1. Introduction
Lung abnormalities, including ARDS, lung cancer, pneumothorax, and pneumonia, pose significant health challenges across all age groups. The emergence of the SARS coronaviruses, particularly the COVID-19 virus (SARS-CoV-2), has further exacerbated respiratory issues, causing lung lesions that impede regular lung function [
1,
2,
3]. The accurate and timely detection of COVID-19 infections has become paramount, given the ongoing global pandemic. Although various diagnostic tools, such as antigen and antibody testing, are available, they may suffer from lengthy turnaround times and limited availability, hindering effective disease management [
4].
Recent developments in radiology imagery have shown that specific scans, such as chest X-rays, can provide essential information for COVID-19 detection. However, the shortage of qualified specialists in remote areas calls for innovative automated detection solutions [
5]. Deep learning (DL) models, particularly Convolutional Neural Networks (CNNs), have demonstrated remarkable capabilities in image analysis tasks and present a promising avenue for automated COVID-19 detection using chest X-ray scans [
6].
In this research, we propose a novel DL model for automated COVID-19 detection from chest X-ray images [
7]. Our model aims to perform extensive binary diagnostics, distinguishing COVID-19 findings from non-COVID-19 findings. To train the model, we utilize a large dataset of publicly accessible chest radiograph images, which includes clinical results consistent with COVID-19, along with verified cases of COVID-19 for creating the test dataset [
8]. By leveraging the power of CNN architecture, our DL model can effectively learn to detect abnormalities in chest X-ray scans and aid healthcare professionals in triaging high-risk patients, especially in inpatient settings.
This paper serves as a comprehensive exploration of cutting-edge deep learning techniques utilized for the detection of COVID-19 from chest X-ray images. It not only showcases the power of this technology in enhancing diagnostic speed and precision but also underscores its broader impact on healthcare systems. By accelerating the identification of COVID-19 cases, these techniques play a pivotal role in optimizing resource allocation, enabling faster isolation and treatment of affected individuals, and ultimately reducing the risk of transmission. Through this research, we aim to contribute to the growing body of knowledge that leverages AI-driven solutions to combat the ongoing pandemic, thus fostering more resilient healthcare systems for the future.
The structure of the paper is as follows:
Section 2 provides an overview of related research in the field and highlights the unique contributions of our work.
Section 3 presents the detailed processing steps of our proposed approach. In
Section 4, we present the experimental findings and performance comparisons. Finally,
Section 5 concludes the paper with a summary of our research findings and outlines potential future research directions.
2. Related Work
The healthcare domain has witnessed significant advancements in image processing, machine learning (ML), deep learning (DL), and computer vision over the past two decades. The outbreak of the COVID-19 pandemic further accelerated research in these fields, leading to a surge in studies related to COVID-19 detection and diagnosis using medical imaging [
1,
9,
10,
11,
12,
13].
During the past decades, many healthcare diagnostic analyses, including X-ray, CT scan, or MRI research, were confined to image processing [
14,
15,
16,
17,
18,
19,
20,
21,
22] and ML [
9]. Recent developments in DL, especially Convolutional Neural Networks (CNNs) [
23,
24,
25,
26], have shown promising results in various medical imaging tasks, including COVID-19 detection from chest X-ray images [
27,
28,
29,
30,
31].
Authors in [
26] aimed to classify and detect objects in X-ray baggage imagery using Convolutional Neural Networks (CNNs) with support vector machines (SVM). This study highlighted that CNN-based feature extraction outperformed handcrafted Bag-of-Visual-Words features. The identification of anomalies in chest X-rays, including the detection of the COVID-19 virus, is presented in [
1].
A framework for early-phase identification of COVID-19 using chest X-ray imaging features is developed in [
9]; this research offered an innovative approach to early COVID-19 detection using machine learning. A comprehensive overview of diagnostic methods for COVID-19, including CT scans, NAAT, RT-PCR, and CRISPR is introduced in [
27]. In the domain of deep learning for medical imaging, Mohan et al. [
25] explored the potential of deep learning in COVID-19 detection, utilizing CNNs and SVMs. Interestingly, their study compared local descriptors with deep learning, noting that while local descriptors were effective, they consumed more time.
The authors of [
24] investigated the use of stationary wavelets for data augmentation and abnormality detection in CT scans. Their study proposed a three-phase method for detecting abnormalities in lung CT scans, classifying them into COVID-positive and COVID-negative cases. In addition, a comparative analysis of deep learning models, including Inception V3, Xception, and ResNeXt, for COVID-19 detection based on chest X-rays is presented in [
23].
In response to the urgent need for effective COVID-19 diagnostic tools, COVID-Net, a deep Convolutional Neural Network designed for detecting COVID-19 cases from chest X-ray images, was introduced in [
32]. Another knowledge transfer and distillation (KTD) framework using CNNs for on-device COVID-19 patient triage and follow-up is proposed in [
33]. Finally, a deep learning-based methodology for detecting coronavirus-infected patients using X-ray images was developed in [
34].
Despite the progress, challenges remain in automated COVID-19 detection, such as text in chest X-rays, dimension mismatch, data imbalance, and the need for large and diverse datasets. In this research, we propose a novel DL-based COVID-19 diagnostic method using state-of-the-art architectures. We address the challenges of preprocessing chest X-ray images from various sources to build a robust classification model.
This study introduces innovative methods in image preprocessing to enhance the accuracy of COVID-19 detection. Specifically, we employ the Canny edge detection algorithm, a technique widely used in computer vision, to effectively reduce noise and emphasize critical edges within X-ray images. This process results in a more refined and informative image representation.
Following the Canny edge filter, we apply the Gradient-weighted Class Activation Mapping (Grad-CAM) method. This approach offers significant advantages by highlighting essential regions within the X-ray image. The Grad-CAM-generated map serves as an intermediary step that empowers the neural network to identify infected areas with greater precision, ultimately boosting the accuracy of COVID-19 detection.
The contributions of this work are stated below:
We develop a deep learning COVID-19 diagnostic method using state-of-the-art architectures, with a focus on addressing the unique challenges posed by chest X-ray images.
A comprehensive analysis of various methods used for COVID-19 detection is presented, highlighting the strengths and limitations of each approach.
Chest X-rays from different sources are collected to build a robust classification model.
The developed diagnostic model provided efficient results for an enormous variety of input images.
The results produced in this work are validated with existing state-of-the-art works.
By leveraging the capabilities of DL and addressing the research gaps in current methods, our approach contributes to advancing the field of automated COVID-19 detection from chest X-ray images.
