Diversity of Ectomycorrhizal Fungal Communities in Four Types of Stands in Pinus massoniana Plantation in the West of China
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sample Collection and Processing
2.3. Molecular Identification of Ectomycorrhizal Fungi
2.4. Analysis of ECM Species Composition
2.5. Calculations
2.6. Statistical Analysis
3. Results
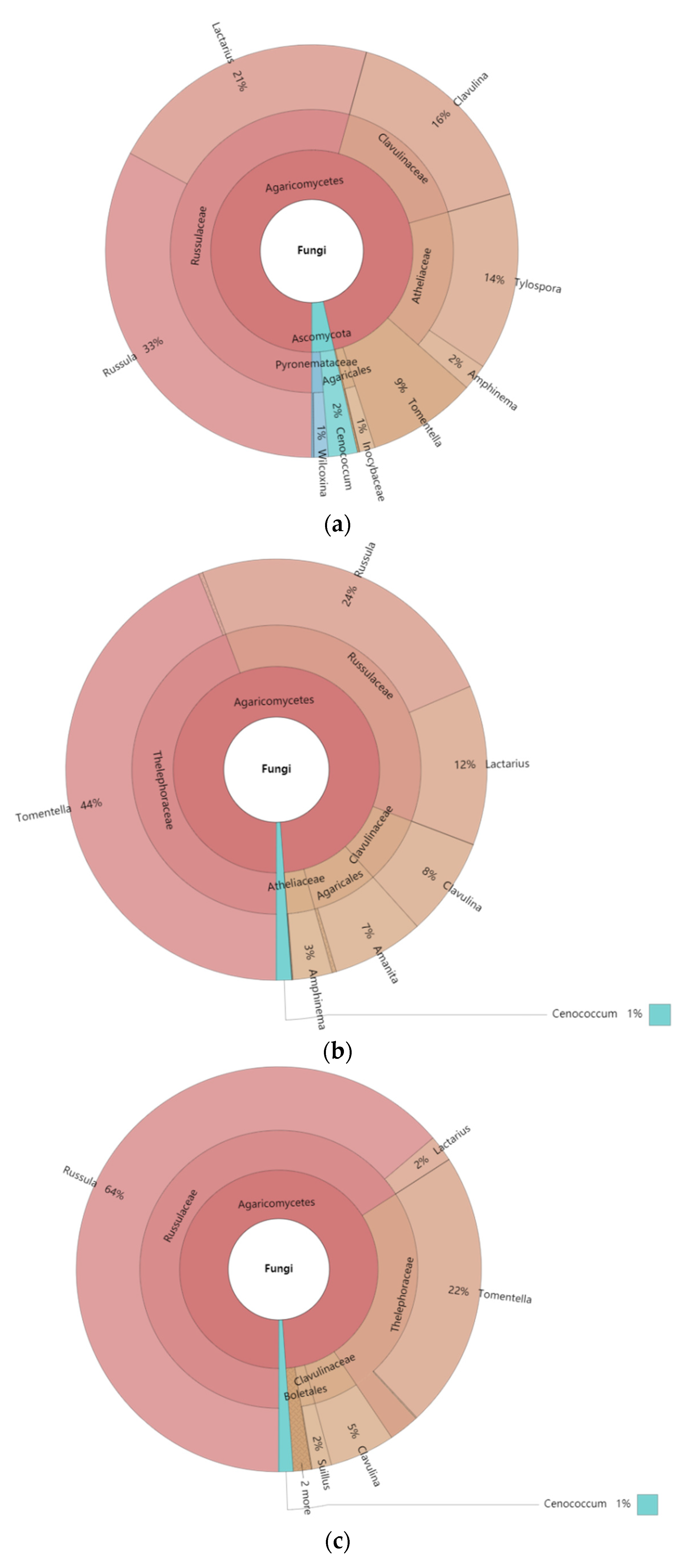
3.1. Composition of ECM Fungi in Four Types of Pinus massoniana Forests
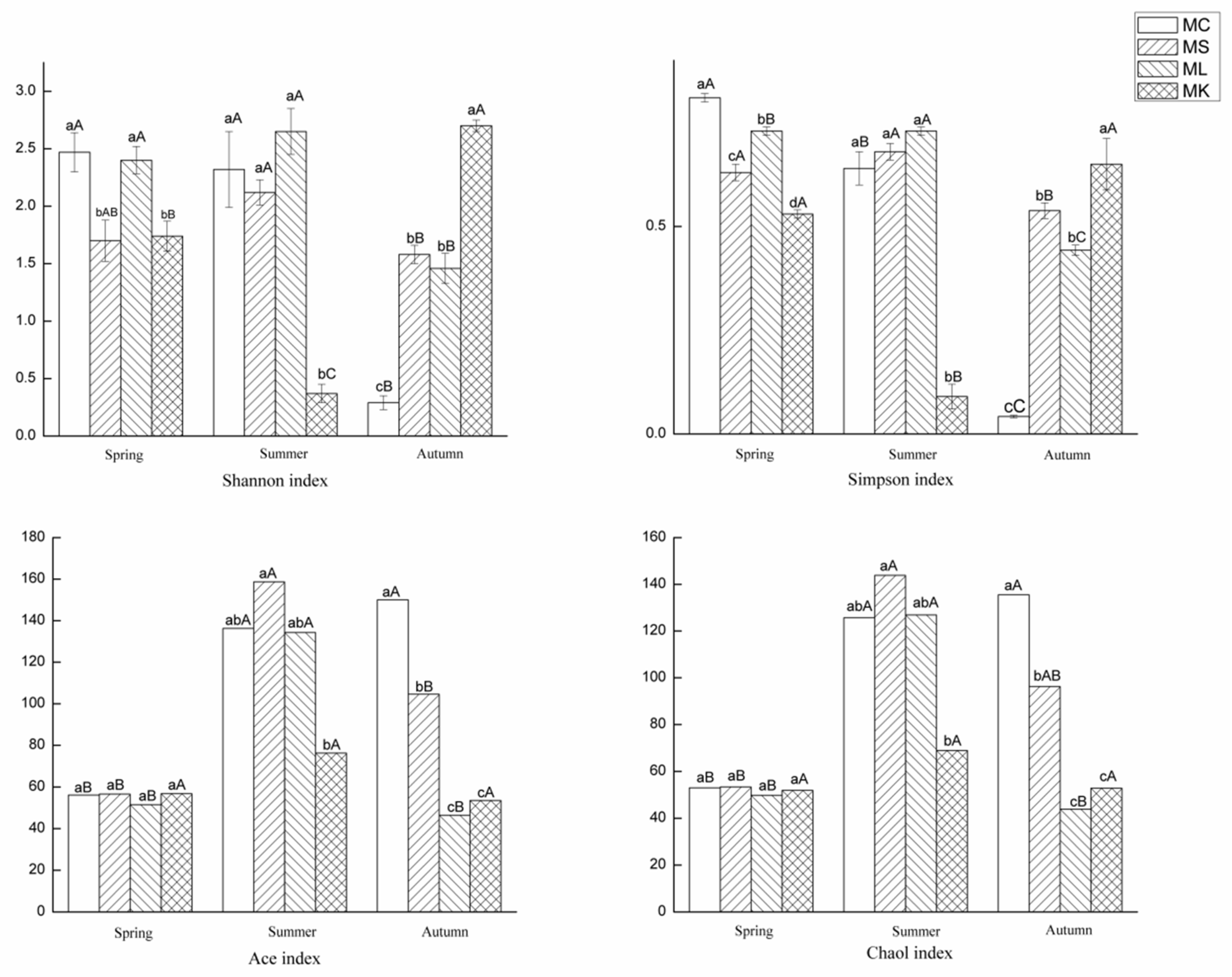
3.2. Alpha Diversity of ECM Fungi in P. massoniana
3.3. Principal Coordinate Analysis of ECM Fungal Community in P. massoniana
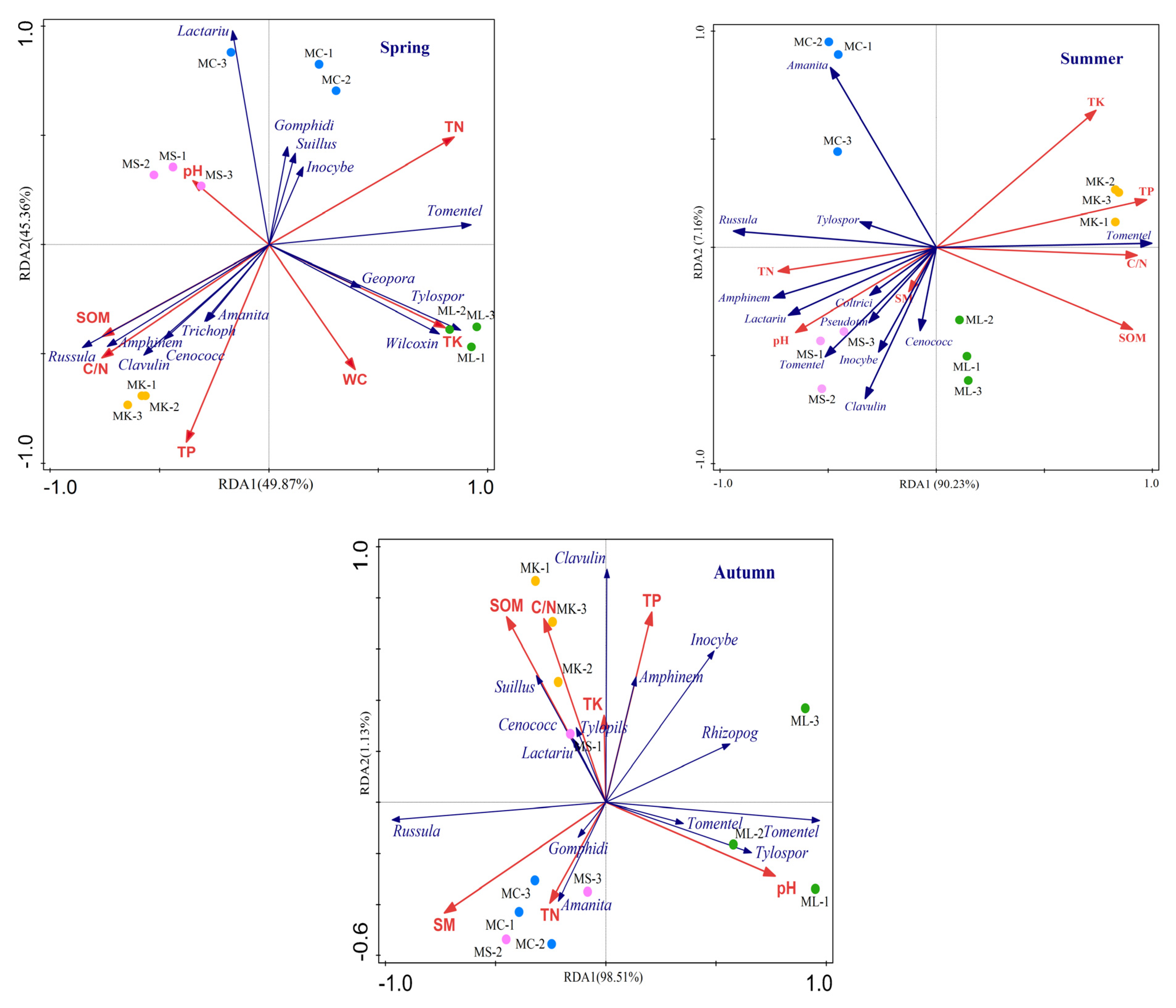
3.4. Effects of Soil Physical and Chemical Properties on ECM in P. massoniana
4. Discussion
4.1. Composition of ECM Fungi in the Four Types of P. massoniana Forest
4.2. Alpha Diversity of ECM Fungi
4.3. Correlation between Soil Physical and Chemical Properties and the ECM Communities
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Zhang, J.; He, Y. Research Progress on Ectomycorrhizal Fungi of Pinus massoniana. Farm. Cultiv. 2016, 66–68. [Google Scholar] [CrossRef]
- Mo, J.M.; Peng, S.L.; Brown, S. Response of community biomass production of Pinus massoniana forest to human disturbance in Dinghushan, Chin. Acta Ecol. Sin. 2004, 24, 193–200. [Google Scholar]
- Smith, M.E.; Henkel, T.W.; Aime, M.C.; Fremier, A.K.; Vilgalys, R. Ectomycorrhizal fungal diversity and community structure on three co-occurring leguminous canopy tree species in a Neotropical rainforest. New Phytol. 2011, 192, 699–712. [Google Scholar] [CrossRef]
- Lee, S.H.; Calvo-Polanco, M.; Chung, G.C.; Zwiazek, J.J. Role of aquaporins in root water transport of ectomycorrhizal jack pine (Pinus banksiana) seedlings exposed to NaCl and fluoride. Plant Cell Environ. 2010, 33, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Bonito, G.; Brenneman, T.; Vilgalys, R. Ectomycorrhizal fungal diversity in orchards of cultivated pecan (Carya illinoinensis; Juglandaceae). Mycorrhiza 2011, 21, 601–612. [Google Scholar] [CrossRef] [PubMed]
- Brundrett, M.; Bougher, N.; Dell, B.; Grove, T.; Malajczuk, N. Working with Mycorrhizas in Forestry and Agriculture; Australian Centre for International Agricultural Research: Canberra, Australia, 2012. [Google Scholar]
