Abstract
The Acer truncatum Bunge is a particular forest tree species found in the north of China. Due to the recent discovery that its seeds contain a considerable amount of nervonic acid, this species has received more and more attention. However, there have been no reports of the genome in this species. In this study, we report on the Acer truncatum genome sequence produced by genome survey sequencing. In total, we obtained 61.90 Gbp of high-quality data, representing approximately 116x coverage of the Acer truncatum genome. The genomic characteristics of Acer truncatum include a genome size of 529.88 Mbp, a heterozygosis rate of 1.06% and a repeat rate of 48.8%. A total of 392,961 high-quality genomic SSR markers were developed and a graphical map of the annotated circular chloroplast genome was generated. Thus far, this is the first report of de novo whole genome sequencing and assembly of Acer truncatum. We believe that this genome sequence dataset may provide a new resource for future genomic analysis and molecular breeding studies of Acer truncatum.
1. Introduction
The Acer truncatum Bunge is a particular forest tree species found in the north of China [1,2]. The seeds contain a considerable amount of nervonic acid (24:1; cis-tetracos-15-enoic acid) and it has been officially classified as an edible oil by the Ministry of Health of China [3]. There are only a few plant species with high amounts of nervonic acid in their seed oil. Nervonic acid is widely present in the sphingolipids of the nervous system of vertebrates and is a core component of brain fibers and nerve cells [4]. Nervonic acid oils have become important targets for pharmaceutical and nutraceutical applications that aim to treat a number of neurological disorders [5]. This tree species is also important in various disciplines, including botany, geography and climatology. Thus far, research into the utilization of Acer truncatum has focused on leaf extracts and there is a distinct lack of genomic information [6,7,8,9,10].
Recently, genome survey sequencing by next generation sequencing (NGS) has emerged as an important and cost-effective strategy for generating a wide range of genetic and genomic information for different plant species [11,12,13,14,15,16,17,18]. It can also identify a large number of molecular markers for breeding [19]. The SSR marker is the most widely used molecular marker system. SSR molecular markers have been developed for many species through NGS [11,13,15,16,18]. An increased molecular marker density will enable better genome-wide association and molecular breeding studies [20,21]. Chloroplasts are plant-specific and important organelles, which are important in photosynthesis and carbon fixation in green plants [22]. The development of NGS technologies has also allowed for the sequencing of entire chloroplast genomes. Many chloroplast genomes of different Sapindales species have already been determined by NGS [23,24,25,26].
In the present study, the Acer truncatum genome sequence produced by genome survey sequencing is reported. The genome was assembled and then used to develop SSRs. After data filtering, we obtained 61.90 Gbp of high-quality data, representing approximately 116x coverage of the Acer truncatum genome. The genomic characteristics of Acer truncatum include a genome size of 529.88 Mbp, a heterozygosis rate of 1.06% and a repeat rate of 48.8%. Furthermore, 392,961 high-quality genomic SSR markers were developed. A graphical map of the annotated circular chloroplast genome was also generated. Thus far, this is the first report of de novo whole genome sequencing and assembly of Acer truncatum. In short, we believe that this genome sequence dataset may provide a new resource for future functional genomic and evolutionary analysis studies of Acer truncatum as well as for studies focusing on its molecular breeding.
2. Materials and Methods
2.1. Plant Materials
Fresh leaves from a single individual Acer truncatum tree (approximately 60 years old) were collected from the testing grounds of Northwest A&F University in Yangling Country, Shannxi Province, China (Figure 1). The genomic DNA was isolated from the fresh leaves with the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA).

Figure 1.
The adult tree (A), seeds (B) and leaves (C) of Acer truncatum in Yangling, Shaanxi province, China.