3. Proposed Methodologies
In various fields, such as malware detection, medicine, information retrieval, and many others, the use of DL models has seen rapid growth. Recently, a new DL methodology based on Convolutional Neural Networks (CNNs) [
35,
36] has shown significant promise in various image analysis tasks. DL algorithms enable computational models to learn data representations across multiple abstraction layers, leading to improved performance in classification tasks. DL models have demonstrated higher accuracy and have the potential to enhance human performance in various domains [
15,
37,
38,
39,
40,
41,
42,
43].
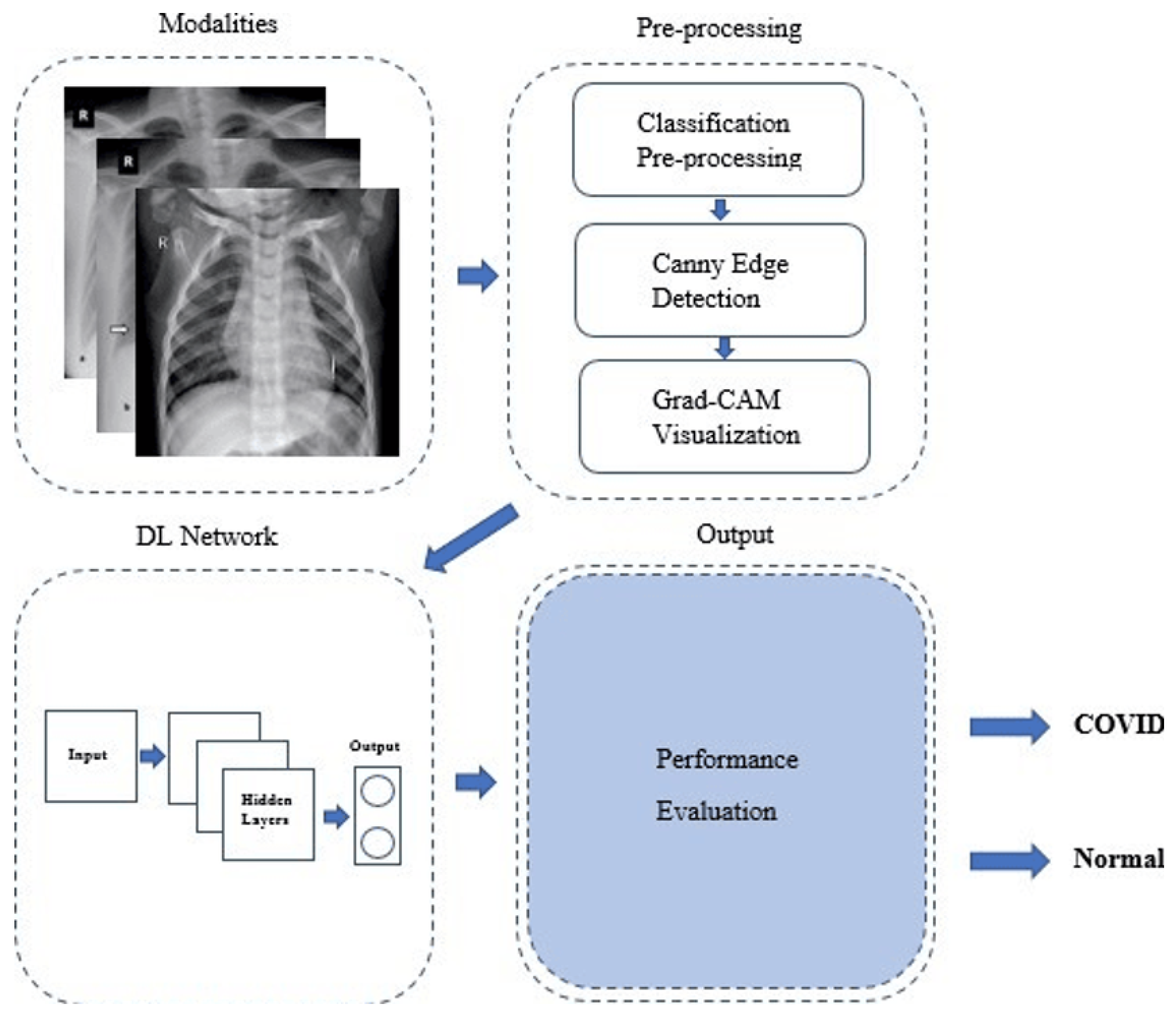
Our proposed CNN-based COVID-19 detection method consists of a sequence of phases, as depicted in
Figure 1.
The following five phases summarize the COVID-19 detection process:
Data Collection and Pre-processing: In this phase, a dataset containing chest X-ray scans of COVID-19 patients and healthy individuals is collected. The images may contain noise and artifacts, and thus, pre-processing techniques are applied to enhance the quality of the images. Additionally, Canny edge detection is employed to identify edges in the X-ray images, which can help in feature extraction. In this study, we utilized a dataset sourced from the Kaggle repository [
44], a recognized platform for medical image datasets. The dataset consists of 6432 chest X-ray images, including cases of COVID-19, normal chest X-rays, and pneumonia. To ensure a balanced representation, we divided the dataset into training and validation sets. The training set comprises 1345 normal cases, 490 COVID-19 cases, and 3632 pneumonia cases, while the validation set includes 238 normal cases, 86 COVID-19 cases, and 641 pneumonia cases. We downscaled the PA (posteroanterior) view scans of COVID-19-affected patients to
pixels to optimize model training and prevent overfitting.
Feature Extraction with Gradient-weighted Class Activation Mapping (Grad-CAM): Grad-CAM is a powerful technique that highlights the regions of the image that are crucial for the model’s prediction. By applying Grad-CAM, we can visualize the regions in the chest X-ray that contribute the most to the COVID-19 detection decision. This not only helps in understanding the model’s decision-making process but also provides insights into relevant areas of the X-ray for COVID-19 identification.
Data Splitting: The original dataset is divided into two sets: a training set and a validation set. The training set is used to train the CNN model, while the validation set is used to assess the model’s performance and fine-tune hyperparameters.
Training the CNN Model: The CNN model is trained on the training dataset using the extracted features and Grad-CAM visualizations. During training, the model learns to identify patterns and features that distinguish COVID-19 cases from healthy cases in the X-ray images.
Performance Evaluation: The trained CNN model’s performance is evaluated on the validation dataset to measure its accuracy and other relevant metrics. This evaluation helps determine the model’s effectiveness in detecting COVID-19 cases from chest X-ray images.
The proposed method aims to leverage the power of CNNs and Grad-CAM visualization to improve the accuracy and interpretability of COVID-19 detection from chest X-ray images. By following these phases, our approach aims to provide an efficient and reliable tool for assisting medical professionals in screening and diagnosing COVID-19 cases.