- Dixon, R.K.; Pallardy, S.G.; Garrett, H.E.; Cox, G.S.; Sander, I.L. Comparative water relations of container-grown and bare-root ectomycorrhizal and nonmycorrhizal Quercus velutina seedlings. Botany 1983, 61, 1559–1565. [Google Scholar] [CrossRef]
- Warren, J.M.; Brooks, J.R.; Meinzer, F.C.; Eberhart, J.L. Hydraulic redistribution of water from Pinus ponderosa trees to seedlings: Evidence for an ectomycorrhizal pathway. New Phytol. 2008, 178, 382–394. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Huang, J.; Long, D.; Wang, X.; Liu, J. Diversity and community structure of ectomycorrhizal fungi associated with Larix chinensis across the alpine treeline ecotone of Taibai Mountain. Mycorrhiza 2017, 27, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Nara, K.; Zong, K.; Wang, J.; Xue, S.; Peng, K.; Shen, Z.; Lian, C. Ectomycorrhizal fungal communities associated with Masson pine (Pinus massoniana) and white oak (Quercus fabri) in a manganese mining region in Hunan Province, China. Fung. Ecol. 2014, 9, 1–10. [Google Scholar] [CrossRef]
- Huang, J.; Nara, K.; Zong, K.; Lian, C. Soil propagule banks of ectomycorrhizal fungi along forest development stages after mining. Microb. Ecol. 2015, 69, 768–777. [Google Scholar] [CrossRef]
- Gilbert, L.; Johnson, D.W. Plant- plant communication through common mycorrhizal networks. Adv. Bot. Res. 2017, 83, 83–97. [Google Scholar]
- Peay, K.G.; Kennedy, P.G.; Davies, S.J.; Tan, S.; Bruns, T.D. Potential link between plant and fungal distributions in a dipterocarp rainforest: Community and phylogenetic structure of tropical ectomycorrhizal fungi across a plant and soil ecotone. New Phytol. 2010, 185, 529–542. [Google Scholar] [CrossRef] [PubMed]
- Erlandson, S.R.; Savage, J.A.; Cavender-Bares, J.M.; Peay, K.G. Soil moisture and chemistry influence diversity of ectomycorrhizal fungal communities associating with willow along an hydrologic gradient. FEMS Microbiol. Ecol. 2016, 92, fiv148. [Google Scholar] [CrossRef]
- Corrales, A.; Arnold, A.E.; Ferrer, A.; Turner, B.L.; Dalling, J.W. Variationin ectomycorrhizal fungal communities associated with Oreomunnea mexicana (Juglandaceae) in a Neotropical montane forest. Mycorrhiza 2016, 26, 1–17. [Google Scholar] [CrossRef]
- Lilleskov, E.A.; Fahey, T.J.; Horton, T.R.; Lovett, G.M. Belowground ectomycorrhizal fungal community change over a nitrogen deposition gradient in Alaska. Ecology 2002, 83, 104–115. [Google Scholar] [CrossRef]