2.2. Illumina Sequencing Data Analysis and Assembly
The whole Acer truncatum genome sequencing was performed by the Illumina Hiseq Platform (Illumina Inc., San Diego, CA, USA). Clean data were obtained by performing rigorous quality assessments and conducting data filtering on the Illumina sequencing raw data. The clean data with high-quality reads were assembled using the de Bruijn graph-based SOAPdenovo software (version 1.05, BGI, Beijing, China) [27]. High-quality Illumina sequencing reads were submitted to the NCBI Short Read Archive (accession number: SUB4843212).
2.3. Genome Size Estimation, GC Content and Genome Survey
The clean data with high-quality reads were used for K-mer analysis. Based on k-mer (k = 21) frequency distributions, we used GENOMESCOPE to estimate the characteristics of the genome (genome size, repeat content and heterozygosity rate) [28]. The 10-kb non-overlapping sliding windows along the assembled sequence were used to calculate GC content and sequencing depth.
2.4. The Genomic SSR Markers Detection
Using the MISA software (http://pgrc.ipk-gatersleben.de/misa/misa.html), 145,640 scaffolds of Acer truncatum were used for genomic SSR marker detection. The following search parameters were set for identification: Di-, Tri-, Tetra-, Penta- and Hexa-nucleotide motifs with a minimum of 6, 5, 4, 4 and 4 repeats, respectively, as previously described [29,30]. The primers for SSR markers were designed by Premier 5.0 (Premier Biosoft International, Palo Alto, CA, USA).
2.5. Assembly and Analysis of the Chloroplast Genome
Using NOVOPlasty software version 2.6.7 [31], the chloroplast genome was assembled. The resulting assembled chloroplast genome was annotated based on GeSeq [32] and comparisons with the chloroplast genomes of Acer davidii (NC_030331) [23], Acer morrisonense (NC_029371) [24], Acer griseum [25] and Acer miaotaiense (NC_030343) [26]. A graphical map of the annotated circular chloroplast genome was generated using the OGDRAW program [33]. The phylogenetic tree was reconstructed with Maximum Likelihood (ML) algorithms using MEGA6 software [34]. The phylogenetic analysis was based on the coding of gene nucleotides. The number of bootstrap replications was 500 for the phylogeny test. The high-quality annotated complete chloroplast genomic sequence was deposited in GenBank with the accession number MH716034.
3. Results and Discussion
3.1. Genome Sequencing and Sequence Assembly
To obtain a sufficient amount of DNA for sequencing libraries, we isolated DNA from the leaves of Acer truncatum (Figure 1). More than 61.90 Gbp of high-quality data with clean reads (Q30 94.8%) was generated from sequencing library (300 bp) by the Illumina HiSeq sequencing platform. This was an approximately 116x coverage of the Acer truncatum estimated genome size (Table S1). All the high-quality and clean data were used for genome sequence assembly analysis. The high-quality and clean data represented over 30x coverage, which indicates successful genome survey sequencing [27]. The high-quality and clean data were de novo assembled (K-mer = 75) using the de Bruijn graph-based SOAPdenovo software [35]. The Acer truncatum genome assembly consisted of 2,412,582 scaffolds with a total length of 866,062,477 bp and a scaffold N50 length of 735 bp (Table 1). After filtering for scaffolds with a size of <1000 bp, Genscan and Augustus were used to predict genes with parameters trained on Acer truncatum. Genes were annotated based on the following nine databases: NR, NT, Swiss-Prot, KEGG, KOG, Pfam, GO, COG and TrEMBL. The functional annotations for the genes are shown in Table S2. For example, we used BLASTX to perform a similarity analysis and comparison against the NR database. Among a wide range of plants, the assembled Acer truncatum genomic sequence had the highest number of hits against Citrus sinensis (Figure S1). This result is consistent with previous transcriptome sequencing findings [29].

Table 1.
Information of the assembled genome sequences of Acer truncatum.