3.1. X-ray Diagnosis Using Deep Learning
X-ray devices are widely used in medical imaging to visualize internal body structures and detect various conditions, such as tumors, lung infections, bone dislocations, and injuries [
15]. The process involves passing light or radio waves through the body, and the resulting images provide valuable information for diagnosing and monitoring a range of medical conditions.
Deep learning has shown remarkable success in various image analysis tasks, including medical imaging. In the context of X-ray diagnosis, deep learning techniques, particularly Convolutional Neural Networks (CNNs), have been leveraged to enhance the accuracy and efficiency of diagnostic procedures. By training CNN models on large datasets of X-ray images, these models can learn to identify patterns and features indicative of specific conditions, allowing for automated and accurate diagnosis.
One of the significant advantages of using deep learning in X-ray diagnosis is the ability to process large volumes of images quickly and accurately. Deep learning models can analyze X-ray images in real time, providing rapid insights to medical professionals, thereby expediting the diagnostic process and enabling timely interventions.
Moreover, deep learning models can assist in detecting subtle abnormalities or early-stage conditions that might be challenging for human experts to identify. The ability of deep learning models to learn complex representations from data makes them particularly useful in capturing intricate patterns in X-ray images, enhancing diagnostic accuracy.
While CT scans offer more precise images of soft tissues and organs, X-rays remain a preferred choice for certain diagnostic scenarios due to their speed, safety, ease of use, and lower radiation exposure [
45,
46]. Additionally, the application of deep learning to X-ray diagnosis complements traditional radiological interpretations, providing an additional layer of accuracy and consistency to the diagnostic process.
The integration of deep learning techniques into X-ray diagnosis holds tremendous potential for improving healthcare outcomes by providing reliable and efficient diagnostic tools for medical professionals.
3.2. Pre-Processing and Canny Edge Detection
In chest X-ray images, the presence of high-intensity or bright-pixel diaphragm regions, as seen in the posteroanterior view presented in
Figure 2, can pose challenges for accurately differentiating possible disease variations. In the medical context, the letter ’R’ typically denotes the right side of the patient. This labeling convention is commonly used in medical imaging to specify the side or location of the body under examination. Here, ’R’ indicates the right side of the chest. This labeling aids healthcare professionals in accurately interpreting the image, focusing their attention on the specific area of interest, and identifying any potential abnormalities or conditions on the right side of the chest. The use of such conventions facilitates precise analysis and diagnosis, ensuring effective patient care and treatment. To improve the automated diagnosis of lung areas and enhance the detection of disease-related features, a pre-processing step is applied to the X-ray images before further analysis [
29].
In this research, we employ the Canny edge detection algorithm as a pre-processing image technique. Canny edge detection is a popular edge detection algorithm known for its ability to effectively remove noise and preserve essential edge data, thus enhancing the clarity of edges in an image. By accentuating sharp edges and producing a smoother image, the Canny algorithm enables us to extract more meaningful features and information for subsequent processing steps.
Compared to other edge detection methods, such as the Sobel, Prewitt, and Roberts, the Canny algorithm has demonstrated superior performance in maintaining more usable edge information while reducing noise. This is especially beneficial in chest X-ray images, where subtle abnormalities and disease-related features can be critical for accurate diagnosis.
Following the Canny edge detection step, we utilize the GRAD Class Activation Mapping (Grad-CAM) technique on the pre-processed images. Grad-CAM allows us to visualize the regions of the image that the CNN model focuses on during the decision-making process. By overlaying the model’s attention on the image, Grad-CAM provides insights into the areas that contribute the most to the COVID-19 detection decision. This interpretable visualization not only aids in understanding the model’s predictions but also enhances trust and confidence in the diagnostic results.
Figure 2 illustrates the output image after pre-processing with the Canny edge algorithm. The enhanced edges and reduced noise in the image contribute to a clearer representation of relevant features, ultimately assisting the deep learning model in detecting COVID-19-related patterns with higher accuracy.
3.3. COVID-19 Infection Detection Using Deep Learning
Deep learning (DL) is a field of artificial intelligence focused on teaching computers to understand and interpret data using mathematical models. In our COVID-19 detection model, DL techniques are employed to classify chest X-ray scans into two categories: COVID-19 positive and COVID-19 negative, based on the features learned through the learning process [
22].
The core architecture of our model consists of convolutional layers responsible for capturing local features indicative of abnormalities present in the X-ray scans of patients. Normalization techniques are applied to enhance the processing of the extracted features obtained from the parallel 3 × 3 Convolutional Neural Network layers. Pooling layers are then utilized to downsample and standardize distinctive characteristics, reducing computational complexity and preventing overfitting of the CNN architecture [
47,
48,
49].
To address overfitting, dropout layers are incorporated into our model. Dropout randomly deactivates a fraction of neurons during training, promoting generalization and preventing the model from relying too heavily on specific features. This ensures that the model can generalize well to unseen data.
Concatenation layers are employed to improve learning accuracy and enable the discovery of new architectural features. By combining features learned from different parts of the network, concatenation facilitates the model’s ability to capture complex patterns and relationships within the data.
The final CNN architecture is depicted in
Figure 3. After the pooling layer, all the layers in our model are flattened to generate the output, which comprises two neurons representing the binary classification of COVID-19 positive and COVID-19 negative cases.
TensorFlow is used to implement the 1D convolution, achieved by wrapping the 2D convolution in reshaped layers to add an H dimension before the 2D convolution and remove it later, adapting the 2D Convolutional Neural Network for 1D data.
The overall methodology combines the power of DL techniques, including normalization, pooling, dropout, and concatenation layers, to extract and learn meaningful features from chest X-ray images, enabling accurate and efficient detection of COVID-19 cases.
3.3.1. ReLU Activation Function
The Rectified Linear Unit (ReLU) is a popular activation function commonly used in deep learning models, including Convolutional Neural Networks (CNNs). It plays a vital role in introducing non-linearity to the network, enabling it to learn complex representations from the input data [
23].
The ReLU activation function is defined as half rectified, meaning that when the input vector
x is less than zero, the output
is set to zero, and, when
x is greater than or equal to zero, the output
is equal to the input
x. The mathematical representation of the ReLU activation function is shown in Equation (
1).
ReLU activation serves several critical purposes in the CNN architecture for COVID-19 detection from chest X-ray images. Firstly, it introduces non-linearity, allowing the model to learn and represent complex patterns and relationships in the data. This non-linearity is essential for capturing the intricate features that distinguish COVID-19 infections from normal cases in the X-ray scans.