- Bao, S.D. Soil Agrochemical Analysis; China Agriculture Press: Beijing, China, 2000. [Google Scholar]
- Makita, N.; Hirano, Y.; Yamanaka, T.; Yoshimura, K.; Kosugi, Y. Ectomycorrhizal-fungal colonization induces physio-morphological changes in Quercus serrata leaves and roots. J. Plant Nutr. Soil Sci. 2012, 175, 900–906. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved selection of internal transcribed spacer-specific primers enables quantitative, ultra-high-throughput profiling of fungal communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef]
- Šmilauer, P.; Lepš, J. Multivariate Analysis of Ecological Data using Canono 5, 2nd ed.; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Kirk, P.; Cannon, P.; Minter, D.; Stalpers, J. Dictionary of the Fungi. Mycol. Res. 2009, 113, 908–910. [Google Scholar]
- Yang, Y.; Xu, M.; Zou, X.; Chen, J.; Ma, H.; Yang, L.; Zhang, J. Research Progress of Ectomycorrhizal Fungi in Southern China. J. West China For. Sci. 2019, 048, 131–142. [Google Scholar]
- Haug, I.; Weiss, M.; Homeier, J.; Oberwinkler, F.; Kottke, I. Russulaceae and Thelephoraceae form ectomycorrhizas with members of the Nyctaginaceae (Caryophyllales) in the tropical mountain rain forest of southern Ecuador. New Phytol. 2004, 165, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Sakai, A.; Hattori, M.; Nara, K. Strong effect of climate on ectomycorrhizal fungal composition: Evidence from range overlap between two mountains. ISME J. 2015, 9, 1870–1879. [Google Scholar] [CrossRef]
- Lamit, L.J.; Holeski, L.M.; Flores-Rentería, L.; Whitham, C.A. Tree genotype influences ectomycorrhizal fungal community structure: Ecological and evolutionary implications. Fung. Ecol. 2016, 24, 124–134. [Google Scholar] [CrossRef]
- Tedersoo, L.; Smith, M.E. Lineages of ectomycorrhizal fungi revisited: Foraging strategies and novel lineages revealed by sequences from belowground. Fung. Biol. Rev. 2013, 27, 83–99. [Google Scholar] [CrossRef]
- Tedersoo, L.; Nilsson, R.H.; Abarenkov, K.; Jairus, T.; Sadam, A.; Saar, I.; Bahram, M.; Bechem, E.; Chuyong, G.; Kõljalg, U. 454 Pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytol. 2010, 188, 291–301. [Google Scholar] [CrossRef]