3.2. Genomic Characteristics
For the 21-mer frequency distribution, the number of k-mers was 47,661,885,129 and the peak of the depth distribution was at 75x. The estimated genome size of Acer truncatum was 529.88 Mbp (Figure 2). The position at a point that is half of the height of the main peak (~38x) indicates the heterozygosis rate, with the heterozygosis rate in this genome found to be approximately 1.06%. This is a relatively high heterozygosis rate. The repeat peak at the position of the integer multiples of the main peak indicates about 48.8% of the Acer truncatum genome (Figure 2). A scatterplot of the genomic GC content and sequencing depth can provide information on sequencing data bias (Figure S2). The GC content of Acer truncatum genome was 35.04%. This is a mid-GC content. As shown in the GC content analysis in Figure S2, there is another dense area (red area), which may be caused by the high rate of heterozygosity (1.06%).
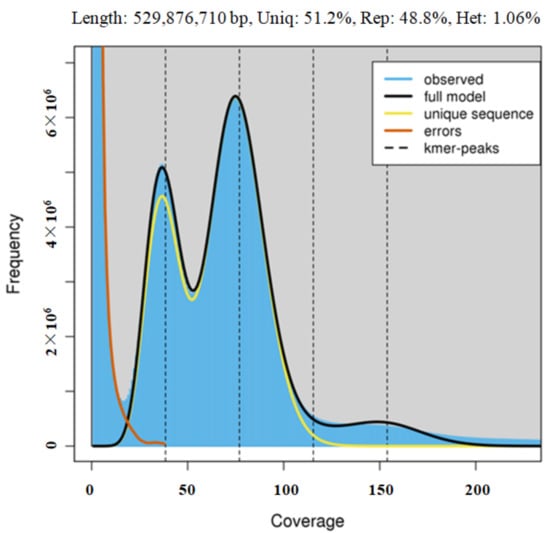
Figure 2.
k-mer distribution as calculated by Genomescope. Blue bars represent the observed k-mer distribution; black line represents the modelled distribution without the k-mer errors (red line) and up to a maximum k-mer coverage specified in the model (yellow line). length, estimated genome length; uniq, unique portion of the genome (nonrepetitive elements); het, genome heterozygosity; rep, repetitive portion of the genome.
3.3. Genomic SSR Marker Development
The assembled scaffolds were searched for the presence of SSR markers by using the MISA software (http://pgrc.ipk-gatersleben.de/misa/misa.html). A total of 392,961 putative SSR markers from 145,640 scaffolds were identified (Table S3). Among the identified SSR markers, the Di-nucleotide was the most abundant SSR marker, accounting for 69.25% of the total SSR markers, which was followed by Tri- (21.36%), Tetra- (6.56%), Penta- (1.71%) and Hexa- (1.21%) nucleotide SSR markers (Figure 3A). There was a large proportion of both Di-nucleotide and Tri-nucleotide SSR markers while the rest amounted to less than 10%.
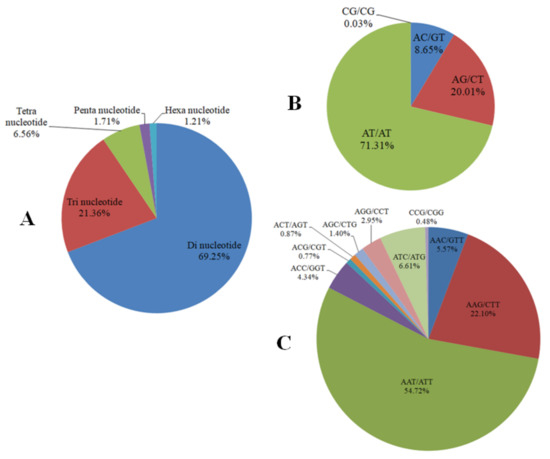
Figure 3.
Characteristics of SSR markers. (A) Frequency of different SSR markers; (B) Frequency of different Di-nucleotide SSR markers; (C) Frequency of different Tri-nucleotide SSR markers.