Secondly, ReLU activation helps mitigate the vanishing gradient problem that can occur in deep neural networks during back-propagation. The vanishing gradient problem hampers the learning process by causing the gradients of the lower layers to become extremely small, resulting in slow or stalled learning. By setting negative values to zero and only propagating positive gradients during forward and backward passes, ReLU activation helps maintain more informative gradients, speeding up convergence and improving training efficiency.
Moreover, ReLU activation has been empirically found to be less computationally expensive than other activation functions, such as sigmoid and tanh. The simple and efficient nature of ReLU makes it a preferred choice for deep learning models, contributing to faster training times and lower memory consumption.
Overall, the ReLU activation function is a critical element in the CNN architecture for COVID-19 detection, enabling the model to learn powerful representations from the chest X-ray images and improving the overall accuracy and performance of the detection system.
3.3.2. SoftMax Activation Function
The SoftMax v 5.4 function is a widely used activation function in the output layer of deep learning models, including the Convolutional Neural Network (CNN) architecture [
23]. It plays a crucial role in converting the final layer’s raw scores into probability scores, allowing the model to make predictions based on class probabilities.
The SoftMax activation function operates by calculating the probabilities of all the neurons in the output layer and producing a vector of probabilities. The sum of these probabilities across all classes in the vector is equal to 1, ensuring that the final output represents a valid probability distribution over the possible classes. The mathematical representation of the SoftMax function for the
i-th element in the output vector is shown in Equation (
2).
where
is the
i-th input vector element,
is the standard exponential function, and
is the normalization term.
In the context of our COVID-19 detection model, the SoftMax activation function is applied to the output layer to obtain the probability scores for each class, i.e., COVID-19 positive and COVID-19 negative. The model then classifies the chest X-ray image as COVID-19 positive if the corresponding SoftMax probability score is higher than a certain threshold, and COVID-19 negative otherwise.
By using SoftMax, the model can make confident and calibrated predictions, providing clear probabilities for each class and enabling more robust decision making. This activation function is especially crucial in the final layer of the CNN architecture, as it allows the model to produce meaningful and interpretable output probabilities, aiding medical professionals in making accurate diagnoses.
3.3.3. Adam Optimizer
The Adam optimizer, short for Adaptive Moment Estimation, is an efficient optimization algorithm widely utilized for classification-based problems in deep learning [
36]. It combines the benefits of two other popular optimization techniques, namely “RMSProp” and “gradient descent with momentum” [
35,
36].
The core idea behind the Adam optimizer lies in estimating the first-order moments (mean) and second-order moments (uncentered variance) of the gradients. These moments, denoted as
and
, respectively, are initialized as zeros and then updated during each iteration using exponential moving averages. Equations (
3) and (
4) illustrate the update rules for
and
.
Here, and are the decay rates of the gradients, typically set to and . These decay rates control the exponential moving averages and help the optimizer to remain regulated and unbiased.
To mitigate the bias introduced by the initialization of
and
as zeros, bias-corrected estimates
and
are computed as shown in Equations (
5) and (
6).
With the bias-corrected estimates, the Adam optimizer updates the model’s weights using the general equation presented in Equation (
7):
where
denotes the weights at a given point in time
t;
denotes the weights at a given point in time
;
represents the learning rate, typically set to
; and
is a small constant (e.g.,
) to avoid division by zero when
approaches zero.
The Adam optimizer efficiently combines the benefits of RMSProp and gradient descent with momentum, leading to faster convergence and better generalization in various deep learning tasks, including COVID-19 detection from chest X-ray images.
3.3.4. Heatmap Using GRAD Class Activation Mapping
Gradient-weighted Class Activation Mapping (Grad-CAM) is a powerful visualization technique that enhances the interpretability of deep learning models, particularly Convolutional Neural Networks (CNNs) [
31]. Grad-CAM enables us to gain visual insights into the regions of the X-ray scans most relevant for COVID-19 infection detection, thereby offering transparency and evidence-based information about the model’s predictions [
17,
31].
The Grad-CAM technique operates by capturing the gradients of each target class during the model’s forward pass. It then uses these gradients to produce a coarse localization map by traversing through the last convolutional layer. This localization map highlights the significant regions in the X-ray image that contribute most to the model’s prediction of COVID-19 infection. An example of the Grad-CAM visualization’s impact is shown in
Figure 4. The arrow marks or arrows added to a chest X-ray image serve as annotations, which are commonly used to highlight or draw attention to specific structures, abnormalities, or areas of interest within the X-ray image. They can be employed to emphasize particular abnormalities such as tumors, nodules, fractures, or other conditions that are of significance for clinical analysis and diagnosis.
Integrating Grad-CAM with our CNN-based COVID-19 detection model, we generate high-resolution visualizations of class discrimination, providing precise insights into the regions affected by the COVID-19 infection in X-ray scans. By superimposing the Grad-CAM heatmap onto the original X-ray image, medical professionals and researchers can gain valuable insights into the areas of abnormality detected by the model [
17].
This visualization technique offers several advantages for our COVID-19 detection approach. First, it enhances the model’s transparency, providing visual evidence of the regions that the model relies on to make its predictions. This transparency is especially critical in medical applications, where the interpretability of AI-based systems is essential for building trust among healthcare practitioners.
Second, the Grad-CAM visualization aids in the fine-grained interpretation of the CNN model’s decisions. By focusing on the localized regions in the X-ray images, medical experts can gain deeper insights into the specific features that the model identifies as indicative of COVID-19 infection. This can lead to a better understanding of the disease and improve the model’s diagnostic accuracy.
Incorporating Grad-CAM into our COVID-19 detection system enhances its interpretability, accuracy, and potential for further research in the medical domain. The ability to visualize and understand the model’s decision-making process fosters confidence in the system’s performance and supports medical professionals in making well-informed decisions.
3.4. Evaluation Metrics
To evaluate the performance of the proposed deep learning-based COVID-19 detector, we employ several essential evaluation metrics: accuracy, precision, recall (also known as sensitivity), and F-measure.
Accuracy measures the overall performance of the model across all classes. It is calculated as the ratio of correctly predicted samples (true positives and true negatives) to the total number of samples in the dataset. The accuracy can be computed using Equation (
8).
Precision is a metric that assesses the percentage of positive predictions that are correct. It quantifies the model’s ability to avoid false positives and is calculated using Equation (
9).
Recall (also known as sensitivity) evaluates the model’s ability to identify all positive instances, thereby measuring its performance in terms of false negatives. Recall is computed as the ratio of true positives to the sum of true positives and false negatives, as shown in Equation (
10).
F-measure, also known as the F1-score, provides a balanced view of the model’s precision and recall. It is the harmonic mean of precision and recall and is given by Equation (
11).