- Tedersoo, T.W.; May, M.; Toots, A.G.; Diédhiou, T.L.; Henkel, R.; Kjoller, M.H.; Morris, K.; Nara, E.; Nouhara, K.; Peay, S.; et al. Towards global patterns in the diversity and community structure of ectomycorrhizal fungi. Mol. Ecol. 2012, 21, 4160–4170. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef]
- Rinaldi, A.C.; Comandini, O.; Kuyper, T.W. Ectomycorrhizal fungal diversity: Seperating the wheat from the chaff. Fung. Divers. 2008, 33, 1–45. [Google Scholar]
- Tedersoo, L.; Nara, K. General latitudinal gradient of biodiversity is reversed in ectomycorrhizal fungi. New Phytol. 2010, 185, 351–354. [Google Scholar] [CrossRef] [PubMed]
- Nara, K. Ectomycorrhizal networks and seedling establishment during early primary succession. New Phytol. 2006, 169, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Manjarrez, J.; Villegas-Ríos, M.; Garibay-Orijel, R.; Contreras-Pacheco, M.; Kõljalg, U. Tomentella brunneoincrust the first described species of the Pisonieae-associated Neotropical Tomentella clade, and phylogenetic analysis of the genus in Mexico. Mycol. Prog. 2016, 15, 10. [Google Scholar] [CrossRef]
- Fernandez, C.W.; Mccormack, M.L.; Hill, J.M.; Pritchard, S.G.; Koide, R.T. On the persistence of Cenococcum geophilum ectomycorrhizas and its implications for forest carbon and nutrient cycles. Soil Biol. Biochem. 2013, 65, 141–143. [Google Scholar] [CrossRef]
- Hawksworth, D.L. The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycol. Res. 2001, 105, 1422–1432. [Google Scholar] [CrossRef]
- Fernandez, C.W.; Koide, R.T. The function of melanin in the ectomycorrhizal fungus Cenococcum geophilum under water stress. Fung. Ecol. 2013, 6, 479–486. [Google Scholar] [CrossRef]
- Deslippe, J.R.; Hartmann, M.; Mohn, W.W.; Simard, S.W. Long-term experimental manipulation of climate alters the ectomycorrhizal community of Betula nana in Arctic tundra. Glob. Chang. Biol. 2011, 17, 1625–1636. [Google Scholar] [CrossRef]
- Walker, J.F.; KMiller, O.; Horton, J.L. Hyperdiversity of ectomycorrhizal fungus assemblages on oak seedlings in mixed forests in the southern Appalachian Mountains. Mol. Ecol. 2005, 14, 829–838. [Google Scholar] [CrossRef]
- Lu, N.; Xu, X.; Wang, P.; Zhang, P.; Ji, B.; Wang, X. Succession in arbuscular mycorrhizal fungi can be attributed to a chronosequence of Cunninghamia lanceolata. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Dumbrell, A.J.; Ashton, P.D.; Aziz, N.; Feng, G.; Nelson, M.; Dytham, C.; Fitter, A.H.; Helgason, T. Distinct seasonal assemblages of arbuscular mycorrhizal fungi revealed by massively parallel pyrosequencing. New Phytol. 2011, 190, 794–804. [Google Scholar] [CrossRef] [PubMed]