In the Di-nucleotide SSR markers, the AT/AT repeat motifs accounted for 71.31%, AG/CT accounted for 20.01%, AC/GT accounted for 8.65% and CG/CG only accounted for 0.03% (Figure 3B). The predominant Tri-nucleotide SSR markers, the AAT/ATT repeat motifs, the AAG/CTT repeat motifs and the ATC/ATG repeat motifs accounted for 54.72%, 22.10% and 6.61%, respectively (Figure 3C).
The SSR markers categorized by the number of repeat motifs were summarized (Figure 4). The Di- and Tri-nucleotide SSR markers were far more prevalent than the other SSR markers. The number of SSR markers decreased with an increased repeat motif length.
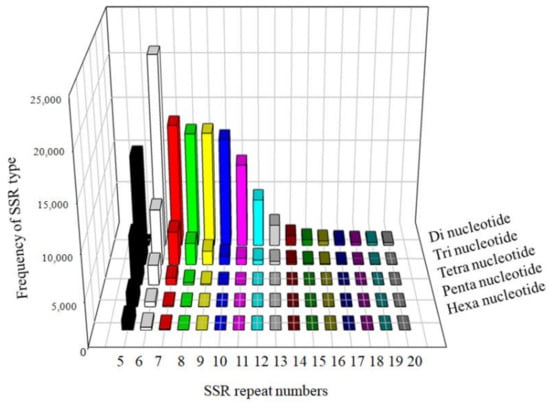
Figure 4.
The distribution and frequency of SSR motif repeat numbers.
3.4. Assembly of Chloroplast Genome
The complete annotated chloroplast genome of Acer truncatum was a double-stranded circular DNA with a length of 156,241 bp. The structure of the chloroplast genome is similar to that of other plant species, including two inverted repeat (IRA and IRB) regions (52,172 bp), a large single-copy (LSC) region (85,972 bp) and a small single-copy (SSC) region (18,097 bp) (Figure 5). In the Acer truncatum chloroplast genome, 132 functional genes were predicted, including 87 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Most of these genes appear in a single copy while 17 gene types appear as double copies or more, including six protein-coding gene species (rps7, rps12, rpl2, rpl23, ycf2 and ndhB), seven tRNA gene species (trnA-UGC, trnE-UUC, trnL-CAA, trnM-CAU, trnN-GUU, trnR-AC and GtrnV-GAC) and four rRNA gene species (rrn23, rrn16, rrn5 and rrn4.5). The overall GC content in the chloroplast genome was approximately 37.9%. Contractions and expansions of the IR regions at the borders are common evolutionary events and represent the main reasons for the size variation among chloroplast genomes. A detailed comparison of four junctions, which were namely LSC/IRA, LSC/IRB, SSC/IRA and SSC/IRB, between the two IRs (IRA and IRB) and the two single-copy regions (LSC and SSC), completed the relative Acer miaotaiense chloroplast genome (Figure S3).
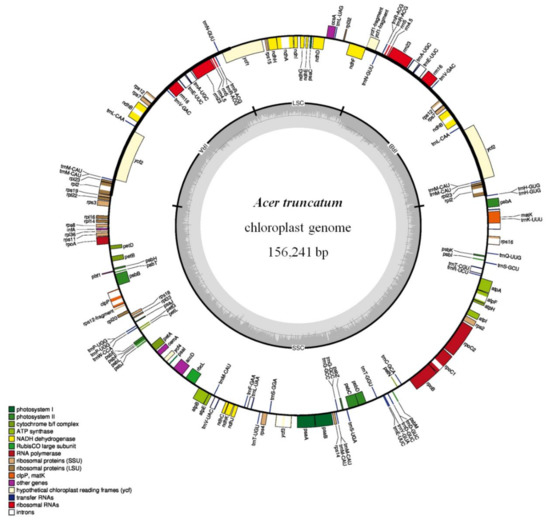
Figure 5.
Physical map of the circular chloroplast genome of Acer truncatum.