These metrics are computed based on the values of True Positive (), False Positive (), True Negative (), and False Negative () obtained during the evaluation of the model.
The confusion matrix, as depicted in
Figure 5, visually summarizes the performance of the detector model. It shows the count of true positive, false positive, true negative, and false negative predictions, providing insights into the model’s strengths and weaknesses.
The results obtained from the evaluation metrics and the confusion matrix play a crucial role in understanding the performance of the proposed COVID-19 detector. By analyzing these metrics, medical practitioners and researchers can assess the model’s accuracy, reliability, and potential for real-world application in COVID-19 detection from chest X-ray images.
4. Experimental Result Analysis
4.1. Datasets of Chest X-ray Images
In this paper, we utilized three datasets containing chest X-ray images to conduct our study. These datasets were sourced from publicly available medical archives and reviewed in collaboration with renowned institutes. The datasets consist of soft copies of X-ray images, specifically the posteroanterior (PA) chest view, which provides crucial visual information for accurate diagnosis.
Although CT scans offer better accuracy and efficiency in certain cases [
24,
26,
30,
31], the potential long-term effects of exposure to ionizing radiation, such as an increased risk of cancer, particularly for young individuals, led us to focus on X-ray scans for our study. The combined dataset consists of a total of 3474 2D X-ray scans [
21]. Among these scans, 415 were confirmed COVID-19 cases, 2880 were standard (healthy) cases, and 179 showed signs of pneumonia.
To ensure consistency and ease of processing, all images in the dataset were resized to a standardized dimension of pixels. To assess the performance of the proposed COVID-19 detection model, we randomly split the dataset, using of the data for training and the remaining for validation.
Figure 6 presents representative X-ray chest images from the dataset, illustrating both healthy (standard) cases and cases affected by COVID-19 and pneumonia.
The utilization of multiple datasets enhances the diversity and generalizability of our model, enabling it to make accurate predictions across different scenarios. The resized images provide a consistent input size for the deep learning model, facilitating efficient training and evaluation. By randomly splitting the dataset into training and validation sets, we ensure an unbiased assessment of the model’s performance during the evaluation phase.
4.2. Experimental Results
For our experiments, we employed a dataset generated from three publicly available chest X-ray datasets [
26,
30]. The combined dataset contains four different labels: Normal, Bacterial, Non-COVID Viral, and COVID-19. As the main goal of our study was to identify positive COVID-19 cases, we binarized the labels as either positive or negative. The entire dataset was then used to develop a COVID-19 detection model based on Convolutional Neural Networks (CNN).
All chest X-ray scans in the dataset were resized to a standardized dimension of pixels. The first layer of the CNN model served as the input layer, with a size of , representing RGB images. The subsequent layers consisted of Convolutional ReLU and Max Pooling layers, which were trained on thousands of images from the dataset. We then applied transfer learning by using pre-trained models on the COVID-19 dataset. The flattened layer was employed to convert the tensor to a vector, which was then passed to a fully connected neural network classifier.
To prevent overfitting, a dropout of was applied. The output layer used the Adam Optimizer for continued optimization of the model. The original dataset was randomly divided into training and testing datasets using random sampling. Data augmentation was carried out during training, which included random rotations, translations, and flips of the training images. The model was implemented using the PyTorch toolbox. The batch size was set to 32, and each model was trained for 50 epochs, with these values determined empirically. The learning rate for the Adam optimizer was also determined empirically.
The performance of the COVID-19 detection model was evaluated using a confusion matrix, which was obtained from testing over 200 accurate X-ray scans. The confusion matrix indicated that 147 scans were correctly classified as COVID-19 positive, and 45 scans were correctly classified as COVID-19 negative. The CNN-based COVID-19 detection model achieved an overall weighted F-measure of , showcasing its effectiveness in identifying positive COVID-19 cases.
The Rectified Linear Unit (ReLU) activation function was used for the model, which is beneficial due to its non-saturation property, leading to faster convergence during training compared to sigmoid or tanh functions. The ReLU function and its derivative are both monotonic, providing an output in the range of zero to infinite, making it suitable for classification tasks.
The training parameters, including the number of layers, the shape of each layer, and the types of convolutional layers used, are summarized in
Table 1.
Additionally, we compared the performance of our COVID-19 detection model with other currently developed methods in
Table 2. The model exhibited superior recall, precision, and F-measure metrics, making it stable and effective in identifying COVID-19 cases. This development contributes to providing expert radiologists in health centers with a valuable second opinion, improving diagnostic accuracy and aiding in the timely identification and management of COVID-19 cases.
The performance of the developed CNN-based COVID-19 detection model is depicted in
Figure 7. The figure shows the accuracy and loss trends during the training process. As observed, the accuracy of the model steadily increases with each epoch, demonstrating that the model effectively learns from the training data. On the other hand, the loss decreases significantly during the initial epochs, indicating that the model is reducing its prediction errors. The convergence of accuracy and loss further validates the effectiveness of the model in learning and generalizing from the training data. Overall, the performance of the CNN model is highly satisfactory, achieving a weighted F-measure of
, which confirms its potential to accurately identify COVID-19 cases from chest X-ray images.
In
Figure 8, we present the performance metrics results of our proposed CNN model. This figure includes key metrics such as sensitivity, specificity, accuracy, and the F1 score, which are essential for a comprehensive evaluation of the diagnostic tool. These metrics offer insights into the model’s ability to correctly identify COVID-19 cases and its ability to accurately classify non-COVID-19 cases.
In contrast to using pre-trained models, which often demand significant computational resources due to their deep and complex architectures, we opted to design a customized Convolutional Neural Network (CNN) for our COVID-19 detection system. This customized CNN is intentionally lighter in structure, resulting in faster processing times and reduced computational demands. During testing, our proposed model demonstrated promising results, outperforming computationally expensive alternatives. Detailed discussions and comparative analyses are provided in the following section. We recommend our customized model for its efficient performance and streamlined architecture.
4.3. Discussion
The experimental results demonstrate the successful development of a deep learning-based COVID-19 detection model using chest X-ray images. The model achieved high accuracy, precision, recall, and F-measure metrics, indicating its ability to effectively distinguish between COVID-19 positive and negative cases. The use of multiple publicly available datasets enhanced the diversity of the training data, leading to a more robust and generalizable model.
The decision to work with X-ray scans instead of CT scans was driven by concerns about the potential long-term effects of ionizing radiation, especially for young individuals. X-ray scans are widely available and cost-effective, making them a practical choice for large-scale COVID-19 screening and diagnosis.
