2.1. Dataset Description
This research utilized surrogate data in the absence of adequate prostate cancer data. More specifically, the surrogate data generation was based on the clinical study by [
1], which was a pilot urological health promotion program that took place in Ireland. In that study, 660 subjects between 18 and 67 years of age were recruited, and no patient had clinical evidence of PCa. PSA values were evaluated and, in the entire cohort, the mean PSA level was 1.7 with a median of 0.9. The mean age of the patients was 58 (range 25–70). The reason for this cutoff in the age range was justified by the fact that beyond age 70, the PSA measurement may not be beneficial because of the fact that in a patient aged 75 or 80, PCa is rarely life-threatening (shorter life expectancy until the tumor progresses). Moreover, in patients aged 80 and above, there is a high probability that malignant PCa is indeed present. Of course, the above statement also depends on the overall physical condition and state of health of an individual. On the other hand, below 40 years of age, the PSA measurement mainly reflects benign conditions such as prostatitis.
In order to produce surrogate PSA values, we utilized the fact the PSA data reported in [
1] approximately followed the lognormal distribution. In Probability Theory, the lognormal distribution is a continuous probability distribution of a random variable whose logarithm is normally distributed. Thus, if a random variable
X follows the lognormal distribution,
Y =
ln(
X) follows the normal distribution [
41]. Equivalently, if
Y follows the normal distribution,
X =
exp(
Y) follows the lognormal distribution. A random variable that follows the lognormal distribution takes only positive real values. We sought to find the values of the
μ and
σ2 parameters for the variable
Y, where
Y =
logX ~ Normal (
μ,
σ2) with mean value
μ and variance
σ2 [
42]. So, the PSA values are represented by the variable
X and, given that the mean value for the lognormal distribution is
Θ =
Ε(
Χ) =
exp (
μ + σ2/2) [
42], we can derive that
μ = −0.105. Moreover, the median value for the lognormal distribution is given by
median =
exp (
μ) = 0.9, where, in combination with
Θ = 1.7, we obtain
μ = −0.105 and
σ = 1.12. Furthermore, with the use of the
numPy library in Python 3.9 [
43], we used the
lognormal (
μ,
σ2, 660) function to produce random numbers that follow the lognormal distribution with mean
θ = 1.7 and
median 0.9 for 660 subjects. With this procedure, we acquired surrogate PSA values in the desired range. The minimum PSA value was 0.12 ng/mL, and the maximum value was 9.99 ng/mL. In
Table 1, we list the PSA values and the probability of PCa. The PSA index alone cannot rule out the possibility of PCa, and additional indexes should be evaluated.
Additionally, we considered the age of the subjects, as there is a close relationship between age and PCa. According to epidemiological findings, 16% of men over 50 years of age in the USA will develop PCa, and the risk increases after 40 years of age. In our dataset, the minimum age was 35 and the maximum was 70 with a mean age of 54.7 years and a standard deviation of 7.9. The match between age and PSA values was random to reflect the fact that an older man could have a low PSA result and a younger man with PCa could have a higher PSA value.
An additional very important variable is the PSA density, which was utilized as an additional feature of our data (PSAD). It was obtained by dividing the PSA value by the prostatic volume in mL, i.e., PSAD =
. In clinical procedures, the prostatic volume is determined with the help of Transrectal ultrasound (TRUS) and even with the simple, non-invasive procedure known as abdominal ultrasound. In the absence of real data, we generated random values between 14 mL and 80 mL to make sure that we accounted for not only small, normal prostate glands but also hypertrophic glands [
45]. So, the advantage of this index is that medical doctors may be able to distinguish between the situation called Prostate Gland Hypertrophy and PCa. A very important research study [
46] analyzed a total of 5.291 men with PSA ≥ 3 ng/mL along with measurement of the prostate volume. The results showed that omitting prostate biopsy in men with PSAD ≤ 0.07 (ng/mL
2) would ultimately result in 19.7% fewer biopsies, while 6.9% of clinically significant prostate cancers would be missed. Using PSAD values of 0.10 and 0.15 ng/mL
2 as a threshold resulted in the detection of 77% and 49%, respectively, of prostate cancers with Gleason Score ≥ 7, in other words, clinically significant PCa [
46]. Furthermore, in another major study [
47] involving 2162 men, of which 56% were African American with PSA values from 4 to 10 ng/mL who eventually underwent into biopsy, it was found that a threshold of PSAD = 0.08 ng/mL
2 has a 95–96% NPV (Negative Predicted Value), i.e., it correctly predicts participants who do not have PCa when the biopsy is negative. For those reasons, in our system, we also chose a threshold limit of PSAD = 0.08 ng/mL
2, with some small tolerance, as other studies chose a PSAD limit of 0.1 ng/mL
2. Moreover, it is worth noting that the PSAD index is more significant for smaller-sized glands, as a larger-sized prostate usually produces more prostate antigen, and as a result, prostate cancer may be hidden in an apparently normal PSAD [
47,
48]. On the other hand, in a prostate gland with a smaller volume, a value greater than 0.08 or 0.09 should alert the treating physician to carry out further investigation. In addition to PSAD, the PSA-TZ (Transition Zone PSA), which is defined as total PSA to the volume of the transition zone of the prostate, is sometimes estimated. In a study spanning 1997 to 1998 [
48,
49] including 273 men with PSA values from 2.5 to 4 ng/mL, it was found that with threshold values of 0.1 ng/mL
2, a sensitivity of 93.181% and a specificity of 17.985% were acquired. In conclusion, the PSA-TZ is less effective. For the purposes of our study, we use only the PSAD with a minimum value of 0.007 ng/mL
2, a maximum value of 0.356 ng/mL
2, a mean value of 0.066 ng/mL
2, and a standard deviation of 0.047 ng/mL
2, as they were obtained by surrogate data.