In order to investigate the placement of Acer truncatum chloroplast genome within the order Sapindales, we performed a phylogenetic analysis based on 75 chloroplast protein-coding genes for 12 species with the Arabidopsis thaliana chloroplast genome as an outgroup (Figure 6). The topology of the phylogenetic tree is basically consistent with the traditional taxonomy of the order Sapindales. The results suggest that Acer truncatum is closely related to the four congeners Acer miaotaiense P. C. Tsoong, Acer davidii Franch, Acer morrisonense Hayata and Acer griseum Pax. These four taxa together with Dipteronia sinensis Oliv. and Dipteronia dyeriana Henry form a monophyletic clade in the family Aceraceae.
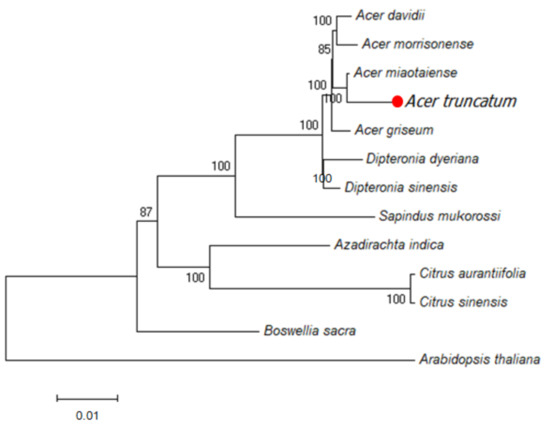
Figure 6.
Maximum likelihood (ML) phylogenetic tree inferred from 13 chloroplast genomes.
4. Conclusions
In this present study, the Acer truncatum genome sequence produced by genome survey sequencing was reported. The genomic characteristics of Acer truncatum include a genome size of 529.88 Mbp, a heterozygosis rate of 1.06% and a repeat rate of 48.8%. A total of 392,961 high-quality genomic SSR markers were developed and a graphical map of the annotated circular chloroplast genome was generated. These results and dataset may provide a new resource for future genomic analysis and molecular breeding studies of Acer truncatum.
Supplementary Materials
The following are available online at http://www.mdpi.com/1999-4907/10/2/87/s1. Figure S1: Species distribution of the top BLAST hits in the NR database, Figure S2: GC content and average sequencing depth of the Acer truncatum genome data, Figure S3: Comparison of the LSC, SSC and IR regions in chloroplast genomes of Acer truncatum and Acer miaotaiense, Table S1: Statistics of Acer truncatum sequencing data, Table S2: Statistics of gene functional annotation, Table S3: The SSR types detected in the Acer truncatum sequences.
Author Contributions
R.W., P.C., L.Z. and M.Z. performed the experiments. L.L. and J.F. analyzed the data, prepared figures and tables, wrote the paper and reviewed drafts of the paper. All authors read and approved the final manuscript.