The CNN architecture, with its convolutional layers, ReLU activation function, and pooling layers, proved to be effective in extracting meaningful features from the X-ray images. The use of transfer learning further boosted the model’s performance by leveraging pre-trained weights from relevant datasets.
Data augmentation techniques, such as random rotations and flips, contributed to preventing overfitting and improving the model’s ability to generalize to unseen data. The Adam optimizer, with its adaptive learning rate and momentum, facilitated efficient optimization and convergence during training.
Comparing our model’s performance with other state-of-the-art methods, our COVID-19 detection model demonstrated competitive results, highlighting its potential as a valuable tool in the fight against the pandemic. By providing a second opinion to radiologists, the model can assist in speeding up the diagnosis process and aid in early detection and patient management.
However, there are certain limitations to our study. The performance of the model heavily relies on the quality and diversity of the training data. As such, access to larger and more diverse datasets would further improve the model’s accuracy and generalization capabilities. Additionally, the current study focuses on binary classification (COVID-19 positive or negative), and future research could explore the possibility of multi-class classification to distinguish between different respiratory conditions, including other viral infections and bacterial pneumonia.
Another limitation of the proposed work is the relatively small size of the dataset used for training and validation. The size of the dataset significantly influences the reliability and generalization of the model. We acknowledge that a larger dataset would have enhanced the robustness of our findings and strengthened the generalizability of the proposed model.
While the focus of this study is on binary classification, distinguishing COVID-19-positive cases from negatives, it is important to clarify our motivation for this choice. During the initial stages of the COVID-19 pandemic, the urgent need for accurate and rapid diagnosis of COVID-19 cases prompted our decision to develop a reliable binary classification model. This binary approach serves as a fundamental building block, allowing us to rigorously evaluate our model’s performance in this critical task. We acknowledge the potential for multi-class classification to distinguish between various respiratory diseases, and we intend to explore this avenue in our future research, thereby extending the scope of our diagnostic framework.
In addition, while our study has achieved promising results in distinguishing between positive and negative cases in chest X-ray images using the available dataset, we acknowledge the limited scale and scope of the dataset. It is important to note that our primary objective was to develop a robust binary classification model. We recognize the potential benefits of incorporating external datasets to further validate and enhance the generalizability of our model. In future research, we plan to collaborate with medical institutions to access larger and more diverse datasets, allowing us to extend the scope of our work and address the concerns regarding dataset scale and representativeness.
In conclusion, our deep learning-based COVID-19 detection model has shown promising results and has the potential to be a valuable tool in the medical field. With further advancements in technology and access to more comprehensive datasets, AI-driven diagnostic systems can play a critical role in pandemic management and public health.
5. Conclusions and Future Work
In this research, we have presented a Convolutional Neural Network (CNN)-based deep learning approach for the detection of COVID-19 infections from chest X-ray imagery. The proposed model achieved a high accuracy of up to , demonstrating its potential as an effective tool to assist doctors and lab technicians in improving the screening of probable COVID-19 patients. The CNN model utilizes an explanatory approach, which allows it to learn and detect significant features indicative of COVID-19 infection, leading to accurate predictions.
However, it is important to note that the developed model is not a fully deployable solution and requires further refinement and evaluation in clinical settings. Future work should focus on conducting extensive validation studies with diverse and larger datasets to ensure the model’s robustness and generalizability across different populations and imaging settings. Additionally, it is essential to integrate the proposed method with real-world clinical practices and workflows to assess its practical utility in real-time COVID-19 screening.
Moreover, ongoing research and developments in deep learning and medical imaging should be leveraged to enhance the COVID-19 infection detection system continuously. Incorporating domain knowledge and expert annotations can further refine the model’s performance and improve its diagnostic accuracy. Furthermore, considering the evolving nature of the COVID-19 virus and potential variations in X-ray image patterns over time, the model should be regularly updated and retrained to adapt to new challenges and emerging variants.
Looking ahead, the proposed approach could also be extended for risk detection in survival analysis, predicting patient hospitalization times, and assessing individualized treatment plans. By integrating this technology into healthcare systems, it has the potential to assist in population control, early intervention, and personalized patient care. However, it is essential to acknowledge a major limitation of the proposed work: the unavailability of a large number of images for COVID-19, which could have aided in the training of the pre-trained network architectures used.
In future research, it would be beneficial to explore the applicability of the proposed method to detect other lung disorders, such as ARDS, lung cancer, pneumothorax, and pneumonia. This would expand the utility of the developed model and make it a versatile tool for diagnosing a wide range of pulmonary conditions.
Furthermore, we are committed to conducting comprehensive clinical trials in collaboration with healthcare professionals and institutions. These trials will provide a robust evaluation of the model’s sensitivity, specificity, and practical utility in aiding healthcare providers. We believe that this crucial step will further enhance the reliability and applicability of our diagnostic tool.
In conclusion, this study demonstrates the effectiveness of deep learning-based approaches in COVID-19 detection, and, with continuous research and refinement, it holds promise to significantly impact medical diagnostics and patient care in the fight against the ongoing COVID-19 pandemic and beyond.
6. Ethical Statement
This study strictly adhered to ethical standards and guidelines, particularly in the handling of anonymized data. The research was conducted in full compliance with the principles outlined in the Declaration of Helsinki. Ethical approval was meticulously obtained from the Ethics Committee of Samsung Medical Center, Sungkyunkwan University (Ethical Approval Number: 2023-02-078).
The Ethics Committee conducted a thorough review of the study protocol and the informed consent procedures to ensure the research was conducted with utmost safety and in complete accordance with ethical norms. Informed consent was thoughtfully obtained from all study participants, and, in the case of minors, their legal guardians also provided consent.
This rigorous ethical framework underscores our commitment to safeguarding the rights, privacy, and well-being of all individuals involved in this research.