A crucial diagnostic indicator is the PSA velocity (PSAV), which is defined as the rate at which the PSA value changes between two consecutive measurements, with a relative distance of one year. Its measurement unit is ng/mL/year. It can highlight an incipient prostate cancer, as a significant increase from the previous measurement may hide cancer (of course, an increase may also be due to inflammation). A particularly important indicator is the so-called doubling time, i.e., the time in which the PSA value doubles from the previous measurement. Thus, certain threshold values were set, which should trigger the physician to complete further testing. The PSAV is mathematically defined as follows:
PSAV =
± 100%, where PSA
f stands for the PSA final measurement and PSA
i stands for the PSA initial measurement, within a distance of a year. Again, in a random way, we generated either an increase or a decrease in a previous PSA measurement (which we obtained from the lognormal distribution generator) to show that PSAV, according to studies performed over the years, is a great indicator and that having the appropriate threshold values of PSAD can provide important and reliable information for further monitoring of the patient [
47]. The thresholds that we utilized were, as listed in
Table 2, 0.25 ng/mL/year for the range of 35–59 years and 0.50 ng/mL/year for the age range of 60–70 years old. The minimum value was −0.85 ng/mL/year, which can be justified by the fact that a patient could have an inflammation of the prostate at the initial measurement and, at the final measurement, the patient might be free of inflammation. On the other hand, the maximum value was 4.54 (or 454%), which could be due to inflammation but could also be a sign of cancer. The mean value for PSAV was 0.141 ng/mL/year, and the standard deviation was 0.332 ng/mL/year.
The prostatic antigen (PSA) is bound to other proteins [
51]. PSA detected in the blood is usually bound to the protein a1-chymotrypsin (a proteolytic enzyme); however, more than 23% of serum PSA is in free form (free PSA). Prostate hyperplasia is more associated with an increase in free PSA, whereas PCa is more associated with a decrease in the free form of PSA [
51]. The reason why this situation occurs is not clear, but one theory is that prostate cancer not only produces more PSA but also the proteins that PSA is attached to, resulting in a decrease in free PSA [
51]. Thus, a very valuable indicator is a feature variable of our system known as the PSA ratio, which is the ratio of free PSA to total serum PSA. The PSA ratio is naturally derived without units and is used either as a decimal number or as a percentage. The threshold value of 0.24 (or 24%) is an important prognostic indicator. Especially for PSA values > 4–10 ng/mL, a free PSA value > 0.24 is probably an indication of benignity (of course, the gland should be evaluated as a whole). On the other hand, when the PSA ratio is <0.19 (quite a strict limit, in other studies, this limit may be lower than 0.15), there is a significant chance of prostate cancer [
50]. Typically, values from 0.19 to 0.24 are reported as a gray zone, which increases the necessity for active monitoring of the patient. We also referred to a related table showing the probability of prostate cancer depending on age and free PSA value [
44]. The minimum value in our dataset was 0.09, the maximum value was 0.39, the mean value was 0.266, and the standard deviation was 0.050. These values are summarized in
Table 3.
Our last feature variable was the Digital Rectal Exam (DRE), which is carried out always by a qualified Urologist when an acute bacterial inflammation of the prostate is absent because of the risk of sepsis [
52]. In our surrogate data, we chose to give the DRE variable a value of 0 or 1, where 0 denotes negative findings, whereas 1 denotes a suspicious lump. Again, in a random way, we produced the binary values of the DRE. In our dataset, we had 624 cases of 0 and 36 cases of 1, meaning that in 36 cases out of 660 (5.45%), the DRE showed a suspicious lump, whereas in 624 (94.5%), the DRE showed no findings, though we should note that no findings in the DRE do not necessarily mean the absence of PCa.
Since we dealt with a binary classification problem, related to whether a patient should or should not proceed to prostate biopsy, our target variable was also in binary form, such as “yes” or “no”, which can also be codified as 1 or 0. So, we used two different target variables (that represent the same result), and we called them “Target” and “Class” according to the model we used to predict the target variable. For example, in the Random Forest Classifier, we use the target variable (dependent variable) with the name Class (“yes” or “no”), whereas in the Neural Network that we utilized, the target variable had the name Target (values 0 and 1). In our surrogate data, Target (and Class) had 170 patients positive to proceed with a biopsy of the prostate (25.75%) and 490 patients (74.24%) where no further action was needed (again, the final decision of course is in the hands of the clinical doctor).