- Newton, A.C. Towards a functional classification of ectomycorrhizal fungi. Mycorrhiza 1992, 2, 75–79. [Google Scholar] [CrossRef]
- Clemmensen, K.E.; Bahr, A.; Ovaskainen, O.; Dahlberg, A.; Ekblad, A.; Wallander, H.; Stenlid, J.; Finlay, R.D.; Wardle, D.A.; Lindahl, B.D. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science 2013, 339, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Averill, C.; Turner, B.L.; Finzi, A.C. Mycorrhiza-mediated competition between plants and decomposers drives soil carbon storage. Nature 2014, 505, 543–545. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: New York, NY, USA, 2008. [Google Scholar]
- Ekblad, A.; Wallander, H.; Carlsson, R.; Huss-Danell, K. Fungal biomass in roots and extramatrical mycelium in relation to macronutrients and plant biomass of ectomycorrhizal Pinus sylvestris and Alnus incana. New Phytol. 1995, 131, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Ju, H.; Qi, J.; Zhou, B. Effect of ectomycorrhizal fungi on seedling growth of Mongol Scotch Pine. J. Fung. Res. 2007, 142–145. [Google Scholar] [CrossRef]
- Smith, F.W.; Rae, A.L.; Hawkesford, M.J. Molecular mechanisms of phosphate and sulphate transport in plants. Biochim. Biophys. Acta 2000, 1465, 236–245. [Google Scholar] [CrossRef]
- Bucher, M.R.; Rausch, C.; Daram, P. Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant Nutr. Soil Sci. 2001, 164, 209–217. [Google Scholar] [CrossRef]


| Altitude (m) | Aspect | Breast Diameter (cm) | Height (m) | Crown Density | Type of Mix | Mixture Ratio | |
|---|---|---|---|---|---|---|---|
| MC | 690 | Southeast | 17.8 | 13.8 | 0.7 | - | - |
| MS | 776 | South | 18.1 | 14.3 | 0.7 | Interline | 1:1 |
| ML | 630 | South | 18.6 | 14.8 | 0.7 | Interline | 1:1 |
| MK | 647 | South | 18.4 | 14.6 | 0.7 | Interline | 1:1 |
| Spring | Summer | Autumn | |
|---|---|---|---|
| MC | 54.79 ± 1.07 bA | 56.75 ± 0.23 bcA | 55.50 ± 0.79 bA |
| MS | 60.83 ± 2.17 aA | 63.99 ± 1.16 aA | 59.79 ± 1.01 aA |
| ML | 58.98 ± 1.61 abA | 59.47 ± 1.38 bA | 56.32 ± 1.65 abA |
| MK | 54.96 ± 0.99 bA | 53.65 ± 1.38 cA | 57.66 ± 1.05 abA |
| Name | MC | MS | ML | MK | |
|---|---|---|---|---|---|