Author Contributions
Conceptualization, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K.; Data Curation, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K.; Writing—Original Draft, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K.; Methodology, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K.; Review and Editing, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K.; Supervision, S.M., D.S., S.K.S., A.R., B.A., V.C.G. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ng, M.Y.; Lee, E.Y.P.; Yang, J.; Yang, F.; Li, X.; Wang, H.; Sze Lui, M.M.; Lo, C.S.Y.; Leung, B.; Khong, P.L.; et al. Imaging Profile of the COVID-19 Infection: Radiologic Findings and Literature Review. Radiol. Cardiothorac. Imaging 2020, 2, e200034. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.K.; Mohanta, G.C.; Kumar, V.; Gupta, K. Diagnostic Tools for Rapid Screening and Detection of SARS-CoV-2 Infection. Vaccines 2022, 10, 1200. [Google Scholar] [CrossRef] [PubMed]
- Vernikou, S.; Lyras, A.; Kanavos, A. Multiclass sentiment analysis on COVID-19-related tweets using deep learning models. Neural Comput. Appl. 2022, 34, 19615–19627. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Zhong, Z.; Zhao, W.; Zheng, C.; Wang, F.; Liu, J. Chest CT for Typical Coronavirus Disease 2019 (COVID-19) Pneumonia: Relationship to Negative RT-PCR Testing. Radiology 2020, 296, E41–E45. [Google Scholar] [CrossRef]
- Antoniou, A.; Storkey, A.J.; Edwards, H. Data Augmentation Generative Adversarial Networks. arXiv 2017, arXiv:1711.04340. [Google Scholar]
- Livieris, I.E.; Kanavos, A.; Tampakas, V.; Pintelas, P.E. An Ensemble SSL Algorithm for Efficient Chest X-ray Image Classification. J. Imaging 2018, 4, 95. [Google Scholar] [CrossRef]
- Fiszman, M.; Chapman, W.W.; Aronsky, D.; Evans, R.S.; Haug, P.J. Research Paper: Automatic Detection of Acute Bacterial Pneumonia from Chest X-ray Reports. J. Am. Med. Inform. Assoc. 2000, 7, 593–604. [Google Scholar] [CrossRef]
- Rai, P.; Kumar, B.K.; Deekshit, V.K.; Karunasagar, I.; Karunasagar, I. Detection Technologies and Recent Developments in the Diagnosis of COVID-19 Infection. Appl. Microbiol. Biotechnol. 2021, 105, 441–455. [Google Scholar] [CrossRef]
- Bernheim, A.; Mei, X.; Huang, M.; Yang, Y.; Fayad, Z.A.; Zhang, N.; Diao, K.; Lin, B.; Zhu, X.; Li, K.; et al. Chest CT Findings in Coronavirus Disease-19 (COVID-19): Relationship to Duration of Infection. Radiology 2020, 295, 685–691. [Google Scholar] [CrossRef]
- Fang, Y.; Zhang, H.; Xie, J.; Lin, M.; Ying, L.; Pang, P.; Ji, W. Sensitivity of Chest CT for COVID-19: Comparison to RT-PCR. Radiology 2020, 296, E115–E117. [Google Scholar] [CrossRef]
- Kay, F.U.; Abbara, S. The Many Faces of COVID-19: Spectrum of Imaging Manifestations. Radiol. Cardiothorac. Imaging 2020, 2, e200037. [Google Scholar] [CrossRef] [PubMed]
- Saeed, M.; Ahsan, M.; Saeed, M.H.; Rahman, A.U.; Mehmood, A.; Mohammed, M.A.; Jaber, M.M.; Damaševičius, R. An Optimized Decision Support Model for COVID-19 Diagnostics Based on Complex Fuzzy Hypersoft Mapping. Mathematics 2022, 10, 2472. [Google Scholar] [CrossRef]
- Vardhana, M.; Arunkumar, N.; Lasrado, S.; Abdulhay, E.W.; Ramírez-González, G. Convolutional Neural Network for Bio-medical Image Segmentation with Hardware Acceleration. Cogn. Syst. Res. 2018, 50, 10–14. [Google Scholar] [CrossRef]
- Abbas, A.; Abdelsamea, M.M.; Gaber, M.M. Classification of COVID-19 in Chest X-ray Images using DeTraC Deep Convolutional Neural Network. Appl. Intell. 2021, 51, 854–864. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, S.; Panigrahi, B.K.; Dey, N.; Rajinikanth, V.; Gandhi, T.K. Deep Transfer Learning-based Automated Detection of COVID-19 from Lung CT Scan Slices. Appl. Intell. 2021, 51, 571–585. [Google Scholar] [CrossRef]
- Allioui, H.; Mohammed, M.A.; Benameur, N.; Al-Khateeb, B.; Abdulkareem, K.H.; Garcia-Zapirain, B.; Damaševičius, R.; Maskeliūnas, R. A Multi-Agent Deep Reinforcement Learning Approach for Enhancement of COVID-19 CT Image Segmentation. J. Pers. Med. 2022, 12, 309. [Google Scholar] [CrossRef]
- Gupta, H.; Jin, K.H.; Nguyen, H.Q.; McCann, M.T.; Unser, M. CNN-Based Projected Gradient Descent for Consistent CT Image Reconstruction. IEEE Trans. Med. Imaging 2018, 37, 1440–1453. [Google Scholar] [CrossRef]
- John, S.J.; Sharmila, T.S. A Neutrosophic Set Approach on Chest X-rays for Automatic Lung Infection Detection. Inf. Technol. Control. 2023, 52, 37–52. [Google Scholar]
- Livieris, I.E.; Kanavos, A.; Tampakas, V.; Pintelas, P.E. A Weighted Voting Ensemble Self-Labeled Algorithm for the Detection of Lung Abnormalities from X-rays. Algorithms 2019, 12, 64. [Google Scholar] [CrossRef]
- Livieris, I.E.; Kanavos, A.; Pintelas, P.E. Detecting Lung Abnormalities From X-rays Using an Improved SSL Algorithm. Electron. Notes Theor. Comput. Sci. 2019, 343, 19–33. [Google Scholar] [CrossRef]
- Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Appl. Sci. 2020, 10, 3233. [Google Scholar] [CrossRef]
- Zebin, T.; Rezvy, S. COVID-19 Detection and Disease Progression Visualization: Deep Learning on Chest X-rays for Classification and Coarse Localization. Appl. Intell. 2021, 51, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Alshazly, H.A.; Linse, C.; Barth, E.; Martinetz, T. Explainable COVID-19 Detection Using Chest CT Scans and Deep Learning. Sensors 2021, 21, 455. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Sohail, A.; Zahoora, U.; Qureshi, A.S. A Survey of the Recent Architectures of Deep Convolutional Neural Networks. Artif. Intell. Rev. 2020, 53, 5455–5516. [Google Scholar] [CrossRef]
- Mohan, A.; Poobal, S. Crack Detection using Image Processing: A Critical Review and Analysis. Alex. Eng. J. 2018, 57, 787–798. [Google Scholar] [CrossRef]