2.3. Hybrid Classifiers
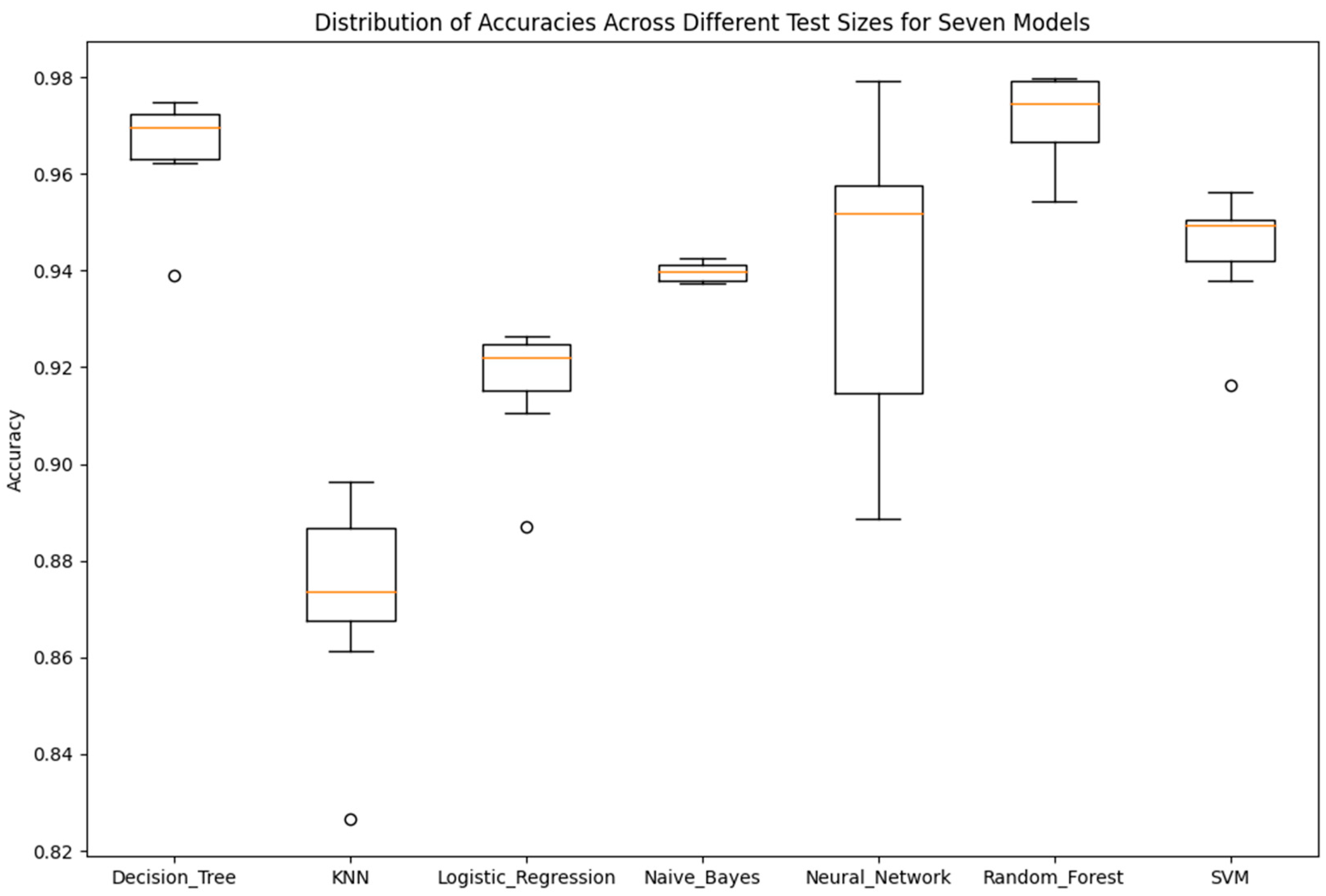
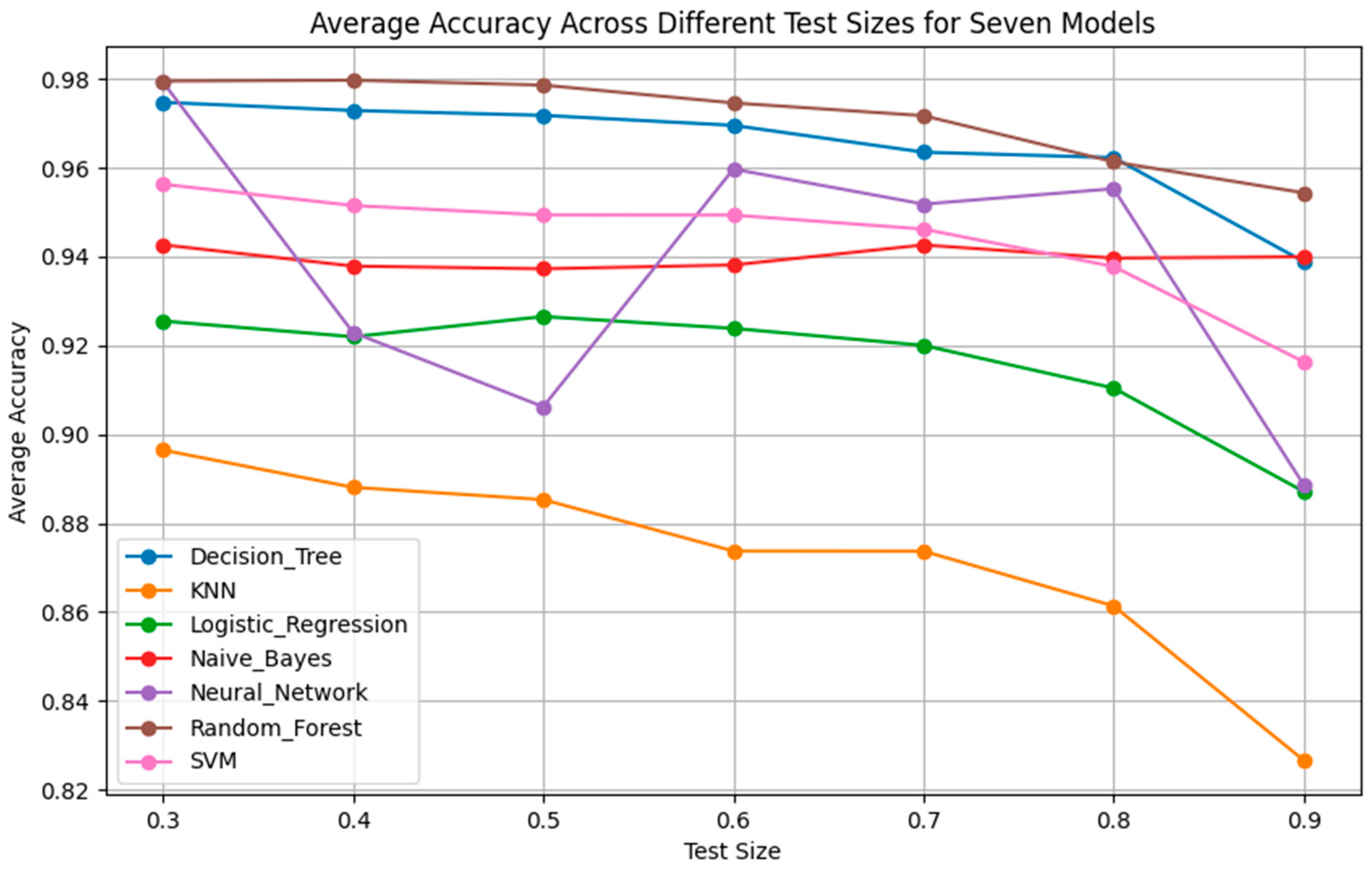
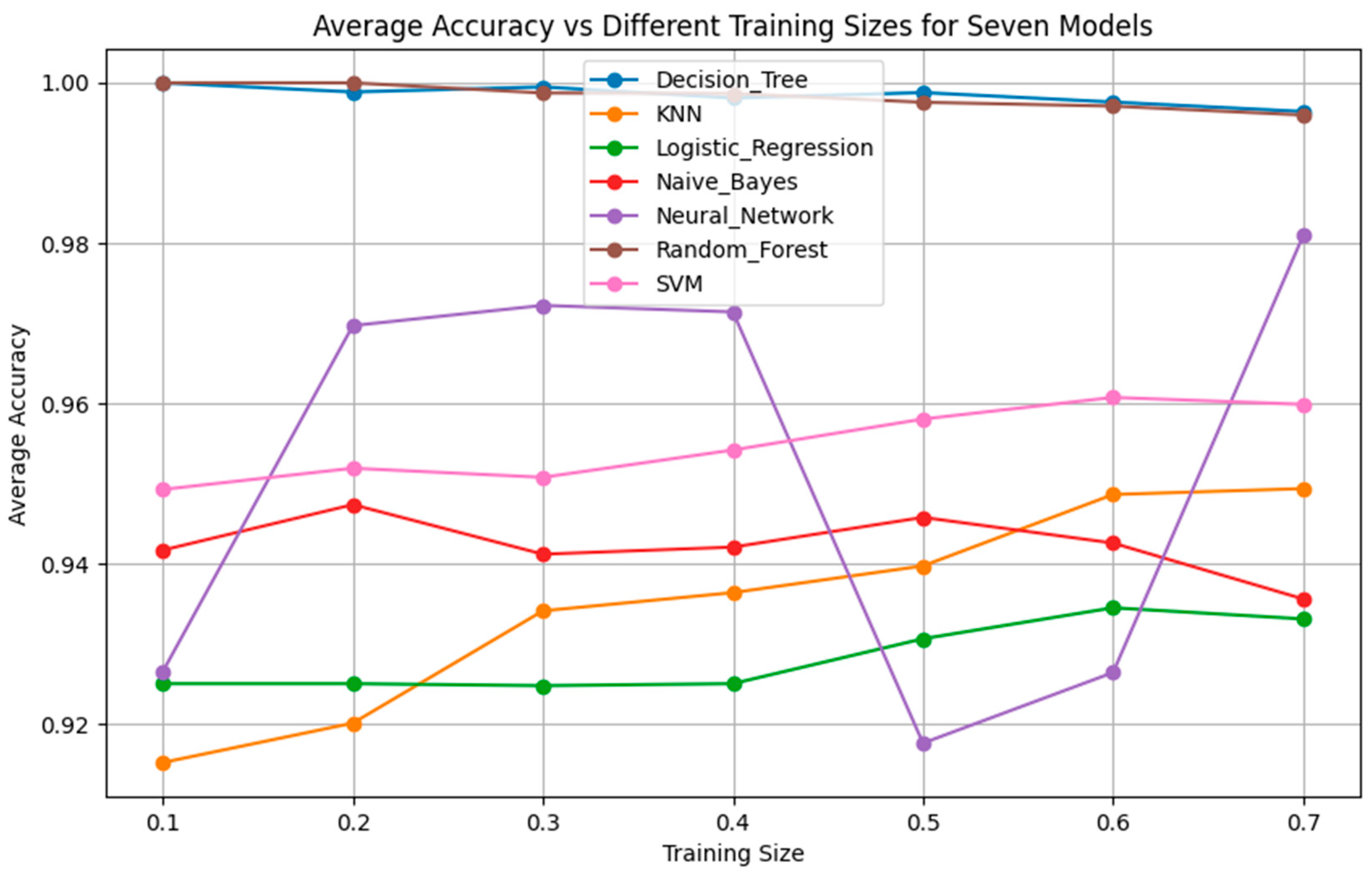
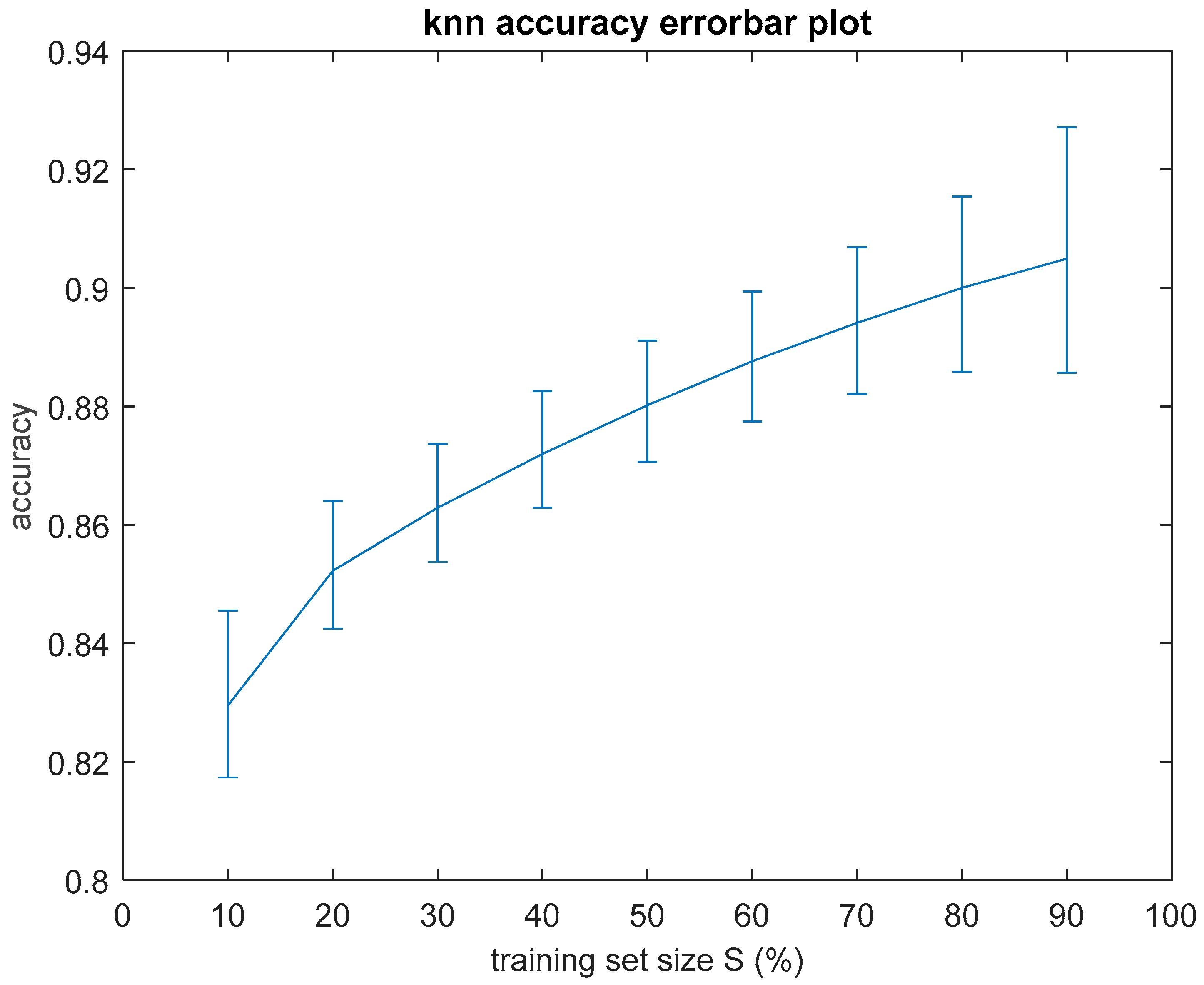
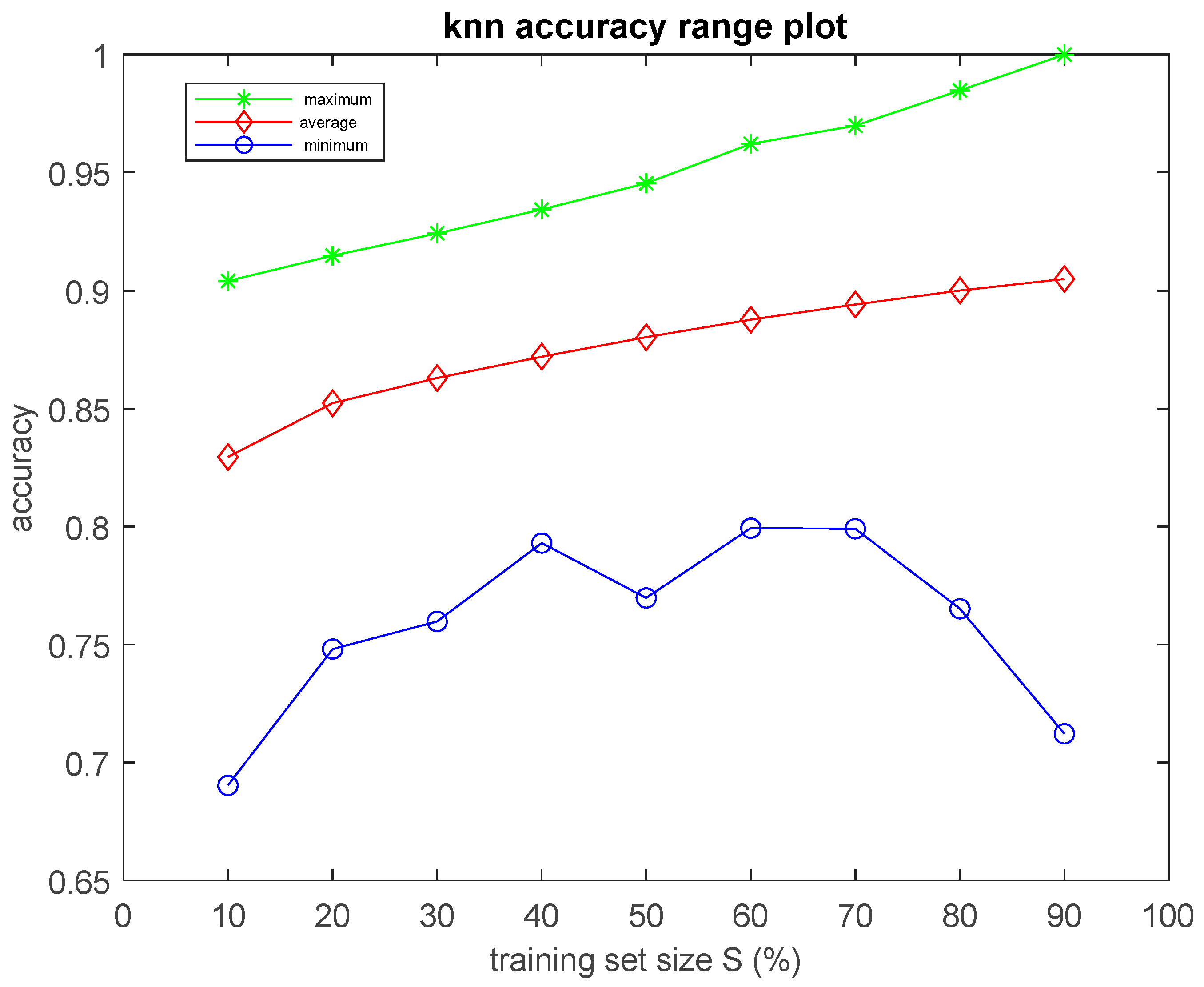
As it will be demonstrated in the Results Section, the classification performance of the seven simple algorithms depends on the size of the training set. In order to investigate the dependence of the performance of the classifiers on the size of the training set, it was considered appropriate to develop an optimization method to search for optimal results with respect to classification performance and training set size. Such an optimization method has two objectives as follows: (a) to optimize the training set, i.e., to search for the smallest training set that the machine learning classifier can use to maximize classification performance on the data of the test set, which remains unseen during the training procedure, and, at the same time, (b) to achieve accuracy equal to the maximum value, ideally equal to 1 (100%). In order to search for solutions that fulfill both the aforementioned objectives, multi-objective, generational Genetic Algorithms (GAs) were implemented asthe optimization method [
40], given that the search space was huge. Specifically, if
S is the size of the training set in number of cases (patients) and
N is the total number of cases in the dataset (
N = 660 in the present study), then there are
distinct ways (combinations) to select
S out of total
N cases in order to construct the training set. For example, according to the previous formula, there exist more than 10
120 ways to choose 110 out of 660 cases. Considering that the training set could have, at its minimum, one case, and, at its maximum,
N − 1 cases, the total number of distinct ways to construct the training set is given by the sum
of possible combinations.