| Phylum | Basidiomycota | 98 | 99.87 | 94.91 | 90.85 |
| Ascomycota | 2 | 0.13 | 5.09 | 9.15 | |
| Class | Agaricomycetes | 98 | 99.87 | 94.91 | 90.85 |
| Dothideomycetes | 1.46 | 0.03 | 3.1 | 4.11 | |
| Pezizomycetes | 0.55 | 0.1 | 1.99 | 5.04 | |
| Family | Russulaceae | 74.72 | 71.84 | 32.68 | 26.18 |
| Thelephoraceae | 15.65 | 1.89 | 8.68 | 24.61 | |
| Clavulinaceae | 5.52 | 22.49 | 9.71 | 31.76 | |
| Gloniaceae | 1.46 | 0.03 | 3.1 | 4.11 | |
| Atheliaceae | 0.79 | 3.62 | 43.64 | 3.57 | |
| Inocybaceae | 0.72 | 0.02 | 0.18 | 4.73 | |
| Pyronemataceae | 0.55 | 0.1 | 1.99 | 5.04 | |
| Suillaceae | 0.55 | 0 | 0 | 0 | |
| Gomphidiaceae | 0.05 | 0 | 0 | 0 | |
| Amanitaceae | 0 | 0 | 0 | 0.01 | |
| Genus | Lactarius | 45.31 | 30.06 | 10.05 | 0.44 |
| Russula | 29.41 | 41.78 | 22.63 | 25.74 | |
| Tomentella | 15.65 | 1.89 | 8.68 | 24.61 | |
| Clavulina | 5.52 | 22.49 | 9.71 | 31.76 | |
| Cenococcum | 1.46 | 0.03 | 3.1 | 4.11 | |
| Suillus | 0.55 | 0 | 0 | 0 | |
| Amphinema | 0.44 | 3.45 | 0.19 | 3.39 | |
| Tylospora | 0.34 | 0.17 | 43.45 | 0.18 | |
| Wilcoxina | 0.04 | 0.1 | 0.12 | 4.73 | |
| Geopora | 0 | 0 | 0.48 | 0 | |
| Inocybe | 0.13 | 0 | 0 | 0 | |
| Gomphidius | 0.05 | 0 | 0 | 0 | |
| Amanita | 0 | 0 | 0 | 0.01 | |
| Unclassified | 1.1 | 0.02 | 1.59 | 5.04 |
| Name | MC | MS | ML | MK | |
|---|---|---|---|---|---|
| Phylum | Basidiomycota | 99.7 | 98.2 | 95.28 | 99.36 |
| Ascomycota | 0.3 | 1.8 | 4.72 | 0.64 | |
| Class | Agaricomycetes | 99.7 | 98.2 | 95.28 | 99.36 |
| Dothideomycetes | 0.3 | 1.8 | 4.72 | 0.64 | |
| Family | Russulaceae | 62.68 | 68.35 | 39.26 | 0.8 |
| Thelephoraceae | 7.33 | 5.05 | 33.36 | 98.24 | |
| Clavulinaceae | 5.25 | 11.89 | 21.13 | 0.28 | |
| Amanitaceae | 19.4 | 0.17 | 0.16 | 0 | |
| Atheliaceae | 5.04 | 11.97 | 0.05 | 0.03 | |
| Gloniaceae | 0.3 | 1.8 | 4.72 | 0.64 | |
| Inocybaceae | 0.01 | 0.76 | 1.32 | 0.01 | |
| Hymenochaetaceae | 0 | 0.02 | 0 | 0 | |
| Genus | Tomentella | 7.31 | 3.71 | 33.36 | 98.24 |
| Russula | 43.63 | 42.11 | 23.93 | 0.63 | |
| Lactarius | 19.04 | 26.2 | 15.3 | 0.18 | |
| Clavulina | 5.25 | 11.89 | 21.13 | 0.28 | |
| Amanita | 19.4 | 0.17 | 0.16 | 0 | |
| Amphinema | 4.76 | 11.88 | 0.04 | 0.03 | |
| Cenococcum | 0.3 | 1.8 | 4.72 | 0.64 | |
| Inocybe | 0.01 | 0.76 | 1.32 | 0.01 | |
| Tomentellopsis | 0.01 | 1.32 | 0 | 0.01 | |
| Tylospora | 0.25 | 0.08 | 0 | 0 | |