- Myles-Worsley, M.; Johnston, W.A.; Simons, M.A. The Influence of Expertise on X-ray Image Processing. J. Exp. Psychol. Learn. Mem. Cogn. 1988, 14, 553. [Google Scholar] [CrossRef]
- Maier, A.; Syben, C.; Lasser, T.; Riess, C. A Gentle Introduction to Deep Learning in Medical Image Processing. Z. Med. Phys. 2019, 29, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Singh, K.; Koundal, D. A novel fusion based convolutional neural network approach for classification of COVID-19 from chest X-ray images. Biomed. Signal Process. Control. 2022, 77, 103778. [Google Scholar] [CrossRef]
- Wang, G.; Li, W.; Zuluaga, M.A.; Pratt, R.; Patel, P.A.; Aertsen, M.; Doel, T.; David, A.L.; Deprest, J.; Ourselin, S.; et al. Interactive Medical Image Segmentation Using Deep Learning With Image-Specific Fine Tuning. IEEE Trans. Med. Imaging 2018, 37, 1562–1573. [Google Scholar] [CrossRef]
- Webb, S.M. The MicroAnalysis Toolkit: X-ray Fluorescence Image Processing Software. In Proceedings of the AIP Conference Proceedings, Chicago, IL, USA, 9 September 2011; Volume 1365, pp. 196–199. [Google Scholar]
- Wu, G.; Masiá, B.; Jarabo, A.; Zhang, Y.; Wang, L.; Dai, Q.; Chai, T.; Liu, Y. Light Field Image Processing: An Overview. IEEE J. Sel. Top. Signal Process. 2017, 11, 926–954. [Google Scholar] [CrossRef]
- Wang, L.; Lin, Z.Q.; Wong, A. Covid-net: A Tailored Deep Convolutional Neural Network Design for Detection of COVID-19 Cases from Chest X-ray Images. Sci. Rep. 2020, 10, 19549. [Google Scholar] [CrossRef]
- Li, X.; Li, C.; Zhu, D. COVID-MobileXpert: On-Device COVID-19 Screening using Snapshots of Chest X-ray. arXiv 2020, arXiv:2004.03042. [Google Scholar]
- Sethy, P.K.; Behera, S.K.; Ratha, P.K.; Biswas, P. Detection of Coronavirus Disease (COVID-19) Based on Deep Features and Support Vector Machine. 2020. Available online: https://pdfs.semanticscholar.org/9da0/35f1d7372cfe52167ff301bc12d5f415caf1.pdf (accessed on 19 October 2023).
- Apostolopoulos, I.D.; Mpesiana, T.A. COVID-19: Automatic Detection from X-ray Images Utilizing Transfer Learning with Convolutional Neural Networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef] [PubMed]
- Goel, T.; Murugan, R.; Mirjalili, S.; Chakrabartty, D.K. OptCoNet: An Optimized Convolutional Neural Network for an Automatic diagnosis of COVID-19. Appl. Intell. 2021, 51, 1351–1366. [Google Scholar] [CrossRef]
- Dilshad, N.; Song, J. Dual-Stream Siamese Network for Vehicle Re-Identification via Dilated Convolutional layers. In Proceedings of the IEEE International Conference on Smart Internet of Things (SmartIoT), Jeju, Republic of Korea, 13–15 August 2021; pp. 350–352. [Google Scholar]
- Dilshad, N.; Ullah, A.; Kim, J.; Seo, J. LocateUAV: Unmanned Aerial Vehicle Location Estimation via Contextual Analysis in an IoT Environment. IEEE Internet Things J. 2022, 10, 4021–4033. [Google Scholar] [CrossRef]
- Hasan, N.; Bao, Y.; Shawon, A.; Huang, Y. DenseNet Convolutional Neural Networks Application for Predicting COVID-19 Using CT Image. SN Comput. Sci. 2021, 2, 389. [Google Scholar] [CrossRef] [PubMed]
- Hira, S.; Bai, A.; Hira, S. An Automatic Approach based on CNN Architecture to Detect COVID-19 Disease from Chest X-ray Images. Appl. Intell. 2021, 51, 2864–2889. [Google Scholar] [CrossRef] [PubMed]
- Kanavos, A.; Kounelis, F.; Iliadis, L.; Makris, C. Deep Learning Models for Forecasting Aviation Demand Time Series. Neural Comput. Appl. 2021, 33, 16329–16343. [Google Scholar] [CrossRef]
- Mahmood, A.; Ospina, A.G.; Bennamoun, M.; An, S.; Sohel, F.; Boussaïd, F.; Hovey, R.; Fisher, R.B.; Kendrick, G.A. Automatic Hierarchical Classification of Kelps Using Deep Residual Features. Sensors 2020, 20, 447. [Google Scholar] [CrossRef]
- Tuyen, D.N.; Tuan, T.M.; Son, L.H.; Ngan, T.T.; Giang, N.L.; Thong, P.H.; Hieu, V.V.; Gerogiannis, V.C.; Tzimos, D.; Kanavos, A. A Novel Approach Combining Particle Swarm Optimization and Deep Learning for Flash Flood Detection from Satellite Images. Mathematics 2021, 9, 2846. [Google Scholar] [CrossRef]
- Chest X-ray (COVID-19 & Pneumonia). Available online: https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia (accessed on 4 October 2023).
- Jiang, Z.; Hu, M.; Gao, Z.; Fan, L.; Dai, R.; Pan, Y.; Tang, W.; Zhai, G.; Lu, Y. Detection of Respiratory Infections Using RGB-Infrared Sensors on Portable Device. IEEE Sensors J. 2020, 20, 13674–13681. [Google Scholar] [CrossRef]
- Song, Y.; Zheng, S.; Li, L.; Zhang, X.; Zhang, X.; Huang, Z.; Chen, J.; Wang, R.; Zhao, H.; Chong, Y.; et al. Deep Learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) With CT Images. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 18, 2775–2780. [Google Scholar] [CrossRef] [PubMed]
- Creswell, A.; White, T.; Dumoulin, V.; Arulkumaran, K.; Sengupta, B.; Bharath, A.A. Generative Adversarial Networks: An Overview. IEEE Signal Process. Mag. 2018, 35, 53–65. [Google Scholar] [CrossRef]
- Jin, Q.; Lin, R.; Yang, F. E-WACGAN: Enhanced Generative Model of Signaling Data Based on WGAN-GP and ACGAN. IEEE Syst. J. 2020, 14, 3289–3300. [Google Scholar] [CrossRef]
- Mehta, N.; Shukla, S. Pandemic Analytics: How Countries are Leveraging Big Data Analytics and Artificial Intelligence to Fight COVID-19? SN Comput. Sci. 2022, 3, 54. [Google Scholar] [CrossRef]
- Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit. Lett. 2020, 138, 638–643. [Google Scholar] [CrossRef]
- Ghoshal, B.; Tucker, A. Estimating Uncertainty and Interpretability in Deep Learning for Coronavirus (COVID-19) Detection. CoRR 2020, abs/2003.10769. [Google Scholar]
| Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).