The implemented GAs evolved populations of individuals with a genome consisting of 660 genes (bits), with each one of these genes corresponding to a patient (case) of the dataset. Therefore, the size of the search space was 2N, which, for N = 660, results in a search space size on the order of 10198. If the value of a gene is 1, this means that the corresponding case is included in the training set (so, it is excluded from the test set and it is unseen during the training procedure). Conversely, if the value of a gene equals 0, this means that the corresponding case is not included in the training set (therefore, it is included in the test set). To demonstrate the genome encoding, let us consider a dataset constructed of n = 10 cases. Considering that the cases in the dataset are numbered from 1 up to 10 and the length of the genome is defined to be 10, then each one gene corresponds to one particular case of the dataset. For example, if the genome of an individual of the GA is 0101001000, then the training set includes the cases that are numbered in the dataset as 2, 4, and 7, and the corresponding test set includes the cases that are numbered in the dataset as 1, 3, 5, 6, 8, 9, and 10. In this demonstration example, the training set is the 30% of the dataset and the test set is the 70% of the dataset and, of course, the cases of the test set are unseen to the classification algorithms during the training procedure. When the training procedure is completed, the trained algorithm is evaluated with respect to its classification performance on the cases of the test set, which were unseen until that point.
Based on the above, the fitness function, with which the individuals in the GA population are evaluated, is given by:
where
acc is the classification accuracy achieved on a specific test set defined by the genome of a specific individual, after the completion of the training procedure, where the corresponding training set of that individual was used. In addition,
alpha is a weight coefficient of
S with respect to
acc. The higher the value of
alpha, the smaller training set size
S the GA searches for, probably at the expense of accuracy. That is, the GA may achieve a very small