Acknowledgments
This research was supported by the Key Research and Development Program of Shaanxi Province (2018ZDXM2-01), Industry Technology Innovation Key Technology Research and Demonstration Project of Acer truncatum Bunge (ZYZB2018-DY1170), Yangling Demonstration Zone Science and Technology Program (2018NY-26) and Young Talent fund of University Association for Science and Technology in Shaanxi, China (20160107). Sample Availability: Samples of the Acer truncatum Bunge are available from the authors.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guo, X.; Wang, R.Q.; Chang, R.Y.; Liang, X.Q.; Wang, C.D.; Luo, Y.J.; Yuan, Y.F.; Guo, W.H. Effects of nitrogen addition on growth and photosynthetic characteristics of Acer truncatum seedlings. Dendrobiology 2014, 72, 151–161. [Google Scholar] [CrossRef]
- More, D.; White, J.; More, D.; White, J. Cassell’s trees of Britain and Northern Europe. Cassells Trees Br. North. Eur. 2003. [Google Scholar]
- Wang, X.Y.; Fan, J.S.; Wang, S.Y.; Sun, R.C. A new resource of nervonic acid from purpleblow maple (Acer truncatum) seed oil. For. Prod. J. 2006, 56, 147–150. [Google Scholar]
- Poulos, A. Very long chain fatty acids in higher animals-a review. Lipids 1995, 30, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sargent, J.R.; Coupland, K.; Wilson, R. Nervonic acid and demyelinating disease. Med. Hypotheses 1994, 42, 237–242. [Google Scholar] [CrossRef]
- Guo, X.; Wang, R.Q.; Wang, C.D.; Xu, F.; Zhao, S.; Guo, W.H. Acer truncatum seedlings are more plastic than Quercus variabilis seedlings in response to different light regimes. Dendrobiology 2016, 76, 35–49. [Google Scholar] [CrossRef]
- Li, L.; Wang, X.; Niu, J.; Cui, J.; Zhang, Q.; Wan, W.; Liu, B. Effects of elevated atmospheric O3 concentrations on early and late leaf growth and elemental contents of Acer truncatum Bung under mild drought. Acta Ecol. Sin. 2017, 37, 31–34. [Google Scholar] [CrossRef]
- Yang, L.; Yin, P.; Fan, H.; Xue, Q.; Li, K.; Li, X.; Sun, L.; Liu, Y. Response Surface Methodology Optimization of Ultrasonic-Assisted Extraction of Acer Truncatum Leaves for Maximal Phenolic Yield and Antioxidant Activity. Molecules 2017, 22, 232. [Google Scholar] [CrossRef]
- Zhang, L.; Tu, Z.C.; Xie, X.; Lu, Y.; Wang, Z.X.; Wang, H.; Sha, X.M. Antihyperglycemic, antioxidant activities of two Acer palmatum cultivars, and identification of phenolics profile by UPLC-QTOF-MS/MS: New natural sources of functional constituents. Ind. Crop. Prod. 2016, 89, 522–532. [Google Scholar] [CrossRef]
- Ma, X.; Wu, L.; Ito, Y.; Tian, W. Application of preparative high-speed counter-current chromatography for separation of methyl gallate from Acer truncatum Bunge. J. Chromatogr. A 2005, 1076, 212–215. [Google Scholar] [CrossRef]
- Jiao, Y. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics 2012, 13, 201. [Google Scholar] [CrossRef]
- Zhou, W.; Hu, Y.; Sui, Z.; Fu, F.; Wang, J.; Chang, L.; Guo, W.; Li, B. Genome survey sequencing and genetic background characterization of Gracilariopsis lemaneiformis (Rhodophyta) based on next-generation sequencing. PLoS ONE 2013, 8, e69909. [Google Scholar] [CrossRef] [PubMed]
- Xin, W.; Linhai, W.; Yanxin, Z.; Xiaoqiong, Q.; Xiaoling, W.; Xia, D.; Jing, Z.; Xiurong, Z. Development of simple sequence repeat (SSR) markers of sesame (Sesamum indicum) from a genome survey. Molecules 2014, 19, 5150–5162. [Google Scholar]
- He, Y.; Xiao, H.; Deng, C.; Xiong, L.; Nie, H.; Peng, C. Survey of the genome of Pogostemon cablin provides insights into its evolutionary history and sesquiterpenoid biosynthesis. Sci. Rep. 2016, 6, 26405–26415. [Google Scholar] [CrossRef]
- An, J.; Yin, M.; Zhang, Q.; Gong, D.; Jia, X.; Guan, Y.; Hu, J. Genome Survey Sequencing of Luffa Cylindrica L. and Microsatellite High Resolution Melting (SSR-HRM) Analysis for Genetic Relationship of Luffa Genotypes. Int. J. Mol. Sci. 2017, 18, 1. [Google Scholar]