| Coltricia | 0 | 0.02 | 0 | 0 | |
| Pseudotomentella | 0 | 0.01 | 0 | 0 | |
| Unclassified | 0.04 | 0.05 | 0.03 | 0 |
| Name | MC | MS | ML | MK | |
|---|---|---|---|---|---|
| Phylum | Basidiomycota | 99.98 | 96.81 | 99.93 | 83.78 |
| Ascomycota | 0.02 | 3.19 | 0.07 | 16.22 | |
| Class | Agaricomycetes | 99.98 | 96.81 | 99.93 | 83.78 |
| Dothideomycetes | 0.02 | 3.19 | 0.07 | 16.22 | |
| Family | Russulaceae | 93.39 | 70.91 | 13.75 | 33.51 |
| Clavulinaceae | 6.06 | 1.68 | 24.96 | 16.17 | |
| Thelephoraceae | 0.47 | 16.19 | 58.45 | 24.82 | |
| Gloniaceae | 0.02 | 3.19 | 0.07 | 16.22 | |
| Amanitaceae | 0.02 | 0.13 | 0 | 0 | |
| Suillaceae | 0.02 | 7.58 | 0 | 0.08 | |
| Atheliaceae | 0.01 | 0.01 | 2.73 | 1.91 | |
| Gomphidiaceae | 0 | 0.31 | 0 | 0 | |
| Inocybaceae | 0 | 0.01 | 0.02 | 7.18 | |
| Boletaceae | 0 | 0 | 0 | 0.12 | |
| Genus | Russula | 90.12 | 63.22 | 13.73 | 32.95 |
| Clavulina | 6.06 | 1.68 | 24.96 | 16.17 | |
| Lactarius | 3.27 | 7.68 | 0.03 | 0.56 | |
| Tomentella | 0.4 | 1.14 | 57.88 | 24.46 | |
| Cenococcum | 0.02 | 3.19 | 0.07 | 16.22 | |
| Suillus | 0.02 | 7.58 | 0 | 0.08 | |
| Amphinema | 0 | 0.01 | 0.98 | 0.03 | |
| Tylospora | 0 | 0 | 1.75 | 1.87 | |
| Tomentellopsis | 0 | 0 | 0.38 | 0.03 | |
| Inocybe | 0 | 0.01 | 0.02 | 7.18 | |
| Amanita | 0.03 | 0.31 | 0 | 0 | |
| Gomphidius | 0 | 0.12 | 0 | 0 | |
| Rhizopogon | 0 | 0 | 0.01 | 0 | |
| Tylopilus | 0 | 0 | 0 | 0.13 | |
| Unclassified | 0.07 | 15.06 | 0.19 | 0.34 |
| Variable | Season | Forest Type | Season × Forest Type | ||||||
|---|---|---|---|---|---|---|---|---|---|
| df | F | P | df | F | P | df | F | P | |
| Shannon | 2 | 13.614 | 0 | 3 | 7.456 | 0.001 | 6 | 40.674 | 0 |
| Simpson | 2 | 99.241 | 0 | 3 | 52.965 | 0 | 6 | 94.173 | 0 |
| Chao1 | 2 | 24.721 | 0 | 3 | 8.424 | 0.001 | 6 | 4.327 | 0.004 |
| ACE | 2 | 27.386 | 0 | 3 | 9.679 | 0 | 6 | 4.857 | 0.002 |
| Season | Types | SM | pH | SOM (g∙kg−1) | TN (g∙kg−1) | C/N | TP (g∙kg−1) | TK (g∙kg−1) |
|---|---|---|---|---|---|---|---|---|
| Spring | MC | 35.03 ± 1.19 ab | 3.64 ± 0.11 b | 38.10 ± 0.67 b | 1.96 ± 0.04 a | 19.47 ± 0.61 b | 2.74 ± 0.03 d | 11.71 ± 0.12 b |
| MS | 32.61 ± 0.95 b | 4.36 ± 0.07 a | 37.17 ± 1.26 b | 1.77 ± 0.01 b | 20.99 ± 0.83 b | 3.16 ± 0.01 c | 11.34 ± 0.15 b | |
| ML | 37.43 ± 0.40 a | 3.69 ± 0.07 b | 31.34 ± 0.73 c | 2.02 ± 0.02 a | 15.49 ± 0.44 c | 3.43 ± 0.03 b | 12.52 ± 0.11 a | |