S, but this implies that the accuracy may also decrease. Thus,
alpha is a factor that balances the two objectives that the GA searches to achieve in order to maximize
f.
Roulette-wheel selection was implemented for parent selection and
elitism was also activated so that the best two individuals of a generation automatically generated clones in the next generation [
40].
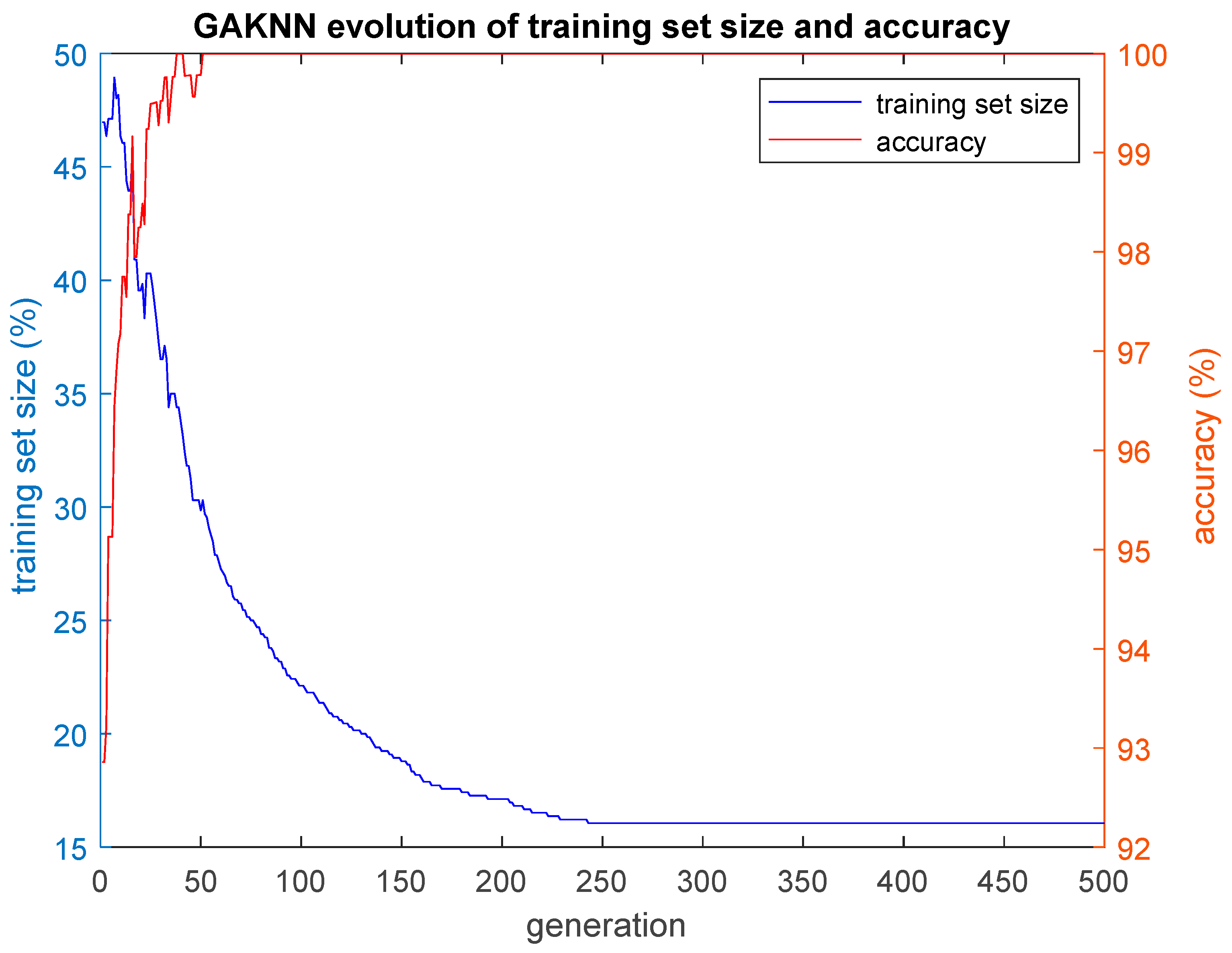
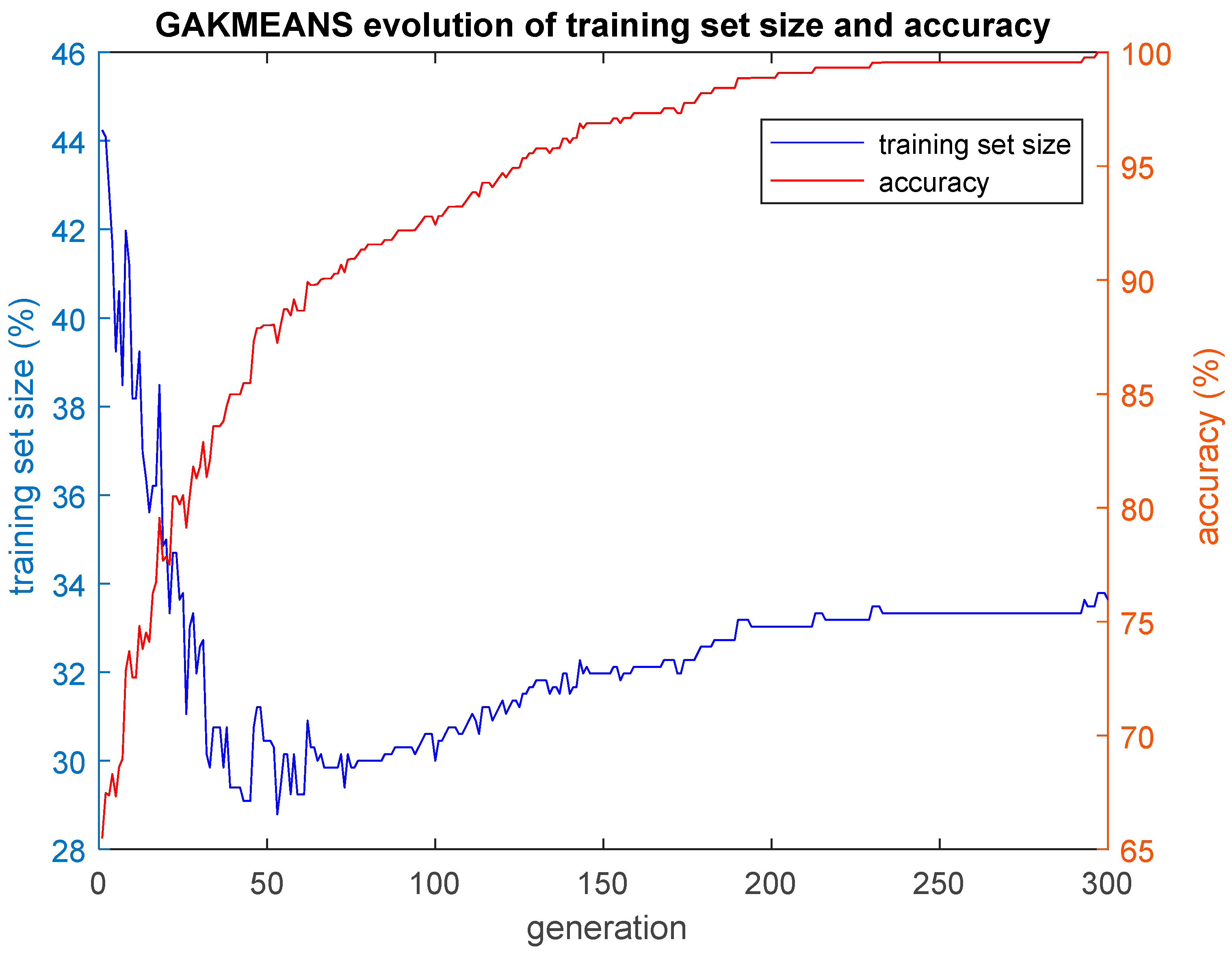
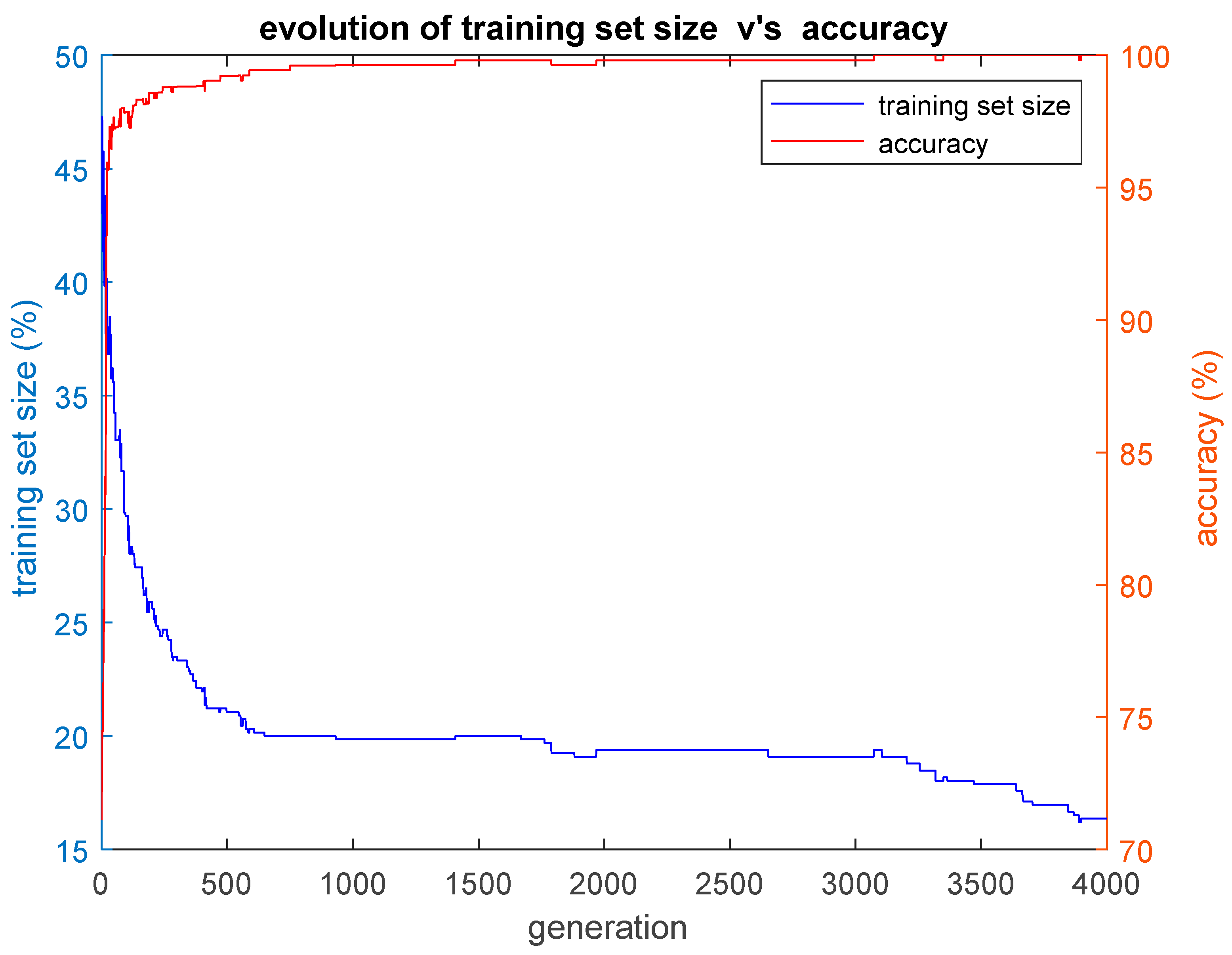
In this paper, we implemented three hybrid classification algorithms. In the first of these algorithms, a GA was combined with the simple classification algorithm K-Nearest Neighbors (GA-KNN). The aim was to find the smallest training set possible for which all the remaining cases that construct the test set are classified by the trained algorithm with an accuracy as high as possible. In other words, the GA-KNN algorithm searched for the smallest set of cases (training set) that could be considered as possible nearest neighbors to any case of the test set in order to classify that case of the test set correctly. The second hybrid algorithm combined a GA with the simple k-means clustering algorithm (GA-KMEANS). In this case, the GA searched for the smallest set of cases (training set) for which the centroids of the two clusters that were formed out of that training set could be considered for the classification of the cases of the test set with an accuracy as high as possible. In the third hybrid algorithm that was implemented, the task was to group the cases into two clusters without the use of the k-means classifier by performing genetic clustering (GACLUST). In this case, the genomes of the individuals of the GA consisted of N = 660 bits, with each gene corresponding to a case in the dataset. Each individual in the GA defined the training set and the test set in the same way as the two aforementioned hybrid algorithms, i.e., a gene with a value of 1 indicated that the corresponding case was included in the training set, whereas a gene with a value of 0 indicated that the corresponding case was included in the test set. The centroids of the two clusters were calculated based on the cases of the training set and the training