- Wang, C.; Yan, H.; Li, J.; Zhou, S.; Liu, T.; Zhang, X.; Huang, L. Genome survey sequencing of purple elephant grass (Pennisetum purpureum Schum ‘Zise’) and identification of its SSR markers. Mol. Breed. 2018, 38, 94–104. [Google Scholar] [CrossRef]
- Lu, M.; An, H.; Li, L. Genome survey sequencing for the characterization of the genetic background of Rosa roxburghii Tratt and leaf ascorbate metabolism genes. PLoS ONE 2016, 11, e0147530. [Google Scholar] [CrossRef]
- Motalebipour, E.Z.; Kafkas, S.; Khodaeiaminjan, M.; Çoban, N.; Gözel, H. Genome survey of pistachio (Pistacia vera L.) by next generation sequencing: Development of novel SSR markers and genetic diversity in Pistacia species. BMC Genomics 2016, 17, 998. [Google Scholar] [CrossRef]
- Moose, S.P.; Mumm, R.H. Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol. 2008, 147, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Davey, J.W.; Hohenlohe, P.A.; Etter, P.D.; Boone, J.Q.; Catchen, J.M.; Blaxter, M.L. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat. Rev. Genet. 2011, 12, 499–510. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Douglas, S.E. Plastid evolution: Origins, diversity, trends. Curr. Opin. Genet. Dev. 1998, 8, 655–661. [Google Scholar] [CrossRef]
- Jia, Y.; Yang, J.; He, Y.L.; He, Y.; Niu, C.; Gong, L.L.; Li, Z.H. Characterization of the whole chloroplast genome sequence of Acer davidii Franch (Aceraceae). Conserv. Genet. Resour. 2016, 8, 141–143. [Google Scholar] [CrossRef]
- Li, Z.H.; Xie, Y.S.; Zhou, T.; Jia, Y.; He, Y.L.; Yang, J. The complete chloroplast genome sequence of Acer morrisonense (Aceraceae). DNA Seq. 2015, 28, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.C.; Chen, S.Y.; Zhang, X.Z. The complete chloroplast genome of the endangered Chinese paperbark maple, Acer griseum (Sapindaceae). Conserv. Genet. Resour. 2017, 9, 1–3. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; Chen, H.; Wang, Y. Characterization of the complete chloroplast genome of Acer miaotaiense (Sapindales: Aceraceae), a rare and vulnerable tree species endemic to China. Conserv. Genet. Resour. 2016, 8, 383–385. [Google Scholar] [CrossRef]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Qi, P.; Liu, Y. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. Gigascience 2012, 1, 18–24. [Google Scholar] [CrossRef]
- Vurture, G.W.; Sedlazeck, F.J.; Nattestad, M.; Underwood, C.J.; Fang, H.; Gurtowski, J.; Schatz, M.C. GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 2017, 33, 2202–2204. [Google Scholar] [CrossRef]
- Wang, R.; Liu, P.; Fan, J.; Li, L. Comparative transcriptome analysis two genotypes of Acer truncatum Bunge seeds reveals candidate genes that influences seed VLCFAs accumulation. Sci. Rep. 2018, 8, 15504–15512. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Liu, Z.; Cui, X.; Zhang, T.; Li, Y.; Zhang, L. Comparative transcriptome sequencing and de novo analysis of Vaccinium corymbosum during fruit and color development. BMC Plant Biol. 2016, 16, 223. [Google Scholar] [CrossRef]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2017, 45, e18. [Google Scholar] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq—Versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef]
- Marc, L.; Oliver, D.; Sabine, K.; Ralph, B. Organellar genome DRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Hamidreza, C.; Yee-Greenbaum, J.L.; Glenn, T.; Mary-Jane, L.; Dupont, C.L.; Badger, J.H.; Mark, N.; Rusch, D.B.; Fraser, L.J.; Gormley, N.A. Efficient de novo assembly of single-cell bacterial genomes from short-read data sets. Nat. Biotechnol. 2011, 29, 915–921. [Google Scholar]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).