [Google Scholar]
- Goudet, J. FSTAT, a Program to Estimate and Test Gene Diversities and Fixation Indices, Version 2.9.3. 2001. Available online: http://www.unil.ch/popgen/softwares/fstat.htm (accessed on 20 June 2021).
- Peakall, R.; Smouse, P.E. Genalex 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Excoffier, L.; Lischer, H. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Belkhir, K.; Borsa, P.; Chikhi, L.; Raufaste, N.; Bonhomme, F. GENETIX (ver.4.02): Logiciel sous Windows TM pour la Genetique des Populations, Laboratoire Genome, Populations, Interactions CNRSUMR 5000; Universite Montpellier II: Montpellier, France, 2004. [Google Scholar]
- Manel, S.; Schwartz, M.K.; Luikart, G.; Taberlet, P. Landscape genetics combining landscape ecology and population genetics. Trends Ecol. Evol. 2003, 18, 189–197. [Google Scholar] [CrossRef]
- Travadon, R.; Sache, I.; Dutech, C.; Stachowiak, A.; Marquer, B.; Bousset, L. Absence of isolation by distance patterns at the regional scale in the fungal plant pathogen Leptosphaeria maculans. Fungal Biol. 2011, 115, 649–659. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F.J. NTSYS-pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.2, 2.1; Department of Ecoloy and Evolution, State University of New York: New York, NY, USA, 2005. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Piry, S.; Luikart, G.; Cornuet, J.M. BOTTLENECK: A computer program for detecting recent reductions in the effective population size using allele frequency data. J. Hered. 1999, 90, 502–503. [Google Scholar] [CrossRef]
- Solórzano, S.; Arias, S.; Dávila, P. Genetics and Conservation of Plant Species of Extremely Narrow Geographic Range. Diversity 2016, 8, 31. [Google Scholar] [CrossRef]
- Yao, M.Z.; Ma, C.L.; Qiao, T.T.; Jin, J.Q.; Liang, C. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genomes 2012, 8, 205–220. [Google Scholar] [CrossRef]
- Ueno, S.; Tomaru, N.; Yoshimaru, H.; Manabe, T.; Yamamoto, S. Genetic structure of Camellia japonica L. in an old-growth evergreen forest, Tsushima, Japan. Mol. Ecol. 2000, 9, 647–656. [Google Scholar] [CrossRef]
- Zhou, P.Y.; Hui, L.X.; Huang, S.-J.; Ni, Z.X.; Yu, F.-X.; Xu, L.A. Study on the Genetic Structure Based on Geographic Populations of the Endangered Tree Species: Liriodendron chinense. Forests 2021, 12, 917. [Google Scholar] [CrossRef]
- Garg, K.M.; Chattopadhyay, B. Gene Flow in Volant Vertebrates: Species Biology, Ecology and Climate Change. J. Indian Inst. Sci. 2021, 101, 165–176. [Google Scholar] [CrossRef]
- Chen, H.; Xuelin, L.U.; Quanqing, Y.E.; Tang, S.Q. Genetic diversity and structure of three yellow Camellia species based on SSR markers. Guihaia 2019, 39, 318–327. [Google Scholar] [CrossRef]
- Li, X.L.; Wang, J.; Fan, Z.Q.; Li, J.Y.; Yin, H.F. Genetic diversity in the endangered Camellia nitidissima assessed using transcriptome-based SSR markers. Trees 2020, 34, 543–552. [Google Scholar] [CrossRef]
- Shi, X.; Wen, Q.; Cao, M.; Guo, X.; Xu, L.A. Genetic Diversity and Structure of Natural Quercus variabilis Population in China as Revealed by Microsatellites Markers. Forests 2017, 8, 495. [Google Scholar] [CrossRef]
- Cristian, T.D.; Eduardo, R.; Fidelina, G.; Glenda, F.; Cavieres, L.A. Genetic Diversity in Nothofagus alessandrii (Fagaceae), an Endangered Endemic Tree Species of the Coastal Maulino Forest of Central Chile. Ann. Bot. 2007, 100, 75–82. [Google Scholar] [CrossRef]
- Miao, C.Y.; Yang, J.; Mao, R.L.; Li, Y. Phylogeography of Achyranthes bidentata (Amaranthaceae) in China’s Warm-Temperate Zone Inferred from Chloroplast and Nuclear DNA: Insights into Population Dynamics in Response to Climate Change During the Pleistocene. Plant Mol. Biol. Rep. 2017, 35, 166–176. [Google Scholar] [CrossRef]
- Peng, G.; Tang, S. Fine-scale spatial genetic structure and gene flow of Camellia flavida, a shadetolerant shrub in karst. Acta Ecol. Sin. 2017, 37, 7313–7323. [Google Scholar] [CrossRef][Green Version]
- Gao, Y.; Ai, B.; Kong, H.H.; Hang, H.W. Geographical pattern of isolation and diversification in karst habitat islands: A case study in the Primulina eburnea complex. J. Biogeogr. 2015, 42, 2131–2144. [Google Scholar] [CrossRef]
- Slatkin, M. A measure of population subdivision based on microsatellite allele frequencies. Genetics 1995, 139, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Slatkin, M. Gene flow and the geographic structure of natural populations. Science 1987, 236, 787–792. [Google Scholar] [CrossRef]
- Bhattacharyya, P.; Kumaria, S. Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Syst. Evol. 2015, 301, 201–210. [Google Scholar] [CrossRef]
- Yang, Z.L.; Ji-Yuan, L.I.; Fan, Z.Q. Effect of Storage Temperature on Pollen Germination of Sect. Camellia Species and C. Japonica Cultivars. J. Zhejiang For. Sci. Technol. 2004, 24, 1–3. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).