procedure of the algorithm. Then, these centroids were used for the classification of the remaining cases, i.e., the cases that construct the test set. According to this procedure, the classification accuracy was evaluated based on the cases of the test set that were unseen during the training procedure. The classification of the cases of the test set was performed in the following manner: the Euclidean distances of each case of the test set with respect to the centroids derived from the training set were calculated; then, the algorithm classifies each specific case of the test set into the group with the smaller distance with respect to the two centroids. All three hybrid algorithms presented above give as output the training set, as well as plots of the evolution of the size of the training set S and the corresponding classification accuracy acc performed on the test set. The plotted values of S and acc correspond to the ones of the best individual of each generation of the GA population, i.e., the individual with the highest value of the fitness function f.
It is reasonable to ask if the GA search for the minimized training set size S and its optimization regarding the cases that it consists of could possibly lead to overfitting if the hybrid algorithms as a side effect. To examine this issue, a second dataset (set 2) was generated independently by the same means that were also used to generate the original dataset (set 1). This gave us the ability to examine if any overfitting effects emerged. This task was accomplished by conducting a large number of computer experiments, where a second round of evaluation took place as follows: the hybrid algorithms were trained using the data of the training set of the original dataset 1 and, in the first round of evaluation, the trained algorithms were applied on the validation subset and the test subset of dataset 1. During the training procedure, the data in the evaluation and test subsets were unseen by the hybrid algorithms. In the second round of evaluation, the trained hybrid algorithms of the first round were tested on the data in the second dataset 2 that were totally unseen by the hybrid algorithms during their training procedure in the first round.