| MK | 36.74 ± 0.80 a | 3.62 ± 0.04 b | 53.66 ± 1.38 a | 1.46 ± 0.05 c | 36.98 ± 0.60 a | 4.45 ± 0.06 a | 11.65 ± 0.16 b | |
| Summer | MC | 38.46 ± 0.41 a | 4.09 ± 0.04 b | 32.79 ± 0.88 d | 2.61 ± 0.06 a | 12.59 ± 0.37 c | 2.70 ± 0.01 c | 13.37 ± 0.18 b |
| MS | 38.30 ± 1.03 a | 4.94 ± 0.11 a | 38.09 ± 0.46 c | 2.32 ± 0.05 b | 16.46 ± 0.48 b | 2.34 ± 0.06 d | 12.18 ± 0.10 d | |
| ML | 39.37 ± 0.38 a | 3.88 ± 0.05 b | 48.89 ± 0.82 b | 2.65 ± 0.04 a | 18.44 ± 0.45 b | 3.03 ± 0.03 b | 12.85 ± 0.07 c | |
| MK | 37.73 ± 1.27 a | 3.87 ± 0.05 b | 51.92 ± 0.94 a | 1.60 ± 0.13 c | 32.73 ± 1.99 a | 3.96 ± 0.03 a | 14.17 ± 0.06 a | |
| Autumn | MC | 38.93 ± 0.40 a | 3.96 ± 0.12 b | 36.31 ± 0.52 b | 1.63 ± 0.04 a | 22.34 ± 0.52 b | 2.48 ± 0.03 c | 16.40 ± 0.03 a |
| MS | 38.29 ± 0.37 a | 3.93 ± 0.09 b | 34.58 ± 0.64 b | 1.64 ± 0.08 a | 21.16 ± 1.33 b | 3.56 ± 0.05 b | 14.35 ± 0.21 c | |
| ML | 34.79 ± 1.02 b | 4.31 ± 0.14 a | 32.57 ± 0.68 c | 1.50 ± 0.09 a | 21.90 ± 1.53 b | 3.76 ± 0.14 b | 15.71 ± 0.13 b | |
| MK | 36.45 ± 1.06 ab | 3.86 ± 0.05 b | 51.76 ± 0.57 a | 1.45 ± 0.08 a | 36.02 ± 2.24 a | 4.50 ± 0.08 a | 16.34 ± 0.12 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Kang, W.; Liu, S.; Yin, H.; Lyu, Q.; Su, Y.; Liu, J.; Liu, J.; Fan, C.; Chen, G.; et al. Diversity of Ectomycorrhizal Fungal Communities in Four Types of Stands in Pinus massoniana Plantation in the West of China. Forests 2021, 12, 719. https://doi.org/10.3390/f12060719
Li X, Kang W, Liu S, Yin H, Lyu Q, Su Y, Liu J, Liu J, Fan C, Chen G, et al. Diversity of Ectomycorrhizal Fungal Communities in Four Types of Stands in Pinus massoniana Plantation in the West of China. Forests. 2021; 12(6):719. https://doi.org/10.3390/f12060719
Chicago/Turabian StyleLi, Xiangjun, Wensi Kang, Size Liu, Haifeng Yin, Qian Lyu, Yu Su, Junjie Liu, Jiangli Liu, Chuan Fan, Gang Chen, and et al. 2021. "Diversity of Ectomycorrhizal Fungal Communities in Four Types of Stands in Pinus massoniana Plantation in the West of China" Forests 12, no. 6: 719. https://doi.org/10.3390/f12060719
APA StyleLi, X., Kang, W., Liu, S., Yin, H., Lyu, Q., Su, Y., Liu, J., Liu, J., Fan, C., Chen, G., Zhao, K., & Li, X. (2021). Diversity of Ectomycorrhizal Fungal Communities in Four Types of Stands in Pinus massoniana Plantation in the West of China. Forests, 12(6), 719. https://doi.org/10.3390/f12060719





